Abstract
Polymicrobial diseases, which can be life threatening, are caused by the presence and interactions of multiple microbes. Peri-implantitis and periodontitis are representative polymicrobial diseases that show similar clinical symptoms. To establish a means of differentiating between them, we compared microbial species and functional genes in situ by performing metatranscriptomic analyses of peri-implantitis and periodontitis samples obtained from the same subjects (n = 12 each). Although the two diseases differed in terms of 16S rRNA-based taxonomic profiles, they showed similarities with respect to functional genes and taxonomic and virulence factor mRNA profiles. The latter—defined as microbial virulence types—differed from those of healthy periodontal sites. We also showed that networks based on co-occurrence relationships of taxonomic mRNA abundance (co-occurrence networks) were dissimilar between the two diseases. Remarkably, these networks consisted mainly of taxa with a high relative mRNA-to-rRNA ratio, with some showing significant co-occurrence defined as interacting core taxa, highlighting differences between the two groups. Thus, peri-implantitis and periodontitis have shared as well as distinct microbiological characteristics. Our findings provide insight into microbial interactions in polymicrobial diseases with unknown etiologies.
Biofilms are matrix-enclosed microbial populations that adhere to hard and soft tissue surfaces and are implicated in over 80% of known infectious diseases1. A technical challenge when studying the pathogenicity of polymicrobial infections by culture-dependent methods2 is the high prevalence of unculturable or fastidious bacteria3. Recent advances in molecular techniques with high-throughput sequencers have enabled the determination of polymicrobial community composition and/or interactions among microbial species in upper respiratory tract infections, osteomyelitis of the jaw, and periodontitis4,5,6.
Periodontitis is a representative polymicrobial biofilm-related disease that occurs in oral cavities. According to a report by the World Health Organization, disease prevalence is 15–20% in middle-aged adults (35–44 years), with 5–15% of these cases resulting in tooth loss (http://www.who.int/mediacentre/factsheets/fs318/en/). Recent evidence suggests that oral infection in periodontitis is associated with systemic diseases such as diabetes7 and cardiovascular diseases8. In addition, dental implant-based reconstruction—which has been adapted to replace conventional fixed or removable partial dentures—has led to the emergence of peri-implantitis as a serious problem in 28–56% of recipients9 and a major cause of implant loss. As such, effective prevention and management of peri-implantitis are essential for improving the quality of life and health of patients.
Peri-implantitis and periodontitis are polymicrobial diseases that present with similar clinical symptoms10. Most periodontitis cases respond favourably to treatment and show long-term stability of periodontal tissues11. However, clinical treatments for peri-implantitis—including those used for periodontal disease—are often ineffective12. In addition, peri-implantitis has been found to progress more rapidly than periodontitis in animal models13. To clarify the cause of differences between the two diseases, studies have characterised their respective microbiomes using culture-independent molecular approaches, including DNA hybridisation and 16S rDNA sequencing14,15,16. However, the causative microbial species in peri-implantitis and periodontitis are disputed. Several studies have reported the predominance of microbial species common to both diseases16, others unique to peri-implantitis sites have also been described10. One possible reason for these conflicting results is the presence of dead and/or inactive microbes in previous studies, which were based on DNA sequencing. Alternatively, microbiome samples from peri-implantitis and periodontitis sites were not obtained from the same oral cavity except in one study15.
To establish a means of differentiating between peri-implantitis and periodontitis, we investigated the microbial species associated with each disease as well as their functions in situ by carrying out a metatranscriptomic analysis at peri-implantitis and periodontitis sites in the same subjects. Our findings reveal that although they share similarities in their mRNA profiles, differences in interacting core taxa of co-occurrence networks account for the distinct etiologies of these diseases.
Results
Clinical characteristics of subjects and summary of sequence reads
A total of 12 patients (five men and seven women) with both peri-implantitis and periodontitis were recruited for this study. The average age was 64.5 years (range: 49–80 years); one patient was a smoker. There were no significant differences in the following clinical parameters for the two disease sites: years in function (for peri-implantitis sites only), probing depth, clinical attachment level, bleeding on probing, suppuration, and radiographic bone loss (Table 1, schematic illustrations in Supplementary Figs S1 and S2, and Supplementary Results).
Table 1. Clinical characteristics of study subjects.
| Peri-implantitis | Periodontitis | P value (two-tailed paired t test) | ||
|---|---|---|---|---|
| Age | 64.5 ± 7.0a | — | ||
| Gender (males/females) | 5/7 | — | ||
| Smoking/non-smoking | 1/11 | — | ||
| Sampled sites | Maxillary anterior | 2 | 1 | 0.34 |
| Maxillary posterior | 5 | 7 | 0.17 | |
| Mandibular anterior | 0 | 1 | 0.34 | |
| Mandibular posterior | 5 | 3 | 0.17 | |
| Years in function | 8.6 ± 7.2a | — | — | |
| PD (mm) | 8.2 ± 2.9a | 8.3 ± 3.1a | 0.95 | |
| CAL (mm) | 8.3 ± 2.8a | 9.9 ± 3.9a | 0.35 | |
| Number of sites with BOP | 12 | 12 | — | |
| Number of sites with SUP | 4 | 3 | 0.34 | |
| Radiographic bone loss (%) | 52.4 ± 20.6a | 60.9 ± 15.6a | 0.29 | |
aValues represent mean ± standard deviation.
BOP, bleeding on probing; CAL, clinical attachment level; PD, probing depth; SUP, suppuration.
Evaluation of microbiome microbial compositions based on 16S rRNA sequences
When short metagenomic reads were analysed, there was no representative microbial composition as previously described17. In this approach, small subunits of the rRNA gene (16S/18S) were reconstructed using a mapping-based algorithm17. To assign microbial compositions with high resolution in situ and identify microbial species responsible for the diseases, we characterised the composition at both disease sites by sequencing total RNA and performing full- or nearly full-length reconstruction of their 16S rRNA regions18. The number of reconstructed 16S rRNAs (hereafter referred to as rc-rRNAs) forming operational taxonomic units (OTUs) was 58.5 ± 21.8 and 62.3 ± 20.3 in peri-implantitis and periodontitis samples, respectively (Supplementary Fig. S3a). Using the Human Oral Microbiome Database (HOMD)19, rc-rRNAs were assigned to 184 microbial taxa at the species level, with 150 and 164 taxa identified in peri-implantitis and periodontitis samples, respectively (Supplementary Table S2). There were no significant differences in alpha diversity, the number of OTUs (P = 0.715), or Shannon index (P = 0.834) between the two diseases (Supplementary Fig. S3a). Results from rarefaction curves indicated that a sufficient number of reads was obtained for 16S rRNA analyses (Supplementary Fig. S3b).
