Abstract
A new multiplex PCR assay was developed to separate the four major Listeria monocytogenes serovars isolated from food and patients (1/2a, 1/2b, 1/2c, and 4b) into distinct groups. The PCR test, which constitutes a rapid and practical alternative to laborious classical serotyping, was successfully evaluated with 222 Listeria strains.
Listeria monocytogenes is a facultative intracellular pathogen that can cause serious illness in susceptible individuals. Persons with specific immunocompromising conditions, pregnant women, newborns, and the elderly are particularly at risk for listeriosis (9, 23). Although rare, listeriosis remains of great public health concern due to its high mortality rate (20 to 30%) (16). Ingestion of contaminated foods is considered to be the primary source of infection for both sporadic and epidemic human listeriosis cases (19). Because of the importance of L. monocytogenes strain characterizations for epidemiological investigations, a number of discriminatory subtyping methods have been described for this organism (2, 4, 5, 18, 20, 24, 25). Pulsed-field gel electrophoresis (PFGE) typing, which has provided the most sensitive strain discrimination up to now, has rapidly become the standard subtyping method to detect listeriosis outbreaks (4, 11). However, this method is labor-intensive and time-consuming and thus for practical purposes is often preceded by serotyping. Since all major outbreaks of the invasive form of listeriosis are due to serovar 4b strains, an infrequent serovar in foods compared to 1/2a strains (6, 9), the procedure adopted for outbreak investigations relies upon serovar characterization to provide valuable information for rapid screening of groups of strains. Indeed, the serovar information allows discrimination between isolates probably belonging to an outbreak and those that are not part of the outbreak and thus decreases the number of strains which need to be characterized by PFGE in order to improve discrimination beyond the serovar level. Moreover, serotyping is widely used for long-term microbiological surveillance of human listeriosis. For the food industry, where the presence of L. monocytogenes is a big concern, tracing contaminating strains within the food chain and the plant environment is of primary importance. Again serotyping is often used as a first-line typing method. Although 13 serovars are described for the species L. monocytogenes, at least 95% of the strains isolated from foods and patients are of serovars 1/2a, 1/2b, 1/2c, and 4b (12, 21, 22). Routine analysis of L. monocytogenes by serotyping with traditional agglutination methods is limited by cost, availability, and the need for technical expertise to perform the assay. Furthermore, the reproducibility of serotyping is not always satisfactory. Schonberg et al. (20) concluded in a multicenter study that a critical need exists for high-quality antisera. A new enzyme-linked immunosorbent assay serotyping format used in conjunction with a commercially available kit to make serotyping more efficient and more accessible was described by Palumbo et al. (17). Recently, a focused Listeria macroarray containing probes representative of specific sequences from three sequenced Listeria genomes, L. monocytogenes serovar 1/2a strain EGDe, L. monocytogenes serovar 4b strain CLIP 80459, and L. innocua serovar 6a strain CLIP 11262, has been constructed and hybridized with genomic DNA of Listeria strains from a diverse collection (8, 10). Based on the variable gene content, the three lineages I, II, and III of L. monocytogenes were further divided into five phylogenetic groups, each correlated with serovars: I.1 (1/2a-3a), I.2 (1/2c-3c), II.1 (4b-4d-4e), II.2 (1/2b-3b-7), and III (4a-4c) (8). Marker genes specifically associated with the four groups mentioned first were identified. Indeed, 19 genes identified in the L. monocytogenes serovar 1/2a strain EGDe were specifically associated with L. monocytogenes lineage I (1/2a-3a-1/2c-3c), and five genes identified in the partial sequence of L. monocytogenes serovar 4b strain CLIP 80459 were specifically associated with L. monocytogenes lineage II (1/2b-3b-7-4b-4d-4e). Furthermore, three genes identified in the L. monocytogenes serovar 4b strain CLIP 80459 were found specific to lineage II.1 (4b-4d-4e). Curiously lineage I.2 (1/2c-3c) was characterized by two genes (lmo1118 and lmo1119) identified in the L. monocytogenes serovar 1/2a strain EGDe, as they hybridized only with the tested serovar group (1/2c-3c) strains, except the serovar 1/2a strain EGDe (8). This last observation indicates that the EGDe strain is an atypical strain of serovar 1/2a. This fact is strengthened by the previous macroarray hybridization results indicating that the global genome structure of strain EGDe is more similar to that of strains of the serovar group 1/2c-3c than to strains of the serovar group 1/2a-3a (8). Alternatively, it is still possible that the presence of these two flanking genes in the serovar 1/2a EGDe strain is the result of a horizontal transfer.
