Abstract
The 16S rRNA gene has previously been used to develop genus-specific PCR primers for identification of enterococci. In addition, the superoxide dismutase gene (sodA) has been identified as a potential target for species differentiation of enterococci. In this study, Enterococcus genus-specific primers developed by Deasy et al. (E1/E2) were incorporated with species-specific primers based upon the superoxide dismutase (sodA) gene for development of a multiplex PCR. This assay provides simultaneous genus and species identification of 23 species of enterococci using seven different reaction mixtures. Accuracy of identification of the multiplex PCR was determined by comparisons to standard biochemical testing, the BBL Crystal kit, VITEK, and API Rapid ID 32 Strep. Isolates from swine feces, poultry carcasses, environmental sources, and retail food were evaluated and, overall, results for 90% of the isolates tested by PCR agreed with results obtained using standard biochemical testing and VITEK. Eighty-five percent and 82% of PCR results agreed with results from the API Rapid ID 32 Strep and BBL Crystal tests, respectively. With the exception of concurrence between identification using standard biochemical testing and VITEK (85%) and between BBL Crystal and VITEK (83%), the percent agreement for PCR was higher than or equal to any other pairwise comparison. Multiplex PCR for genus and species determination of enterococci provides an improved, rapid method for identification of this group of bacteria.
Enterococci are important not only because they are a leading cause of nosocomial infections, but also because they may have a significant role in dissemination and persistence of antimicrobial resistance (10, 11). Correct identification is necessary in order to monitor which species are causing disease, for treatment purposes. Presently, the standard method for identification of enterococci is phenotypic characterization, primarily using biochemical tests (3, 4). Tests are usually performed in test tubes and may require significant amounts of time for preparation and interpretation of results. Furthermore, processing of large numbers of samples is inhibited by phenotypic characterization, as 10 or more tests may be necessary for differentiation of the species.
Commercial identification kits, such as the API Rapid ID 32 Strep and BBL Crystal identification gram-positive ID kits, and automated identification systems, such as the VITEK gram-positive identification system, are available for identifying enterococci to the species level (5, 7, 18). These methods have been developed to allow rapid identification of enterococci based upon reactions to panels of biochemicals. Although the kits are cost-effective and results can be obtained in less than 24 h, there are concerns about the reliability of the kits (2, 5). Possible reasons for these observations include atypical species that do not conform to the present biochemical testing scheme or newer species that have not been routinely encountered using the kits. Also, the majority of commercial kits and the traditional phenotypic characterization have been evaluated using clinical enterococcal strains and not isolates from environmental, agricultural, or animal sources. Enterococcal strains from these sources have not been extensively studied using commercial kits and therefore may also not be correctly identified.
In order to overcome problems associated with biochemical testing, molecular methods for identification have been developed. Genus-specific PCR primers to 16S rRNA have already been designed and found useful for distinguishing strains of Enterococcus (2). PCR amplification followed by sequencing and sequence comparison of target genes has also allowed differentiation of species of enterococci. To date, several genes, such as heat shock protein 60, elongation factor EF-Tu, d-Ala:d-Ala ligase, and manganese-dependent superoxide dismutase appear to have species-specific variable regions which may be useful for further development of methods for species identification (6, 9, 13, 16). Although procedures for genus and species identifications of enterococci have been developed separately, a single technique that would identify both the genus and species simultaneously has yet to be reported. In order to develop this procedure, genus-specific primers were combined with species-specific primers in several different reactions by using multiplex PCR. Because variations in sequences of manganese-dependent superoxide dismutase (sodA) genes appeared to be greater between species and less within species, this gene was used for designing species-specific primers. In addition, since genus primers were included in the reaction, it was not necessary to test species primers against numerous sodA sequences from other gram-positive bacteria. This novel multiplex PCR identifies 23 species of enterococci and greatly simplifies the identification procedure, allowing its use in the most basic laboratories.
MATERIALS AND METHODS
Bacterial strains, isolation, and identification.