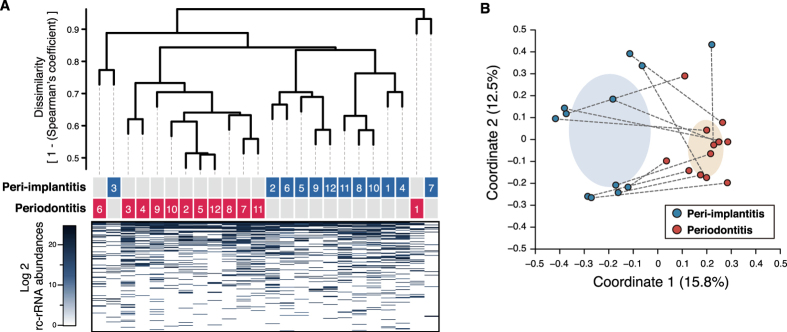
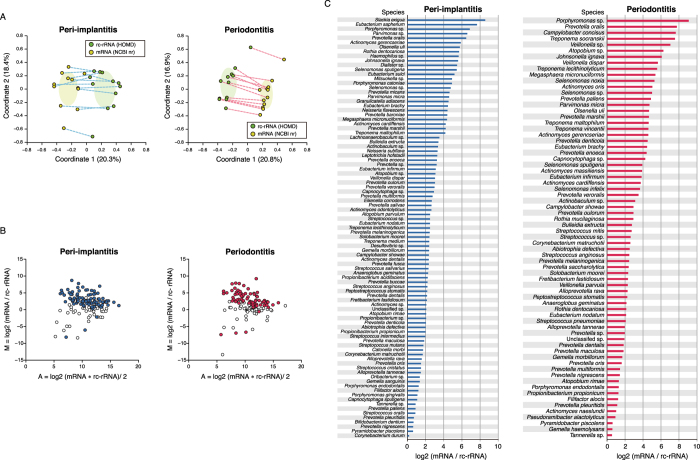
A dendrogram with a heat map and principal coordinates analysis (PCoA) plot (based on 1 − Spearman’s coefficient) was generated to examine differences in beta diversity. Most of the samples formed two clusters (Fig. 1), indicating differences in beta diversity between the two diseases. This was supported by an analysis of similarity (ANOSIM), which revealed that microbial compositions were dissimilar between the two groups (R = 0.399 and P = 1.00E-3). In addition, microbial composition at the genus level was diverse among samples for each disease and between both samples from each individual (Supplementary Fig. S4a). However, the predominant species were similar (Supplementary Fig. S3c and Supplementary Table S2), and no species differed significantly in terms of rc-rRNA abundance between the two diseases in Wilcoxon signed-rank tests.
Figure 1. Dendrogram with a heat map and a PCoA plot of rc-rRNA profiles.
(A) Dissimilarity values (1 − Spearman’s coefficient) were clustered using the average linkage method, as shown in the dendrogram. Disease types and patient numbers are shown under the tree. The heat map shows log2 rc-rRNA abundances for each taxon, as indicated by the colour gradient. (B) PCoA was carried out for the dissimilarity matrix value of 1 − Spearman’s coefficient, and 12 samples from the peri-implantitis (blue circles) and periodontitis (red circles) groups were plotted with two coordinates. The mean and standard deviation in each axis are indicated by an ellipse for each disease group. Dots corresponding to peri-implantitis and periodontitis samples from the same patient are connected by a broken line.
Functional profiles of microbiomes
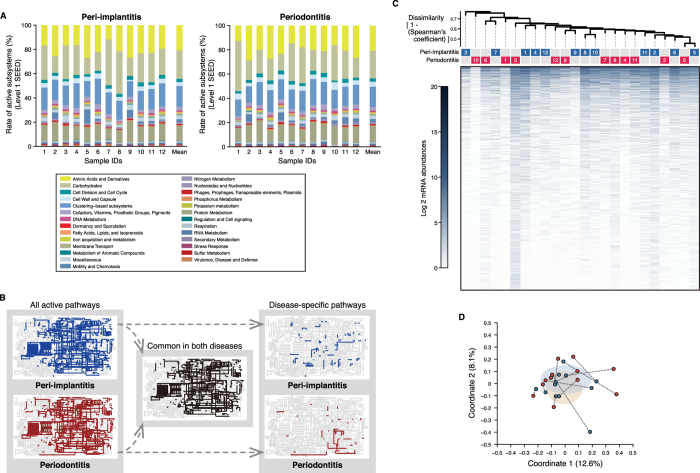
The above analyses revealed dissimilarities in microbial composition between the two diseases; however, examining mRNA profiles can provide greater insight into functional differences between microbiomes20,21. We used the publically available Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST) analysis pipeline to characterise putative mRNA reads in our data. We also used the SEED subsystems database to categorise functional genes into four hierarchical subsystems22. A total of 2461 and 2379 functional genes were assigned to peri-implantitis and periodontitis samples, respectively (Supplementary Table S3). Of these, 2006 were common to both diseases. There was no significant difference in the number of functional genes between the two diseases (P = 0.41). Among level-1 SEED subsystems, ‘carbohydrates’ was predominant at both peri-implantitis (22.1 ± 4.9%) and periodontitis (21.7 ± 5.1%) sites, followed by ‘protein metabolism’ and ‘clustering-based subsystems’ (Fig. 2A). The composition of level-1 subsystems was similar among samples of each disease and between both samples from each individual (Fig. 2A). An ANOSIM showed similarities in the functional profiles (R = 5.26E-5 and P = 0.403), and there were no genes with significantly different mRNA abundance between the two diseases in Wilcoxon tests.
Figure 2. mRNA profiles obtained following assignment with SEED subsystems and KEGG and NCBI nr databases.
(A) Percentage composition of level-1 SEED subsystems are shown for each sample ID, with the corresponding colours shown in the box below. (B) Active KEGG pathways present in any of the 12 samples for each disease (left map) and common and disease-specific pathways (middle and right maps, respectively). (C) Dendrogram constructed based on mRNA abundances during assignments with the NCBI nr database, as described in Fig. 1A. (D) PCoA plot prepared from mRNA profiles assigned with the NCBI nr database, as described in Fig. 1B.
Similar functional compositions were observed in the MG-RAST-processed data using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, in which metabolic pathways are listed hierarchically23. According to this analysis, 1380 and 1291 genes were assigned to peri-implantitis and periodontitis sites, respectively (Supplementary Table S4) and 1134 genes were shared by both diseases. There was no significant difference in the number of functional genes between the two disease groups (P = 0.370), which shared most metabolic pathways (Fig. 2B and Supplementary Table S4). Although there were some disease-specific pathways (Fig. 2B), most were active in only a single sample (Supplementary Table S4). Similar functional profiles were observed in an ANOSIM (R = −1.00E-4 and P = 0.471), and the results of the Wilcoxon tests showed no differences in expression between samples of either disease, in agreement with the SEED subsystems database assignments.