In targeting simultaneously one gene of these four marker gene groups, we developed a new multiplex PCR assay in order to separate the four major serovars (1/2a, 1/2b, 1/2c, and 4b) of L. monocytogenes strains into four distinct groups. The marker genes selected for the multiplex PCR assay were lmo0737 and lmo1118, identified in the sequenced L. monocytogenes serovar 1/2a EGDe strain, and ORF2819 and ORF2110, identified in the partial sequence of L. monocytogenes 4b strain CLIP 80459. The prs gene, specific for strains of the genus Listeria, was targeted for an internal amplification control. Primer sequences used in this study, the putative functions of the selected marker genes, and their serovar specificities are outlined in Table 1. Each PCR product was designed for amplifying distinct fragment sizes between 370 and 906 bp. The specificity and reliability of the PCR method were first evaluated with 12 L. monocytogenes serovar reference strains (Fig. 1) and then confirmed with 222 Listeria strains isolated from humans and foods (Table 2). As we did not possess an L. monocytogenes strain of the 13th serovar described for this species (serovar 4ab) in our collection, containing >90,000 isolates, it was not tested in our study. Strain identification and serotyping were done according to standard methods (1, 21).
TABLE 1.
Nucleotide sequences of primer sets used in this study
| Gene target | Primer sequence (5′-3′)a | Product size (bp) | Serovar specificityb | Protein encoded by the target gene |
|---|---|---|---|---|
| lmo0737 | For: AGGGCTTCAAGGACTTACCC | 691 | L. monocytogenes serovars 1/2a, 1/2c, 3a, and 3c | Unknown, no similarity |
| Rev: ACGATTTCTGCTTGCCATTC | ||||
| lmo1118 | For: AGGGGTCTTAAATCCTGGAA | 906 | L. monocytogenes serovars 1/2c and 3c | Unknown, no similarity |
| Rev: CGGCTTGTTCGGCATACTTA | ||||
| ORF2819 | For: AGCAAAATGCCAAAACTCGT | 471 | L. monocytogenes serovars 1/2b, 3b, 4b, 4d, and 4e | Putative transcriptional regulator |
| Rev: CATCACTAAAGCCTCCCATTG | ||||
| ORF2110 | For: AGTGGACAATTGATTGGTGAA | 597 | L. monocytogenes serovars 4b, 4d, and 4e | Putative secreted protein |
| Rev: CATCCATCCCTTACTTTGGAC | ||||
| prs | For: GCTGAAGAGATTGCGAAAGAAG | 370 | All Listeria species | Putative phosphoribosyl pyrophos- phate synthetase |
| Rev: CAAAGAAACCTTGGATTTGCGG |
For, forward; Rev, reverse.
For the specificity of lmo1118 gene fragment amplification within L. monocytogenes strains of serovar 1/2c or 3c, we note the exception of the serovar 1/2a EGDe strain in which the gene was first identified.
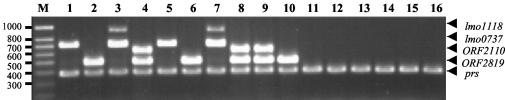
FIG. 1.