Twenty-three enterococcal type strains and Enterococcus ratti (ATCC 700914) obtained from the American Type Culture Collection were used as controls in this study (Table 1). Lactococcus garvieae (ATCC 43921) was used as a negative control. One hundred additional enterococci used in this study were randomly selected from a group of isolates collected during 2000 to 2003 to ensure diversity of source. These enterococci were isolated from poultry carcass rinsates, fruits, vegetables, retail meats, and environmental rinsates or from swine fecal samples collected on-farm. All media used in this study were purchased from Becton Dickinson (Sparks, Md.). One-milliliter aliquots of rinsates were inoculated into BBL Enterococcosel broth and incubated for 24 h at 37°C to enrich for enterococci. Presumptive positive cultures were transferred onto BBL Enterococcosel agar and incubated for 24 h at 37°C. For swine fecal samples, 1 g of fecal sample was diluted 1:10 in phosphate-buffered saline (pH 7.4) and vortexed. One hundred microliters of the diluted sample was inoculated onto BBL Enterococcosel agar and incubated for 24 h at 37°C. A single colony of presumptive enterococcal isolates was subcultured onto slants of brain heart infusion agar (BHIA) for initial storage. From slants, isolates were then subcultured twice onto blood agar (Trypticase soy agar containing 5% defibrinated sheep blood) or blood agar followed by Columbia agar for identification. A single colony of each positive culture was frozen in glycerol medium at −70°C.
TABLE 1.
PCR primers, products, and reference strains
| Strain | Primer | Sequence (5′-3′) | Product size (bp) | Multiplex group |
|---|---|---|---|---|
| E. asini ATCC 700915 | AS1 | GCATCATGACAAGCATCACGC | 365 | 7 |
| AS2 | GGCTTTTTGCCTTCAGATAAA | |||
| E. avium ATCC 14025 | AV1 | GCTGCGATTGAAAAATATCCG | 368 | 5 |
| AV2 | AAGCCAATGATCGGTGTTTTT | |||
| E. casseliflavus ATCC 25788 | CA1 | TCCTGAATTAGGTGAAAAAAC | 288 | 2 |
| CA2 | GCTAGTTTACCGTCTTTAACG | |||
| E. cecorum ATCC 43198 | CE1 | AAACATCATAAAACCTATTTA | 371 | 6 |
| CE2 | AATGGTGAATCTTGGTTCGCA | |||
| E. columbae ATCC 51263 | CO1 | GAATTTGGTACCAAGACAGTT | 284 | 5 |
| CO2 | GCTAATTTACCGTTATCGACT | |||
| E. dispar ATCC 51266 | DI1 | GAACTAGCAGAAAAAAGTGTG | 284 | 3 |
| DI2 | GATAATTTACCGTTATTTACC | |||
| E. durans ATCC 19432 | DU1 | CCTACTGATATTAAGACAGCG | 295 | 1 |
| DU2 | TAATCCTAAGATAGGTGTTTG | |||
| E. faecalis ATCC 19433 | FL1 | ACTTATGTGACTAACTTAACC | 360 | 1 |
| FL2 | TAATGGTGAATCTTGGTTTGG | |||
| E. faecium ATCC19434 | FM1 | GAAAAAACAATAGAAGAATTAT | 215 | 1 |
| FM2 | TGCTTTTTTGAATTCTTCTTTA | |||
| E. flavescens ATCC 49996 | FV1 | GAATTAGGTGAAAAAAAAGTT | 284 | 4 |
| FV2 | GCTAGTTTACCGTCTTTAACG | |||
| E. gallinarum ATCC 49673 | GA1 | TTACTTGCTGATTTTGATTCG | 173 | 2 |
| GA2 | TGAATTCTTCTTTGAAATCAG | |||
| E. gilvus ATCC BAA-350 | GI1 | CTGGCTGGGCTTGGCTAGTGA | 98 | 7 |
| GI2 | ATAATCGGTGTTTTACCGTCT | |||
| E. hirae ATCC 8043 | HI1 | CTTTCTGATATGGATGCTGTC | 187 | 6 |
| HI2 | TAAATTCTTCCTTAAATGTTG | |||
| E. malodoratus ATCC 43197 | MA1 | GTAACGAACTTGAATGAAGTG | 134 | 1 |
| MA2 | TTGATCGCACCTGTTGGTTTT | |||
| E. mundtii ATCC 43186 | MU1 | CAGACATGGATGCTATTCCATCT | 98 | 4 |
| MU2 | GCCATGATTTTCCAGAAGAAT | |||
| E. pallens ATCC BAA-351 | PA1 | TGGCACCAAATGCTGGCGGAA | 160 | 7 |
| PA2 | TGGTGTAGAAGTAATTTCAAG | |||
| E. porcinus/villorum ATCC 700913 | PO1 | TGGTTTCTGATATGGATGCGA | 280 | 7 |
| PO2 | GTAATCGCTAATTTCTCTCCA | |||
| E. pseudoavium ATCC 49372 | PV1 | TCTGTTGAGGATTTAGTTGCA | 173 | 3 |
| PV2 | CCGAAAGCTTCGTCAATGGCG | |||
| E. raffinosus ATCC 49427 | RF1 | GTCACGAACTTGAATGAAGTT | 287 | 6 |
| RF2 | AATGGGCTATCTTGATTCGCG | |||
| E. saccharolyticus ATCC 43076 | SA1 | AAACACCATAACACTTATGTG | 371 | 3 |