We also examined whether there were similarities when using the National Center for Biotechnology Information non-redundant (NCBI nr) protein database for functional assignment24 (Supplementary Fig. S1). In this database, protein functions are not categorised or organised hierarchically, but are archived with descriptions of the taxonomic origins of each function. A total of 95 487 ± 47 017 and 110 868 ± 64 965 clusters25 were formed from the pre-processed reads (i.e., those in which low-quality bases and putative eukaryotic organisms from raw reads were removed) for the peri-implantitis and periodontitis sites, respectively; of these, 22 988 ± 16 581 and 42 831 ± 18 992, respectively, were removed as clusters derived from putative 16S rRNA reads. The remaining clusters were further analysed as mRNA clusters (72 499 ± 48 541 and 68 038 ± 54 801 in the peri-implantitis and periodontitis samples, respectively). Using the NCBI nr database, these were assigned to 22 613 and 21 187 functional genes, respectively, for a total of 30 923 distinct mRNA clusters (Supplementary Table S5). In total, 12 737 genes were common to the two diseases; hypothetical and ribosomal proteins were predominant in both. In the dendrogram and PCoA plot, samples from each disease had similar mRNA profiles for the two diseases (Fig. 2C,D). An ANOSIM revealed similarity between the two groups (R = −1.18E-2 and P = 0.627), and the Wilcoxon tests showed no differences in read abundance of mRNA clusters or mRNA abundances for any of the genes between them. These observations were cosistent even when hypothetical and ribosomal proteins were excluded (data not shown).
Functional profiles of putative virulence factors
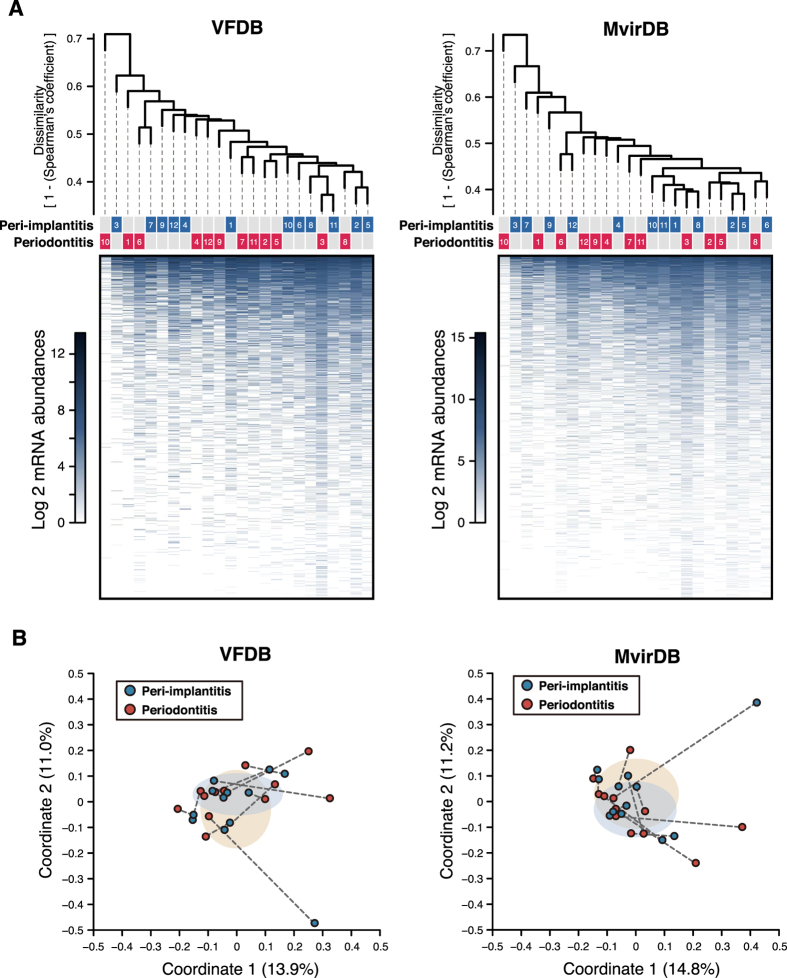
The SEED subsystems, KEGG, and NCBI nr databases were used for comprehensive functional assignment of metatranscriptomic data, whereas virulence factor databases were more informative for generating detailed profiles of virulence genes20,21. Functional assignments for mRNA clusters relied on the Virulence Factors of Pathogenic Bacteria (VFDB; http://www.mgc.ac.cn/VFs/) and MvirDB (http://mvirdb.llnl.gov/) databases26,27. A total of 1579 and 1537 virulence genes in the VFDB were used to assign mRNA clusters found in peri-implantitis and periodontitis samples, respectively (Supplementary Table S6); of 1827 analysed genes, 1289 were common to both diseases. An elongation factor-encoding gene was most prevalent in peri-implantitis and periodontitis (9.89 ± 1.07% and 10.13 ± 2.26%, respectively), followed by glyceraldehyde 3-phosphate dehydrogenase, alkyl hydroperoxide reductase (ahpC), and enolase genes. The mRNA profiles of the two diseases were similar based on dendrograms and PCoA plots (Fig. 3A,B), which was supported by an ANOSIM (R = 3.16E-04 and P = 0.482). Wilcoxon tests revealed no differences in mRNA abundance of any virulence genes between the two diseases.
Figure 3. Functional mRNA profiles for assignments made with VFDB and MvirDB.
(A) Dendrograms constructed as described in Fig. 1A. Disease types and patient numbers are shown under the tree. The heat map shows log2 mRNA abundances for each functional gene. (B) PCoA plots prepared as described in Fig. 1B.
The MvirDB returned 2722 and 2622 virulence genes for the assignment of mRNA clusters in the peri-implantitis and periodontitis samples, respectively (Supplementary Table S7); 2136 genes were shared by both diseases. The predominant genes in terms of mRNA abundance were similar between peri-implantitis and periodontitis, although their rank order differed (Supplementary Table S7). As with assignments made with the VFDB, the similarity in mRNA profiles was apparent in the dendrogram-PCoA plot (Fig. 3A,B) and by ANOSIM (R = −1.82E-2 and P = 0.738), and there was no difference in the mRNA abundance of any gene between the two diseases.
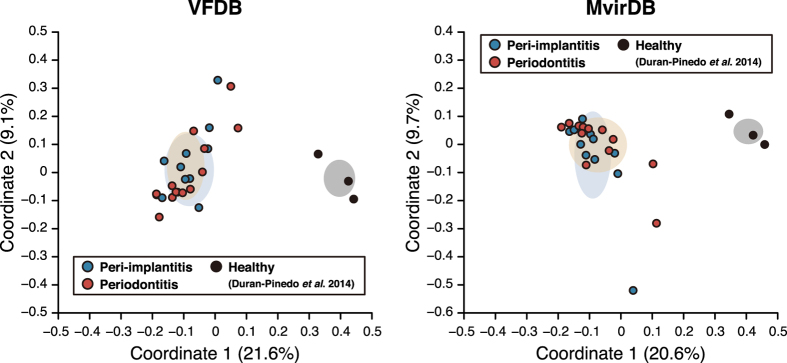
Our metatranscriptomic data were further characterised according to mRNA profiles of virulence factors, which were designated as microbial virulence (MV) types. The MV types of peri-implantitis and periodontitis sites were similar, suggesting that the two diseases were associated with similar virulence factors. This raised the question of whether MV types of diseased and healthy sites differ. MV types were assigned to RNA sequencing (RNA-seq) data from samples of healthy periodontal sites20, which were then compared to our data. The MV types observed in our data were distinct from those of healthy sites, as shown by PCoA plots based on assignments made with the VFDB and MvirDB (Fig. 4 and Supplementary Table S8). This was supported by ANOSIMs: we determined correlations of R = 0.980, P = 3.00E-3 and R = 0.845, P = 3.00E-3 for peri-implantitis and periodontitis, respectively, vs. healthy sites using VFDB-based profiles; and R = 0.867, P = 3.00E-3 and R = 0.783, P = 5.00E-3, respectively using MvirDB-based profiles. These results indicate that MV types of the disease sites differed from those of healthy body sites, despite similarities between the two diseases.