Agarose gel electrophoresis of DNA fragments generated by multiplex PCR with the serotyping reference strains of the species L. monocytogenes and strains of the remaining Listeria species. Lanes 1 to 12, L. monocytogenes reference strains of serovars 1/2a (CIP 104794), 1/2b (CIP 105449), 1/2c (CIP 105448), 4b (CIP 78.38), 3a (CIP 78.34), 3b (CIP 78.34), 3c (CIP 78.36), 4d (CIP 105458), 4e (CIP 105459), 7 (CIP 78.43), 4a (CIP 74908), and 4c (CIP 78.39), respectively; lane 13, L. innocua strain of serovar 6a (CLIP 91918); lane 14, L. welshimeri strain of serovar 4c (CLIP 87073); lane 15, L. ivanovii strain of serovar 5 (CLIP 88111); lane 16, L. seeligeri strain of serovar 1/2b (CLIP 73021); lane M, SmartLadder SF molecular weight markers (Eurogentec). Genes corresponding to the amplified fragments are indicated on the right. CIP, Collection of the Institut Pasteur; CLIP, Listeria Culture Collection of the National Reference Center for Listeria, Institut Pasteur. Molecular sizes are given (in base pairs) at the left.
TABLE 2.
Correlation of multiplex PCR and conventional serotyping results for Listeria tested strains
| Species | Serovar | No. of strains | Multiplex PCR fragment amplification
|
Multiplex PCR serovar (sv) group classification | ||||
|---|---|---|---|---|---|---|---|---|
| lmo1118 (906 bp) | lmo0737 (691 bp) | ORF2110 (597 bp) | ORF2819 (471 bp) | prs (370 bp) | ||||
| L. monocytogenes | 1/2a | 40 | −a | + | − | − | + | L. monocytogenes sv 1/2a, 3a |
| 1/2b | 40 | − | − | − | + | + | L. monocytogenes sv 1/2b, 3b, 7 | |
| 1/2c | 40 | + | + | − | − | + | L. monocytogenes sv 1/2c, 3c | |
| 4b | 40 | − | − | + | + | + | L. monocytogenes sv 4b, 4d, 4e | |
| 3a | 5 | − | + | − | − | + | L. monocytogenes sv 1/2a, 3a | |
| 3b | 5 | − | − | − | + | + | L. monocytogenes sv 1/2b, 3b, 7 | |
| 3c | 5 | + | + | − | − | + | L. monocytogenes sv 1/2c, 3c | |
| 7 | 5 | − | − | − | + | + | L. monocytogenes sv 1/2b, 3b, 7 | |
| 4d | 5 | − | − | + | + | + | L. monocytogenes sv 4b, 4d, 4e | |
| 4e | 5 | − | − | + | + | + | L. monocytogenes sv 4b, 4d, 4e | |
| 4a | 5 | − | − | − | − | + | Listeria species | |
| 4c | 5 | − | − | − | − | + | Listeria species | |
| L. innocua | 6a | 5 | − | − | − | − | + | Listeria species |
| 6b | 5 | − | − | − | − | + | Listeria species | |
| L. ivanovii | 5 | 8 | − | − | − | − | + | Listeria species |
| L. seeligeri | 1/2b | 2 | − | − | − | − | + | Listeria species |
| L. welshimeri | 4c | 1 | − | − | − | − | + | Listeria species |
| 6a | 1 | − | − | − | − | + | Listeria species | |
The L. monocytogenes serovar 1/2a EGDe strain, in which the lmo1118 gene was first identified, was the only strain from the serovar group 1/2a tested to amplify the 906-bp lmo1118 fragment.
As PCR templates, three to five bacterial colonies grown on Columbia agar plates (Bio-Rad Laboratories) were emulsified in 50 μl of an 0.25% sodium dodecyl sulfate-0.05 N NaOH solution and incubated at 99°C for 15 min. Then 100 μl of H2O was added to the mixture, 2 μl of which was used for the PCR. Amplification reactions were performed in a 96-well plate (Abgene) in a final volume of 100 μl containing 2 U of Taq DNA polymerase (Roche, Boehringer), 0.2 mM deoxynucleoside triphosphates (Perkin-Elmer), and 50 mM Tris-HCl-10 mM KCl-50 mM (NH4)2SO4-2 mM MgCl2, pH 8.3. The five primer sets were added at the following final concentrations: 1 μM for lmo0737, ORF2819, and ORF2110; 1.5 μM for lmo1118; and 0.2 μM for prs (Table 1). PCR was performed with an initial denaturation step at 94°C for 3 min; 35 cycles of 94°C for 0.40 min, 53°C for 1.15 min, and 72°C for 1.15 min; and one final cycle of 72°C for 7 min in a thermocycler (Icycler; Bio-Rad Laboratories). Five microliters of the reaction mixture was mixed with 3 μl of loading buffer and separated on a 2% agarose gel in a TBE buffer (90 mM Trizma base, 90 mM boric acid, 2 mM EDTA, pH 8.3). The PCR product was visualized by ethidium bromide staining.