| SA2 | GTAGAAGTCACTTCTAATAAC | |||
| E. seriolicida ATCC 49156 | SE2 | ACACAATGTTCTGGGAATGGC | 100 | 5 |
| SE2 | AAGTCGTCAAATGAACCAAAA | |||
| E. solitarius ATCC 49428 | SO1 | AAACACCATAACACTTATGTGACG | 371 | 2 |
| SO2 | AATGGAGAATCTTGGTTTGGCGTC | |||
| E. sulfureus ATCC 49903 | SU1 | TCAGTGGAAGACTTAATCGCA | 173 | 4 |
| SU2 | CCAAATGTATCTTCGATCGCT |
Identification using standard biochemical testing and commercial kits.
Standard biochemical testing for species identification was performed at the Centers for Disease Control and Prevention (CDC) as previously described (3, 4). Enterococcal species identification was performed in duplicate on isolates from blood agar using the BBL Crystal kit and the BBL Crystal AutoReader (Becton Dickinson) and the API Rapid ID 32 Strep kit (bioMerieux, Durham, N.C.). Duplicate isolates from Columbia agar were identified using the automated VITEK system (bioMerieux). Manufacturers' instructions were followed for all procedures. No additional tests were necessary for determination of species using VITEK.
Primers.
Enterococcal genus primers were as previously published (2). Enterococcal superoxide dismutase (sodA) gene sequences were acquired from the National Center for Biotechnology Information public databases. Additional sequences were generated by amplification of a portion of sodA by using degenerate primers and then sequencing the PCR products at the ARS Regional Sequencing Facility, Southeastern Poultry Research Laboratory, Athens, Ga. (16). Sequences were compared to other sodA gene sequences using NCBI-BLAST analysis and aligned using Align Plus (Scientific and Educational Software, Durham, N.C.). Conserved sequences within species and degenerate regions between species were used to design species-specific primers with the Oligo primer analysis software (Molecular Biology Insights, Inc., Cascade, Colo.). All primers were synthesized by Operon (Alameda, Calif.).
PCR.
Template for PCR was prepared by suspending a single isolated bacterial colony in 100 μl of sterile deionized water. Seven PCR master mixes consisting of different primer sets were prepared. Group 1 was E. durans, E. faecalis, E. faecium, and E. malodoratus; group 2 was E. casseliflavus, E. gallinarum, and E. solitarius; group 3 was E. dispar, E. pseudoavium, and E. saccharolyticus; group 4 was E. flavescens, E. mundtii, and E. sulfureus; group 5 was E. avium, E. columbae, and E. seriolicida; group 6 was E. cecorum, E. hirae, and E. raffinosus; and group 7 was E. asini, E. gilvus, E. pallens, and E. porcinus/villorum. The base master mix consisted of 3 mM MgCl2 (with Ficoll and tartrazine; Idaho Technology, Salt Lake City, Utah), 0.2 mM deoxynucleoside triphosphate mix (Roche, Indianapolis, Ind.), 16 mM (10×) NH4, 3.5 U of Expand high-fidelity PCR system (Roche), and 1.25 μl of each genus primer (16 μM). With the exception of E. faecalis, E. malodoratus, E. gallinarum, E. saccharolyticus, and E. dispar, 1.25 μl of each species primer (16 μM) was added to the base mix as indicated (Table 1). For primers FL1, FL2, MA1, MA2, GA1, GA2, SA1, SA2, DI1, and DI2, 2.5 μl of each primer was used. PCRs were performed in a final volume of 22.5 μl consisting of 20 μl of master mix and 2.5 μl of whole-cell template. Following an initial denaturation at 95°C for 4 min, products were amplified by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C (groups 1, 2, 5, and 6) or 60°C (groups 3, 4, and 7) for 1 min, and elongation at 72°C for 1 min. Amplification was followed by a final extension at 72°C for 7 min. Ten microliters of product was electrophoresed on a 2% 1× Tris-acetate-EDTA agarose gel containing 2 μg of ethidium bromide/ml. DNA molecular weight marker XIV (100 bp; Roche) was used as the standard.