Figure 4. PCoA plots of functional profiles for diseased and healthy sites.
PCoA plots of functional profiles for our sequence data, as well as downloaded data pertaining to healthy sites assigned with the VFDB and MvirDB, were prepared as described in Fig. 1B.
Characterisation of taxonomic mRNA origins and detection of viable taxa with high mRNA abundance
A previous metatranscriptomic study reported that microbial composition determined based on 16S rRNA sequences differed from that based on taxonomic mRNA profiles in active sludge28. Using data that was functionally assigned with NCBI nr, we first assessed the taxonomic origin of each gene for taxonomic assignment of mRNA clusters (see Supplementary Figs S4B and S5A, Supplementary Table S9, and Supplementary Results). PCoA plots and ANOSIMs showed differences in the species compositions of mRNA and read abundances for rc-rRNAs (Fig. 5A; R = 0.496, P = 1.00E-3 in peri-implantitis and R = 0.588, P = 1.00E-3 in periodontitis samples). Taxa detected in both rc-rRNA and mRNA profiles were defined as viable taxa with in situ function (VTiF), of which 146 and 133 were identified in the peri-implantitis and periodontitis samples, respectively. Wilcoxon test results revealed significant differences in read abundance of each VTiF between rc-rRNA and mRNA clusters (96 and 74 taxa in the peri-implantitis and periodontitis samples, respectively; Supplementary Table S10). Log ratio–mean average (MA)-plots showed that these VTiFs mainly included taxa with high relative mRNA-to-rRNA ratio (referred to as active taxa), with mRNA abundances higher than rc-rRNA abundances (93/96 and 70/74 in peri-implantitis and periodontitis samples, respectively; Fig. 5B and Supplementary Table S10). Relative mRNA-to-rRNA ratios were indicators of the viability and functionality of microbial taxa responsible for disease etiology29,30 in the same way that the 16S rRNA-to-16S rDNA ratio has been used as an indicator of current bacterial activity31. In peri-implantitis samples, Slackia exigua and Eubacterium saphenum showed a relative mRNA-to-rRNA ratio >7; in periodontitis samples, Porphyromonas sp., Prevotella oralis, Campylobacter concisus, Treponema socranskii, and Veillonella sp. showed a relative mRNA-to-rRNA ratio >7 (Fig. 5C and Supplementary Table S10).
Figure 5. Differences in rc-rRNA and taxonomic mRNA profiles.
(A) PCoA was performed to analyse rc-rRNA and mRNA abundances, and 12 samples in both the peri-implantitis and periodontitis groups are plotted with two coordinates. The mean and standard deviation in each axis are indicated by an ellipse. Dots corresponding to rc-rRNA and mRNA in the same patient are connected by a broken line. (B) VTiFs shown in MA plots for peri-implantitis and periodontitis samples. The y axis shows difference values [M] of rc-rRNA and mRNA abundances and the x axis shows mean values [A]. Coloured points indicate taxa with statistically significant differences in abundance between rc-rRNAs and mRNAs. (C) mRNA/rc-rRNA abundance ratio calculated for each VTiF shown in (B); predominant taxa (based on mean log2 ratios) for the 12 samples are shown in descending order.
VTiFs in co-occurrence networks and interacting core taxa
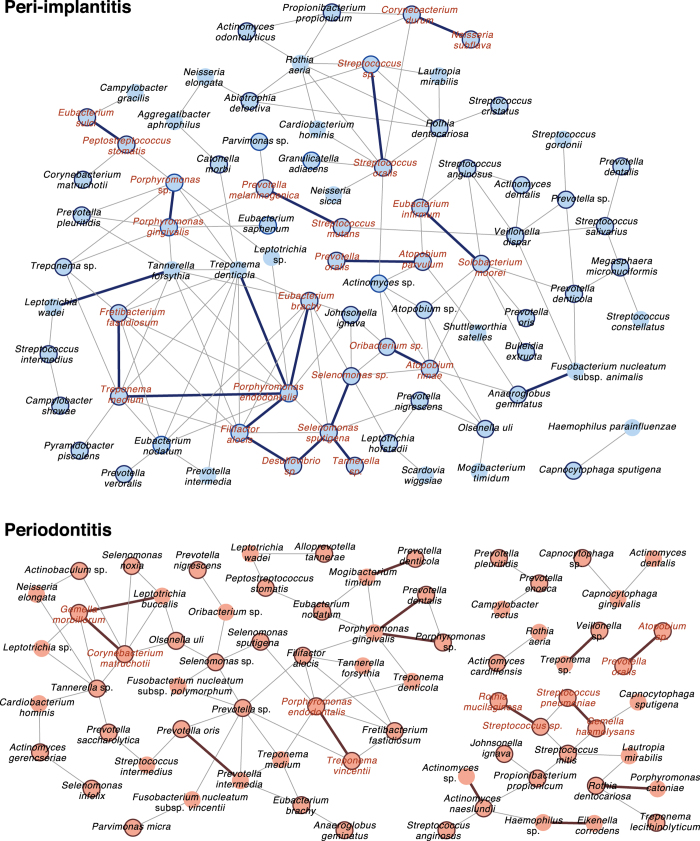
Specific microbial co-occurrence patterns in polymicrobial disease were previously characterised by correlation analysis15, which is a useful tool for identifying representative and important microbial associations in polymicrobial disease32. We analysed co-occurrence relationships in mRNA profiles of VTiFs by constructing network structures (referred to as co-occurrence networks) in which two co-occurring taxa were indicated by nodes and connected by a degree. There were one and two main network(s) with > three nodes in peri-implantitis and periodontitis samples, respectively. Means of 1.75 and 1.21 degrees per node connected 79 peri-implantitis and 71 periodontitis nodes, respectively, with clustering coefficients of 0.221 and 0.165, respectively, in these networks (Fig. 6). Active taxa were prevalent in the networks, with 60/79 and 45/71 such cases found in peri-implantitis and periodontitis samples, respectively (Fig. 6 and Supplementary Tables S10–S12). Most nodes connected by interactions with significant co-occurrence were active taxa and were detected in at least eight patients; these taxa were considered as interacting core taxa (Fig. 6 and Table 2).
Figure 6. Co-occurrence networks of VTiF profiles.
(A) All networks are shown with each microbial taxon and co-occurrence relationship indicated by a node and edge, respectively. Active taxa are indicated with bold circles, and interactions with significant co-occurrence are indicated with bold lines. Interacting core taxa are indicated in red text for peri-implantitis and periodontitis samples.