Figure 1 shows examples of the multiplex PCR amplifications obtained under the above standard conditions with L. monocytogenes strains of different serovars and strains of the other Listeria species. All strains amplified the prs gene fragment (370 bp). The amplifications of the four chosen serovar-specific fragments allowed separation of L. monocytogenes strains into four groups. Group 1 comprised strains of serovars 1/2a and 3a (amplification of only the lmo0737 DNA fragment); group 2 comprised strains of serovars 1/2c and 3c (amplification of both lmo0737 and lmo1118 DNA fragments); group 3 comprised strains of serovars 1/2b, 3b, and 7 (amplification of only an ORF2819 DNA fragment); and group 4 comprised strains of serovars 4b, 4d, and 4e (amplification of both ORF2819 and ORF2110 DNA fragments). The absence of amplifications of the 1/2b serovar-specific fragment in the two L. seeligeri strains (serovar 1/2b) indicates a high specificity of our PCR method for the species L. monocytogenes (Fig. 1). Results summarized in Table 2 show that all isolates of serovars 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4d, 4d, 4e, and 7 of the species L. monocytogenes except the serovar 1/2a EGDe strain were correctly typed to serovar group level with the multiplex PCR, thus confirming the reliability of the test.
The proposed multiplex PCR profiles do not distinguish, within the species L. monocytogenes, serovar 1/2a from 3a, 1/2c from 3c, 1/2b from 3b and 7, or 4b from 4d and 4e. These profiles also do not distinguish L. monocytogenes serovars 4a and 4c from the other Listeria species. However, serovars 3a, 3b, 3c, 4a, 4c, 4e, 4d, and 7 are very infrequent in food and rarely reported as implicated in human listeriosis. The data collected by the National Reference Center in France showed that serovars 1/2a, 1/2b, 1/2c, and 4b, which are separated by our test into four distinct PCR profiles, represent over 98% of the 5,000 isolates collected from food and patients during the last 3 years. These observations suggest the adaptability of this classification method as an alternative to serotyping, in particular to long-term surveillance and epidemiological investigations.
Several PCR-based methods such as restriction enzyme analysis-PCR, PCR-single-strand conformation polymorphism, and mismatch amplification mutation assays-PCR were described in the literature for rapid L. monocytogenes interspecies subdivision typing (7, 13-15). However, these methods subdivided L. monocytogenes strains into only two or three subgroups in which 1/2a and 1/2c serovars and/or 1/2b and 4b serovars were not separated. Recently, Borucki and Call (3) described a PCR method that distinguished the serovars within the species L. monocytogenes as we propose, into five distinct groups. However, Borucki and Call needed to perform two or three independent PCRs to obtain this classification, and out of 122 L. monocytogenes strains tested, 4 strains were not typeable or were incorrectly classified. Moreover, the nonpathogenic L. innocua species seemed to be indistinguishable from L. monocytogenes strains of serovar 4b (3).
Therefore, the method proposed here should prove more powerful, as the targeting of different marker genes than those used by Borucki and Call allowed correct classification of all the strains included in our study in only one multiplex PCR step with easily interpretable results. Our proposed multiplex PCR assay is highly specific for pathogenic L. monocytogenes, providing thus in the same step a species confirmation also. Although an efficient enzyme-linked immunosorbent assay serotyping protocol adaptable to automation that makes serotyping less expensive and more efficient was described by Palumbo et al., a routine PCR technique with more accessible materials should increase the feasibility and accessibility of correct L. monocytogenes serogrouping (17).
Acknowledgments
This work received financial support from the Institut Pasteur.