RESULTS
Primer design.
Using previously designed sodA degenerate primers, a 438-bp internal fragment of the gene was amplified from enterococcal type strains (Table 1). Both strands of the PCR product were sequenced and analyzed to form one contiguous sequence. Multiple sequences were aligned using generated sodA sequences and those available in public databases, allowing conserved regions within species and variable regions between species to be identified (data not shown). Due to the small region sequenced, the number of isolates analyzed, and the close relatedness of some species, some species primers differed from other primers by only a few base pairs, primarily on the 3′ end (Table 1). Differences in only 1 bp were enough to amplify sodA from multiple target species, but not from other enterococcal species (data not shown). For other species, one forward or reverse primer was identical in sequence to another species primer, but the remaining primer in the set was different. These manipulations in conjunction with a proofreading polymerase mix allowed only the correct target sequence to be amplified. Selected sodA enterococcal species primer sequences were not compared to other bacterial species sequences, because genus-specific primers were included in the PCR, verifying the genus of the isolate. Groups of species primers were developed due to the small region of sodA sequenced (Table 1). Each group contained primer sets that would amplify a portion of DNA in four size ranges: 95 to 135 bp, 170 to 215 bp, 280 to 300 bp, or ≥360 bp. These size ranges allowed separation of amplified product while also standardizing amplicon sizes across groups. Repeated attempts to amplify a fragment from E. ratti were unsuccessful, resulting in no primer sets for this species.
Amplification using enterococcal genus and species primers.
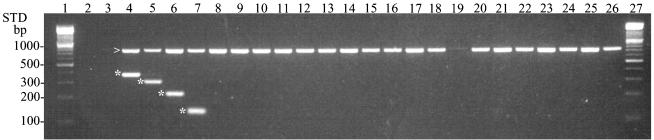
The specificity of the genus and species PCR was determined by testing all species shown in Table 1 against all groups of the multiplex primer sets. All strains reacted with the enterococcal genus primer, indicating that they were members of Enterococcus (Fig. 1). A very weak band was produced with E. seriolicida despite repeated attempts (Fig. 1, lane 19). Only positive control strains of group 1 (E. faecalis, E. durans, E. faecium, and E. malodoratus) reacted with the appropriate group 1 species primers, producing products of 360, 295, 215, and 134 bp, respectively (Fig. 1). This process was repeated for all groups to verify specificity and rule out cross-reactivity. L. garvieae, used as a negative DNA control, did not react with the genus primers or any of the species primers, and no product was produced in control samples in which target DNA was not provided (Fig. 1).
FIG. 1.
Group 1 genus and species multiplex PCR of enterococci. All species of enterococci were tested against group 1 multiplex primers in order to confirm specificity. Genus-specific bands are indicated by the arrow, and species-specific bands are indicated by asterisks. Species positive controls are in lanes 4 to 7 as follows: E. faecalis (360 bp), E. durans (295 bp), E. faecium (215 bp), and E. malodoratus (134 bp). Negative controls in lanes 2 and 3 contained no DNA and L. garvieae, respectively. Lanes 8 to 26: E. solitarius, E. casseliflavus, E. gallinarum, E. mundtii, E. saccharolyticus, E. dispar, E. pseudoavium, E. gilvus, E. flavescens, E. sulfureus, E. raffinosus, E. seriolicida, E. avium, E. columbae, E. cecorum, E. hirae, E. asini, E. porcinus, and E. pallens. Lanes 1 and 27, DNA standard.