Table 2. Interactions of interacting core taxa.
| Specificity to disease groups | Species name | Log2 mRNA/rc-rRNA ratio | Number of samples detected | Species name | Log2 mRNA/rc-rRNA ratio | Number of samples detected | Positive correlation coefficient |
|---|---|---|---|---|---|---|---|
| Specific to peri-implantitis group | Porphyromonas sp. | 6.863 | 12 | Porphyromonas gingivalis | 1.098 | 12 | 0.740 |
| Porphyromonas endodontalis | 1.238 | 12 | Filifactor alocis | 1.207 | 12 | 0.643 | |
| Streptococcus sp. | 2.489 | 12 | Streptococcus oralis | 0.878 | 11 | 0.625 | |
| Porphyromonas endodontalis | 1.238 | 12 | Eubacterium brachy | 4.465 | 11 | 0.605 | |
| Selenomonas sputigena | 5.428 | 10 | Selenomonas sp. | 4.771 | 12 | 0.605 | |
| Streptococcus sp. | 2.489 | 12 | Selenomonas sputigena | 5.428 | 10 | 0.601 | |
| Treponema medium | 2.409 | 9 | Porphyromonas endodontalis | 1.238 | 12 | 0.526 | |
| Treponema medium | 2.409 | 9 | Fretibacterium fastidiosum | 2.154 | 12 | 0.496 | |
| Oribacterium sp. | 1.361 | 11 | Eubacterium sulci | 5.186 | 11 | 0.487 | |
| Solobacterium moorei | 2.421 | 9 | Eubacterium infirmum | 3.113 | 11 | 0.483 | |
| Oribacterium sp. | 1.361 | 11 | Atopobium rimae | 2.072 | 9 | 0.479 | |
| Filifactor alocis | 1.207 | 12 | Desulfovibrio sp. | 2.398 | 10 | 0.443 | |
| Neisseria subflava | 3.314 | 11 | Corynebacterium durum | 0.187 | 8 | 0.431 | |
| Prevotella oralis | 6.469 | 11 | Atopobium parvulum | 2.493 | 9 | 0.413 | |
| Streptococcus mutans | 1.760 | 11 | Prevotella melaninogenica | 2.460 | 11 | 0.392 | |
| Selenomonas sputigena | 5.428 | 10 | Desulfovibrio sp. | 2.398 | 10 | 0.375 | |
| Specific to periodontitis group | Treponema vincentii | 4.598 | 11 | Porphyromonas endodontalis | 1.300 | 11 | 0.595 |
| Streptococcus pneumoniae | 2.080 | 10 | Gemella haemolysans | 0.592 | 11 | 0.468 | |
| Gemella morbillorum | 1.696 | 11 | Corynebacterium matruchotii | 2.583 | 9 | 0.430 | |
| Streptococcus sp. | 2.615 | 10 | Rothia mucilaginosa | 2.928 | 9 | 0.384 | |
| Prevotella oralis | 7.763 | 11 | Atopobium sp. | 6.261 | 12 | 0.355 |
Discussion
Polymicrobial communities are composed of a variety of microorganisms, including bacteria, archaea, fungi, and viruses. The microbiome of oral cavities comprise ~600–1000 species of bacteria alone33. We previously showed by sequencing PCR-amplified 16S rRNA libraries that many microorganisms are responsible for chronic osteomyelitis of the jaw4 and peri-implantitis and periodontitis15. RNA-Seq is a powerful tool for examining the diversity of microbial species and their functional profiles since the obtained data reflect RNA abundance in situ34. We carried out metatranscriptomic analyses of peri-implantitis and periodontitis samples from the same oral cavities to minimise the effects of inter-individual differences. Using this approach, we observed microorganisms common to as well as differing between the two diseases with similar clinical symptoms. Moreover, we found similarities in functional profiles—including metabolic pathways and virulence factors—whereas interacting core taxa were dissimilar between the two diseases.
We first characterised the taxonomic profiles of microbial species in peri-implantitis and periodontitis samples by quantifying 16S rRNA instead of 16S rDNA; viable species were assigned while dead species were excluded35. Consistent with previous reports15,20, we detected high rc-rRNA abundances of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in both diseases (Supplementary Fig. S3C and Supplementary Table S2). These species are collectively known as the red complex and are prevalent in deep periodontal pockets and thus strongly associated with periodontitis36. Although microbial transmission from periodontal lesions to implant-adjacent tissues may underlie the onset of peri-implantitis37, samples from both diseases showed distinct microbial compositions (Fig. 1A,B), despite the similarities reported in our previous DNA-based study15. In addition, species common to both diseases with relative abundances of ≥0.5% observed here and in a previous study15 showed partial overlap (19.64% of 11/56 in this study), including P. gingivalis, T. denticola, T. forsythia, and Eubacterium nodatum (Supplemental Table S2), although both studies used the HOMD for taxonomic assignments. These differences could be due to amplicon bias38 and the detection of dead bacteria in DNA samples35.
It was previously reported that functional microbiome profiles of various healthy body sites were similar in terms of robustness, despite differences in microbial composition39. It is thus possible that peri-implantitis and periodontitis have distinct microbial compositions but similar functional profiles. Functional assignment of putative mRNA reads revealed that mRNA profiles were similar between the two diseases, with no functional gene differing significantly in terms of mRNA abundance between them according to assignments made with the SEED subsystems, KEGG, and NCBI nr databases (although some disease-specific genes were identified). The diseases shared some of the same SEED subsystems (such as ‘carbohydrates’ and ‘protein metabolism’) and KEGG pathways (Fig. 2 and Supplementary Tables S3 and S4) related to functions essential for microbial viability. However, microbial functional categories associated with the progression of both diseases showed low abundance. The level-1 SEED subsystem ‘fatty acids, lipids, and isoprenoids’—which is reportedly associated with periodontitis progression40—showed a prevalence of 2.22 and 2.43% at peri-implantitis and periodontitis sites, respectively, while the ‘virulence, disease, and defense’ subsystem showed a prevalence of 2.22 and 1.84%, respectively, at these sites. These were lower than percentages in environmental sample microbiomes41. We further analysed putative mRNA reads using the virulence-factor databases VFDB and MvirDB (Supplementary Tables S6, S7). Similarities were observed in the mRNA profiles of both disease sites despite being limited to virulence functions (Fig. 3A,B); these included genes encoding elongation factor Tu, glyceraldehyde 3-phosphate dehydrogenase, alkyl hydroperoxide reductase (ahpC), and enolase (Supplementary Table S6). There were no correlations between the mRNA abundance of virulence factors and clinical parameters (data not shown); indeed, the presence of virulence factors may be more important than their relative abundances at disease sites42. Correlations may have been observed if the virulence functions had been highly specific instead of broad.