REFERENCES
- 1.Bille, J., B. Catimel, E. Bannerman, C. Jacquet, M. N. Yersin, I. Caniaux, D. Monget, and J. Rocourt. 1992. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl. Environ. Microbiol. 58:1857-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boerlin, P., and J. C. Piffaretti. 1991. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 57:1624-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borucki, M. K., and D. R. Call. 2003. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 41:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosch, R., M. Brett, B. Catimel, J. B. Luchansky, B. Ojeniyi, and J. Rocourt. 1996. Genomic fingerprinting of 80 strains from the WHO multicenter international typing study of Listeria monocytogenes via pulsed-field gel electrophoresis (PFGE). Int. J. Food Microbiol. 32:343-355. [DOI] [PubMed] [Google Scholar]
- 5.Bruce, J. L., R. J. Hubner, E. M. Cole, C. I. McDowell, and J. A. Webster. 1995. Sets of EcoRI fragments containing ribosomal RNA sequences are conserved among different strains of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 92:5229-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchrieser, C., R. Brosch, B. Catimel, and J. Rocourt. 1993. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can. J. Microbiol. 39:395-401. [DOI] [PubMed] [Google Scholar]
- 7.Comi, G., L. Cocolin, C. Cantoni, and M. Manzano. 1997. A RE-PCR method to distinguish Listeria monocytogenes serovars. FEMS Immunol. Med. Microbiol. 18:99-104. [DOI] [PubMed] [Google Scholar]
- 8.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 11.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 12.Graves, L. M., B. Swaminathan, and S. B. Hunter. 1999. Subtyping Listeria monocytogenes, p. 251-297. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety. Marcel Dekker Inc., New York, N.Y.
- 13.Jinneman, K. C., and W. E. Hill. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 14.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 1998. A rapid method for the identification and partial serotyping of Listeria monocytogenes in food by PCR and restriction enzyme analysis. Int. J. Food Microbiol. 42:207-212. [DOI] [PubMed] [Google Scholar]
- 15.Manzano, M., L. Cocolin, C. Pipan, E. Falasca, G. A. Botta, C. Cantoni, and G. Comi. 1997. Single-strand conformation polymorphism (SSCP) analysis of Listeria monocytogenes iap gene as tool to detect different serogroups. Mol. Cell. Probes 11:459-462. [DOI] [PubMed] [Google Scholar]
- 16.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocourt, J., A. Audurier, A. L. Courtieu, J. Durst, S. Ortel, A. Schrettenbrunner, and A. G. Taylor. 1985. A multi-centre study on the phage typing of Listeria monocytogenes. Zentbl. Bakteriol. Mikrobiol. Hyg. A 259:489-497. [DOI] [PubMed] [Google Scholar]
- 19.Schlech, W. F., III, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 20.Schonberg, A., E. Bannerman, A. L. Courtieu, R. Kiss, J. McLauchlin, S. Shah, and D. Wilhelms. 1996. Serotyping of 80 strains from the WHO multicentre international typing study of Listeria monocytogenes. Int. J. Food Microbiol. 32:279-287. [DOI] [PubMed] [Google Scholar]
- 21.Seeliger, H. P. R., and K. Höhne. 1979. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13:31-49. [Google Scholar]
- 22.Tappero, J. W., A. Schuchat, K. A. Deaver, L. Mascola, J. D. Wenger, et al. 1995. Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts? JAMA 273:1118-1122. [DOI] [PubMed] [Google Scholar]
- 23.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wernars, K., P. Boerlin, A. Audurier, E. G. Russell, G. D. Curtis, L. Herman, and N. van der Mee-Marquet. 1996. The WHO multicenter study on Listeria monocytogenes subtyping: random amplification of polymorphic DNA (RAPD). Int. J. Food Microbiol. 32:325-341. [DOI] [PubMed] [Google Scholar]
- 25.Wesley, I. V., and F. Ashton. 1991. Restriction enzyme analysis of Listeria monocytogenes strains associated with food-borne epidemics. Appl. Environ. Microbiol. 57:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]