Ease of use of the multiplexing PCR was achieved by first circumventing the need for purifying template DNA. Although the PCR performed well with pure DNA, isolated colonies suspended in sterile water worked equally as well and required little processing. The DNA was released from the cells during the initial 95°C denaturation step, and negative results with strains that produced no product with genus and species primers were not due to heat-resistant strains. Results using colonies isolated on blood agar were somewhat problematic in the PCR, but all colonies from BHIA produced the expected amplicons. This suggests that some components present in blood agar may have interfered with the PCR, yielding negative results. An essential requirement for the multiplex PCR was a pure starting culture. Strains streaked less than twice appeared mixed, producing signals in multiple PCR groups. For example, weak bands in group 4 representing E. flavescens and intense bands in group 1 (E. faecalis) were apparent for some isolates from poultry and retail foods. This problem was resolved when the isolates were restreaked onto BHIA and retested (data not shown). Therefore, in order to ensure purity of culture, isolates were routinely streaked onto nonselective medium at least twice before selection of colonies.
Another important feature of the multiplex PCR was the ability to amplify all PCR groups by using either a 55 or 60°C annealing temperature. Higher annealing temperatures (greater than 60°C) were tested but resulted in reduced product yield or negative results. FM1 and FM2 primers appeared to be sensitive to increased annealing temperatures, as the E. faecium positive control did not amplify well at 60°C annealing. Group 2 species primers were the most sensitive to changes in annealing temperature and in-lab variations. Care was taken with reaction components (primers, etc.) to ensure successful amplification of controls. Considerable variation in PCR results was also observed when the supplier of PCR primers was changed. In addition to annealing temperature, all other components of the PCR were standardized between reactions, except primer concentrations. Primer quantities for some species were less than for other species due to primer interference. For example, higher concentrations of E. faecalis and E. durans primers reduced the intensity of the E. faecium band. Reduction in intensity or no product was also observed when all positive controls for a PCR group were mixed in one tube for multiplexing. This could be due to overload of the reaction mixture with target DNA or impurities in the target DNA. The best results were obtained when controls were used in individual reactions.
Identification of amplified products.
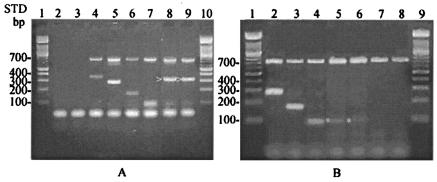
In rare cases, nonspecific products were observed when using the genus- and species-specific multiplex PCR. In order to be identified as an Enterococcus species, amplicons produced by unknown strains were required to be the same size as the genus and species PCR product. If the bands were not the expected size, then they were not identified as a particular species. An example of an errant band present in several multiplexing groups is shown in Fig. 2. Two unknown enterococcal isolates were tested against all seven groups of species primers. A band was obtained for both isolates when using groups 7, 6, and 4 (Fig. 2). Although an intense similar-sized band (300 to 400 bp) was obtained using group 7 primers, this band was slightly larger than the 280-bp E. porcinus/villorum band and slightly smaller than the 365-bp E. asini band (Fig. 2A). When tested with group 6 primers, amplicons for the two isolates were located between the 371-bp band representing E. cecorum and the 287-bp band representing E. raffinosus, indicating that the isolates were not E. cecorum or E. raffinosus (data not shown). But, when tested against group 4 primers, the same-sized band for both isolates and E. mundtii was observed, indicating that the unknown isolates were E. mundtii (Fig. 2B).
FIG. 2.
Nonspecific amplicons of genus and species multiplex PCR. Group 7 primers (A) and group 4 primers (B) are shown. Arrows indicate nonspecific bands, and asterisks indicate specific bands. (A) Lanes 2 and 3, no DNA control and L. garvieae, respectively; lanes 4 to 9, E. asini, E. porcinus/villorum, E. pallens, E. gilvus, unknown species A, and unknown species B. (B) Lanes 2 to 8, E. flavescens, E. sulfureus, E. mundtii, unknown species A, unknown species B, E. faecalis, and E. faecium. Lanes 1 and 10 (A) and lanes 1 and 9 (B) are DNA standards.