Peri-implantitis and periodontitis showed similar mRNA profiles although these were of virulence functions; different functional profiles may occur at healthy periodontal sites. Functional pathways in healthy and periodontitis sites have been compared by metatranscriptomic analysis43. In addition, differences in mRNA abundance between healthy and periodontitis sites have been reported for each microbial taxon and for functional and taxonomic profiles of virulence factors20. When the functional properties were clustered in the same manner as enterotypes for enteric bacteria44, they highlighted the microbiological origin of both diseases as well as their contrast with healthy sites. In this study, we compared the MV types of both diseases with those of healthy sites using metatranscriptomic data from a previous study20, and demonstrated that the MV types of peri-implantitis and periodontitis sites were distinct from those of healthy sites (Fig. 4). In microbiomes showing dysbiosis, there was a reduction in microbial functional robustness in the healthy state following changes in MV type, which led to the diseased states. We therefore propose that MV types determined only by assignment of virulence factors can serve as a means of distinguishing between diseased and healthy states.
It was not always the case that the major species were also functionally predominant or that the taxonomic origins of functional genes were similar between two diseases with similar functional profiles. We therefore determined the taxonomic origins of functional genes using the NCBI nr database. The red complex species were predominant in terms of mRNA abundance and in the rc-rRNA profile, indicating that they are associated with both diseases. However, there were similarities in the taxonomic profile and origin of each mRNA. The taxonomic profiles of rc-rRNAs differed from those of mRNA clusters in both disease groups (Fig. 5A,B); this was consistent with previous results obtained by comparing microbial compositions between 16S rRNA and mRNA using the same database28. Since the results may have been affected by differences in the databases for the assignment of rc-RNA and mRNA clusters, we focused on VTiFs shared by the rc-rRNA and mRNA profiles and evaluated the functional activities of each VTiF by calculating their ratios of mRNA-to-rc-rRNA abundance. The anaerobic Gram-positive rod S. exigua45 and E. saphenum were highly abundant in peri-implantitis samples. S. exigua DNA was detected at slightly higher levels at periodontitis sites than at healthy sites, while E. saphenum DNA was more abundant at the former46. These two taxa were also more abundant at peri-implantitis sites than at healthy implant sites47. Taxa that were active in periodontitis samples (Fig. 5C) were also detected at periodontitis sites in a previous study48. Our analyses also revealed unclassified bacterial taxa whose functions and virulence are unknown but that highlight the dissimilarity between the two diseases. Our results demonstrate that metatranscriptomic analysis—including taxonomic mRNA classification—can detect active taxa in the microbiome.
We also observed that co-occurrence relationships in mRNA-based taxonomic profiles of VTiFs may be dissimilar between peri-implantitis and periodontitis when visualised by network structures (Fig. 6), although this was not apparent in the taxonomic mRNA profiles described above. Microbial network analyses can be used to characterise microbial interactions in environments such as soil and oral-cavity samples32,49. The networks were more complex in the peri-implantitis as compared to the periodontitis microbiome (Fig. 6); in the former, the red complex species P. gingivalis, T. denticola, and T. forsythia were associated with each other, whereas in the latter there were limited connections between P. gingivalis and T. denticola. Furthermore, active taxa were prevalent in the networks of both diseases (Fig. 6). In periodontitis samples, the interacting core taxa included E. nodatum, Streptococcus pneumoniae, and Atopobium sp., which are reportedly more abundant at periodontal sites than at healthy sites affected with periodontitis21. P. gingivalis and Prevotella nigrescens were among the interacting core taxa in peri-implantitis samples and were found to be more abundant16 while Veillonella dispar was less abundant50 at peri-implantitis sites than at healthy implant sites. Previous data showed that P. gingivalis is a major species in oral biofilm, which can lead to dysbiosis through changes in polymicrobial community composition51. In this study, P. gingivalis was included among interacting core taxa in the peri-implantitis but not in the periodontitis microbiome. Moreover, there were no significant interactions common to the two diseases (Table 2). These network dissimilarities are presumed to be associated with differences in disease etiology.
Our analytical methods provide an approach for characterising the interactions between the putative causal agents of diseases based on 16S rRNA and mRNA expression data. We also propose that it could be useful for characterising the MV types. Additional studies with a larger number of analysed samples could help identify differences in the functional profiles of the two diseases and clarify the role of interacting core taxa52.
In summary, we propose that interacting core taxa in co-occurrence networks offer novel insights into polymicrobial diseases with similar mRNA profiles. The observed similarities and dissimilarities could explain the common symptoms and differences in disease progression, respectively. In microbiomes with dysbiosis, we characterised MV types that were distinct from those of healthy sites and found that the breakdown of microbial functional robustness in the healthy state leads to a diseased state. Our findings provide a basis for the development of treatment approaches specific to peri-implantitis or periodontitis as well as a framework for metatranscriptomic studies of other polymicrobial diseases with unknown etiologies. This, in turn, can lead to a deeper understanding of the relationship between healthy and diseased states at various sites in the human body.
Methods
Overview of experimental workflow and statistical analyses
Experiments were performed as shown in the flowchart in Supplemental Fig. S1. Statistical analyses are schematically illustrated in Supplemental Fig. S2 and described in the Supplementary Methods.
Ethical statement
This study was performed in accordance with the Ethical Guidelines for Clinical Studies (2008 Notification number 415 of the Ministry of Health, Labor, and Welfare). Ethical approval was obtained from the Ethics Committee of Tokyo Medical and Dental University (approval no. 661), and all patients provided informed consent before their participation.
Patient selection and clinical assessment
Twelve subjects with at least one dental implant functioning for ≥1 year with peri-implantitis as well as one tooth with periodontitis were recruited for this study at the Tokyo Medical and Dental University Hospital Faculty of Dentistry from 2012 to 2013 (Table 1). The patients were healthy adults who had not received systemic antibiotics or oral anti-inflammatory agents in the 3 months prior to enrollment in the study. For all implants and teeth in each patient, probing depth (PD), clinical attachment level, bleeding on probing (BOP), and suppuration (SUP) were assessed at six sites per implant or tooth (i.e., mesiobuccal, buccal, distobuccal, mesiolingual, lingual, and distolingual sites). Bone loss surrounding each implant/tooth was examined with intra-oral periapical radiographs using Insight Dental Films (Eastman Kodak Co., Tokyo, Japan) obtained using the parallel technique, and was quantified by a single examiner as previously described2,3. Based on clinical data, one implant and one tooth exhibiting PD ≥ 4 mm, BOP and/or pus, and radiographic bone loss were selected for sample collection (Table 1).
Procedure for obtaining Illumina sequence data
Subgingival plaque samples were obtained from the peri-implant and periodontal pockets at the deepest of six sites in each implant/tooth using sterile cotton and were dried to reduce eukaryotic DNA contamination. Ten pieces of paper were inserted into the pocket for 30 s, placed in a sterile tube, and stored at −80 °C. RNA was extracted from each sample, followed by cDNA synthesis and library preparation, and Illumina sequencing (Supplementary Methods). Sequence data were processed as described in the Supplementary Methods.
Reconstruction and taxonomic assignment of putative 16S rRNA reads
Pre-processed, paired-read data were screened with EMIRGE17 to select putative reads derived from 16S rRNA genes and to form rc-rRNAs as OTUs. The rc-rRNAs were assigned by BLASTN against the HOMD19. Alpha and beta diversities were estimated and visualised by a dendrogram-PCoA plot and heat map. Detailed procedures are descried in Supplementary Methods.
Functional assignment of putative mRNA reads with the MG-RAST pipeline
Pre-processed data were further processed with MG-RAST (v.3.3)53 to obtain mRNA profiles with the online BLAT program54, which were then analysed using the SEED subsystems22 and KEGG23 databases (Supplementary Methods).
Formation of mRNA clusters and functional annotation
Clusters were formed from pre-processed data using the Cluster Database at High Identity with Tolerance program25, and putative 16S rRNA reads were removed (Supplementary Methods). The remaining (mRNA) clusters were presumed to be derived from mRNA reads and their abundance was calculated as the number of reads in that cluster. The mRNA clusters were assigned by BLASTX against the NCBI nr, VFDB, and MvirDB databases as described in the Supplementary Methods. A dendrogram-PCoA plot and heat map were prepared as described for rc-rRNA analyses.
Evaluation of differences in taxonomic profiles between rc-rRNAs and mRNAs
The assignment of rc-rRNA and mRNA clusters took into account differences in sequence type (nucleic/amino acid) and sampled environments among the databases used. The taxonomic names of rc-rRNAs in the HOMD were manually adjusted to those of mRNA clusters, which were excluded if they were not in the list of adjusted names for rc-rRNAs, since the breadth of sampled environments was greater in the NCBI nr than in the HOMD. In addition, the % reads per kilobase per million mapped reads (RPKM) values obtained when assigning rc-rRNAs were converted to RPKM values (Supplementary Methods).
The Spearman’s coefficient was calculated for all pairs of each sample during the assignment of rc-rRNAs with HOMD and of mRNA clusters with NCBI nr. A dissimilarity matrix based on the value 1 − Spearman’s coefficient for both disease groups was used for the PCoA. The average and difference of paired log2 values of rc-rRNAs and mRNAs were calculated for each taxon to obtain MA plots for both sample groups55.
Characterisation of VTiF co-occurrence networks
We first removed VTiFs with a relative abundance of <0.1% of the total number of VTiF reads52,56,57. Co-occurrence coefficients were then calculated using the SparCC program58 and the mRNA taxonomic abundances in each disease. Ten iterations were used to estimate the median correlation of each pairwise comparison, and the statistical significance of each correlation was calculated by bootstrapping with 500 iterations56. Taxon pairs with SparCC values ≥0.3 were considered as exhibiting a co-occurrence relationship with a positive correlation. Our criterion for significance testing was more stringent than the previously used value of ≥0.2559. Co-occurrence patterns were drawn using a network structure in which each taxon and co-occurrence was indicated by a node and edge, respectively, for all taxon pairs with a positive correlation. The networks were visualised using Cytoscape software v.2.860.
Additional Information
Accession codes: Illumina read data generated in this study are available in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/) under accession number DRA003492.
How to cite this article: Shiba, T. et al. Distinct interacting core taxa in co-occurrence networks enable discrimination of polymicrobial oral diseases with similar symptoms. Sci. Rep. 6, 30997; doi: 10.1038/srep30997 (2016).
Supplementary Material
Acknowledgments
This study was supported by the Japan Society for the Promotion of Science KAKENHI grants (nos 24792321 and 26463129 to Y.T.; 25670776, 25713060, and 24117508 to F.M.; and 26·2840 and 26861544 to T.W.). Super-computing resources were provided by the Human Genome Center at the Institute of Medical Science (University of Tokyo) (http://sc.hgc.jp/shirokane.html).
Footnotes
Author Contributions T.S., Y.T. and F.M. designed the experiments. T.S., T.K., N.M., Y.T. and Y.I. carried out sample collection. T.S., T.W., H.K., F.M. and I.N. carried out data collection and performed data analysis. H.K. and K.M. provided bioinformatic support. T.S., T.W. and F.M. wrote the manuscript, with contributions from other authors.
References
- Dongari-Bagtzoglou A. Pathogenesis of mucosal biofilm infections: challenges and progress. Expert Rev. Anti Infect. Ther. 6, 201–208, doi: 10.1586/14787210.6.2.201 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Jabra-Rizk M. A., O’May G. A., Costerton J. W. & Shirtliff M. E. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25, 193–213, doi: 10.1128/cmr.00013-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Ludwig W. & Schleifer K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda A., Maruyama F., Michi Y., Nakagawa I. & Harada K. Analysis of the factors affecting the formation of the microbiome associated with chronic osteomyelitis of the jaw. Clin. Microbiol. Infect. 20, O309–317, doi: 10.1111/1469-0691.12400 (2014). [DOI] [PubMed] [Google Scholar]
- Griffen A. L. et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6, 1176–1185, doi: 10.1038/ismej.2011.191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns J. C. et al. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J 9, 1246–1259, doi: 10.1038/ismej.2014.250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preshaw P. M. & Bissett S. M. Periodontitis: oral complication of diabetes. Endocrinol. Metab. Clin. North Am. 42, 849–867, doi: 10.1016/j.ecl.2013.05.012 (2013). [DOI] [PubMed] [Google Scholar]
- Mustapha I. Z., Debrey S., Oladubu M. & Ugarte R. Markers of systemic bacterial exposure in periodontal disease and cardiovascular disease risk: a systematic review and meta-analysis. J. Periodontol. 78, 2289–2302, doi: 10.1902/jop.2007.070140 (2007). [DOI] [PubMed] [Google Scholar]
- Zitzmann N. U. & Berglundh T. Definition and prevalence of peri-implant diseases. J. Clin. Periodontol. 35, 286–291, doi: 10.1111/j.1600-051X.2008.01274.x (2008). [DOI] [PubMed] [Google Scholar]
- Leonhardt A., Renvert S. & Dahlen G. Microbial findings at failing implants. Clin. Oral Implants Res. 10, 339–345 (1999). [DOI] [PubMed] [Google Scholar]
- Lindhe J. & Nyman S. Long-term maintenance of patients treated for advanced periodontal disease. J. Clin. Periodontol. 11, 504–514 (1984). [DOI] [PubMed] [Google Scholar]
- de Waal Y. C., Raghoebar G. M., Meijer H. J., Winkel E. G. & van Winkelhoff A. J. Prognostic indicators for surgical peri-implantitis treatment. Clin. Oral Implants Res., doi: 10.1111/clr.12584 (2015). [DOI] [PubMed] [Google Scholar]
- Lindhe J., Berglundh T., Ericsson I., Liljenberg B. & Marinello C. Experimental breakdown of peri-implant and periodontal tissues. A study in the beagle dog. Clin. Oral Implants Res. 3, 9–16 (1992). [DOI] [PubMed] [Google Scholar]
- Koyanagi T. et al. Comprehensive microbiological findings in peri-implantitis and periodontitis. J. Clin. Periodontol. 40, 218–226, doi: 10.1111/jcpe.12047 (2013). [DOI] [PubMed] [Google Scholar]
- Maruyama N. et al. Intraindividual variation in core microbiota in peri-implantitis and periodontitis. Sci. Rep. 4, 6602, doi: 10.1038/srep06602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibli J. A. et al. Composition of supra- and subgingival biofilm of subjects with healthy and diseased implants. Clin. Oral Implants Res. 19, 975–982, doi: 10.1111/j.1600-0501.2008.01566.x (2008). [DOI] [PubMed] [Google Scholar]