Comparison of PCR to standard biochemical testing and commercial identification methods.
One hundred isolates from four different sources (swine, poultry, environmental, and retail food) were identified using standard biochemical tests, the BBL Crystal Gram-Positive ID kit, VITEK, Rapid ID 32 Strep, and multiplex PCR in order to determine the accuracy of the PCR (Table 2). Standard biochemical tests can identify all enterococcal species, whereas the three kits allowed identification of seven primary species (E. avium, E. casseliflavus, E. durans, E. faecalis, E. faecium, E. gallinarum, and E. hirae). In addition to these seven species, Rapid ID 32 Strep would also identify E. saccharolyticus, whereas BBL Crystal would also identify E. raffinosus and E. solitarius. E. casseliflavus and E. gallinarum were grouped into one species with the BBL Crystal kit, requiring additional tests for differentiation. In contrast to the commercial kits, the multiplex PCR identified 23 enterococcal species.
TABLE 2.
Identification of enterococci by different identification methods
| Source | Sample | Species determination
|
|||||
|---|---|---|---|---|---|---|---|
| Standard | PCR | BBL | Vitek | ID 32 Strep | Consensus ID | ||
| Swine feces | 110 | E. hirae | E. hirae | E. durans | E. hirae | E. hirae | E. hirae |
| 113 | E. hirae | E. hirae | E. durans | E. hirae | E. hirae | E. hirae | |
| 124 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 126 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 127 | E. faecalis | E. faecalis | E. faecium | E. faecalis | E. faecalis | E. faecalis | |
| 188 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 190 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. hirae | E. faecalis | |
| 186 | E. hirae | E. hirae | E. durans | E. hirae | E. gallinarum | E. hirae | |
| 165 | E. durans | E. hirae | E. durans | E. hirae | E. hirae | E. hirae | |
| 166 | E. hirae | E. hirae | E. durans | E. hirae | E. hirae | E. hirae | |
| 170 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 135 | E. durans | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae | |
| 134 | E. faecium | E. faecium | E. faecium | E. faecium | E. gallinarum | E. faecium | |
| 133 | E. faecium | E. faecium | E. faecium | E. faecalis | E. gallinarum | E. faecium | |
| 131 | E. hirae | E. hirae | E. faecium | E. hirae | E. hirae | E. hirae | |
| 130 | E. faecium | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. gallinarum | E. casseliflavus | |
| 125 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 128 | E. faecium | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 129 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 141 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. hirae | E. faecalis | |
| 107 | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | |
| 108 | E. faecium | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 109 | E. faecium | E. faecium | E. faecium | E. faecium | E. gallinarum | E. faecium | |
| 151 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| 152 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| Poultry carcass | A1 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| A2 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A3 | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | |
| A4 | E. faecium | E. faecium | E. hirae | E. faecium | E. faecalis | E. faecium | |
| A5 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A6 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A7 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A8 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A9 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A10 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| A11 | E. hirae | E. hirae | E. durans | E. hirae | E. hirae | E. hirae | |
| A12 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| B1 | E. faecium | E. faecium | E. durans | E. faecium | E. gallinarum | E. faecium | |
| B2 | E. hirae | E. hirae | E. durans | E. hirae | E. hirae | E. hirae | |
| B3 | E. faecium | E. faecalis | E. faecium | E. faecium | E. gallinarum | E. faecium | |
| B4 | E. faecium | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | |
| B5 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| B6 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | |
| B7 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| B8 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| B9 | E. hirae | E. hirae | E. durans | E. hirae | E. hirae | E. hirae | |
| B10 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| B11 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| B12 | E. faecalis | E. faecalis | E. faecalis | E. gallinarum | E. faecalis | E. faecalis | |
| C1 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | |
| Park | |||||||
| Sand | Mem-9 | E. casseliflavus | E. casseliflavus/flav | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Fly strip | Mem-18 | E. casseliflavus | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Metal picnic table | Bish-1 | E. mundtii | E. mundtii | E. faecium | E. faecium | E. casseliflavus | NDa |
| Metal picnic table | Bish-2 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Metal trash can | Bish-3 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Rubber swing | Bish-4 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Ladder bar | Bish-10 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Plastic slide | Bish-12 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Steering wheel | Bish-13 | E. gallinarum | E. gallinarum | E. gallinarum | E. gallinarum | E. gallinarum | E. gallinarum |