- Miller C. S., Baker B. J., Thomas B. C., Singer S. W. & Banfield J. F. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 12, R44, doi: 10.1186/gb-2011-12-5-r44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes da Rocha U. et al. Isolation of a significant fraction of non-phototroph diversity from a desert Biological Soil Crust. Front. Microbiol. 6, 277, doi: 10.3389/fmicb.2015.00277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010, baq013, doi: 10.1093/database/baq013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo A. E. et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J 8, 1659–1672, doi: 10.1038/ismej.2014.23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost S., Duran-Pinedo A. E., Teles R., Krishnan K. & Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7, 27, doi: 10.1186/s13073-015-0153-3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702, doi: 10.1093/nar/gki866 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart F. J., Ulloa O. & DeLong E. F. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ. Microbiol. 14, 23–40, doi: 10.1111/j.1462-2920.2010.02400.x (2012). [DOI] [PubMed] [Google Scholar]
- Li W., Wooley J. C. & Godzik A. Probing metagenomics by rapid cluster analysis of very large datasets. PLos One 3, e3375, doi: 10.1371/journal.pone.0003375 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong Z., Sun L., Yang J. & Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40, D641–645, doi: 10.1093/nar/gkr989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. E. et al. MvirDB–a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 35, D391–394, doi: 10.1093/nar/gkl791 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K. & Zhang T. Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLos One 7, e38183, doi: 10.1371/journal.pone.0038183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D. & Pazos F. Protein functional features are reflected in the patterns of mRNA translation speed. BMC Genomics 16, 513, doi: 10.1186/s12864-015-1734-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keer J. T. & Birch L. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53, 175–183 (2003). [DOI] [PubMed] [Google Scholar]
- Campbell B. J. & Kirchman D. L. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7, 210–220, doi: 10.1038/ismej.2012.93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo A. E., Paster B., Teles R. & Frias-Lopez J. Correlation network analysis applied to complex biofilm communities. PLos One 6, e28438, doi: 10.1371/journal.pone.0028438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas J. A., Paster B. J., Stokes L. N., Olsen I. & Dewhirst F. E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732, doi: 10.1128/jcm.43.11.5721-5732.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E. G., Rajilic-Stojanovic M. & de Vos W. M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615, doi: 10.1136/gut.2007.133603 (2008). [DOI] [PubMed] [Google Scholar]
- Moeseneder M. M., Arrieta J. M. & Herndl G. J. A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol. Ecol. 51, 341–352, doi: 10.1016/j.femsec.2004.09.012 (2005). [DOI] [PubMed] [Google Scholar]
- Socransky S. S., Haffajee A. D., Cugini M. A., Smith C. & Kent R. L. Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998). [DOI] [PubMed] [Google Scholar]
- Heitz-Mayfield L. J. & Lang N. P. Comparative biology of chronic and aggressive periodontitis vs. peri-implantitis. Periodontol. 2000 53, 167–181, doi: 10.1111/j.1600-0757.2010.00348.x (2010). [DOI] [PubMed] [Google Scholar]
- Strom C. M. et al. Development and validation of a next-generation sequencing assay for BRCA1 and BRCA2 variants for the clinical laboratory. PLos One 10, e0136419, doi: 10.1371/journal.pone.0136419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214, doi: 10.1038/nature11234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A. et al. Comparative genome analysis and identification of competitive and cooperative interactions in a polymicrobial disease. ISME J 9, 629–642, doi: 10.1038/ismej.2014.155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale E. A. et al. Functional metagenomic profiling of nine biomes. Nature 452, 629–632, http://www.nature.com/nature/journal/v452/n7187/suppinfo/nature06810_S1.html (2008). [DOI] [PubMed] [Google Scholar]
- Lin L. et al. Virulence genes of Porphyromonas gingivalis W83 in chronic periodontitis. Acta Odontol. Scand. 67, 258–264, doi: 10.1080/00016350902841890 (2009). [DOI] [PubMed] [Google Scholar]
- Jorth P. et al. Metatranscriptomics of the human oral microbiome during health and disease. MBio 5, e01012–01014, doi: 10.1128/mBio.01012-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180, doi: 10.1038/nature09944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade W. G. et al. The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49 Pt 2, 595–600 (1999). [DOI] [PubMed] [Google Scholar]
- Abiko Y., Sato T., Mayanagi G. & Takahashi N. Profiling of subgingival plaque biofilm microflora from periodontally healthy subjects and from subjects with periodontitis using quantitative real-time PCR. J. Periodontal Res. 45, 389–395, doi: 10.1111/j.1600-0765.2009.01250.x (2010). [DOI] [PubMed] [Google Scholar]
- Tamura N., Ochi M., Miyakawa H. & Nakazawa F. Analysis of bacterial flora associated with peri-implantitis using obligate anaerobic culture technique and 16S rDNA gene sequence. Int. J. Oral Maxillofac. Implants 28, 1521–1529, doi: 10.11607/jomi.2570 (2013). [DOI] [PubMed] [Google Scholar]
- Wade W. G., Munson M. A., de Lillo A. & Weightman A. J. Specificity of the oral microflora in dentinal caries, endodontic infections and periodontitis. International Congress Series 1284, 150–157, http://dx.doi.org/10.1016/j.ics.2005.06.097 (2005). [Google Scholar]
- Zhou J., Deng Y., Luo F., He Z. & Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio 2, e00122–00111, doi: 10.1128/mBio.00122-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva E. S. et al. Microbiological diversity of peri-implantitis biofilm by Sanger sequencing. Clin. Oral Implants Res. 25, 1192–1199, doi: 10.1111/clr.12231 (2014). [DOI] [PubMed] [Google Scholar]
- Darveau R. P. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490, doi: 10.1038/nrmicro2337 (2010). [DOI] [PubMed] [Google Scholar]
- Berry D. & Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5, 219, doi: 10.3389/fmicb.2014.00219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386, doi: 10.1186/1471-2105-9-386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J. BLAT-the BLAST-like alignment tool. Genome Res. 12, 656–664, doi: 10.1101/gr.229202. Article published online before March 2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. D. et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2, 15, doi: 10.1186/2049-2618-2-15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milici M. et al. Co-occurrence analysis of microbial taxa in the Atlantic Ocean reveals high connectivity in the free-living bacterioplankton. Front. Microbiol. 7, 649, doi: 10.3389/fmicb.2016.00649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju F., Xia Y., Guo F., Wang Z. & Zhang T. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ. Microbiol. 16, 2421–2432, doi: 10.1111/1462-2920.12355 (2014). [DOI] [PubMed] [Google Scholar]
- Friedman J. & Alm E. J. Inferring correlation networks from genomic survey data. PLos Comput. Biol. 8, e1002687, doi: 10.1371/journal.pcbi.1002687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio I. et al. Oral samples as non-invasive proxies for assessing the composition of the rumen microbial community. PLos One 11, e0151220, doi: 10.1371/journal.pone.0151220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M. E., Ono K., Ruscheinski J., Wang P. L. & Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432, doi: 10.1093/bioinformatics/btq675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.