| Plastic slide | Bish-14 | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae |
| Plastic slide | Bish-15 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Metal picnic table | Bish-17 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Plastic slide | Bish-19 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Plastic slide | Bish-20 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Sand | Bish-21 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Sand | Bish-22 | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae |
| Sand | Bish-23 | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae |
| Sand | Bish-24 | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae | E. hirae |
| Fly strip | Bish-25 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Fly strip | Bish-26 | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium |
| Plastic slide | SC-5 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Plastic slide | SC-6 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Tic-tac-toe wheels | SC-9 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Step vine climber | SC-10 | E. mundtii | E. mundtii | E. faecium | E. faecium | E. casseliflavus | ND |
| Plastic slide | SC-11 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. gallinarum | E. casseliflavus | E. casseliflavus |
| Retail foods | |||||||
| Red potato | BSM 36 | E. casseliflavus | E. casseliflavus/flav | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Chicken | BSM 35 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Red potato | BSM 37 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Beef | BSM 29 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Red potato | BSM 34 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Lettuce | KSM 24 | E. mundtii | E. mundtii | E. casseliflavus/gallinarum | E. faecium | E. casseliflavus | ND |
| Turkey | KSM 28 | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium |
| Turkey | KSM 29 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Lettuce | KSM 25 | E. mundtii | E. mundtii | E. faecium | E. faecium | E. casseliflavus | ND |
| Chicken | KSM 26 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Cucumber | WSM 17 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Beef | WSM 29 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Pork | WSM 32 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Chicken | WSM 26 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| White potato | ISM 42 | E. mundtii | E. mundtii | E. faecium | E. faecium | E. casseliflavus | ND |
| Pork | ISM 30 | E. faecalis | E. faecalis | E. faecalis | E. gallinarum | E. faecalis | E. faecalis |
| Red potato | ISM 32 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Red potato | ISM 36 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Apple | ISM 1 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Turkey | ISM 26 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Pork | FSM 29 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| White potato | FSM 38 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
| Beef | FSM 26 | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium | E. faecium |
| Red potato | FSM 32 | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus | E. casseliflavus |
| Chicken | FSM 23 | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis | E. faecalis |
ND, not determined.
All five methods identified the isolates as belonging to the genus Enterococcus, but not all species identifications agreed (Table 2). Consensus identification was determined by matching results from at least three of the five methods. From swine, poultry, environmental, and retail food samples, five consensus species (E. faecalis, E. faecium, E. casseliflavus, E. hirae, and E. gallinarum) were identified. The predominant species was E. faecalis (n = 53), followed by E. casseliflavus (n = 16) and E. hirae (n = 14). Although some isolates were identified as E. durans or E. mundtii in preliminary biochemical analyses, these species were not the consensus species when results from all identification methods were combined.
Overall, 69% (69 of 100) of the isolates tested agreed in species determination for all five methods, whereas 19% agreed with four of five methods. Consensus identification could not be confidently determined in 5% of the isolates. For these isolates, only two of the five methods agreed in species identification. This was due to certain tendencies of some of the commercial kits. For example, when standard testing, PCR, VITEK, and ID 32 Strep identified an isolate as E. hirae, BBL Crystal identified the same isolate as E. durans. This accounted for 6 of 19 (31.5%) of the isolates for which one test method differed. In addition, ID 32 Strep appeared to identify more E. gallinarum than any other commercial method (Table 2). Two isolates identified as E. gallinarum by ID 32 Strep were identified as E. faecium using the other methods. This kit also identified two isolates as E. hirae while the other four methods identified them as E. faecalis (Table 2). Standard biochemical testing also differed from the other four tests for 5 of 19 (26%) of the isolates. Three isolates identified as E. faecium by standard testing were identified as E. faecalis and E. casseliflavus by the other four methods. Differences in identification between the multiplex PCR and the commercial identification kits also resulted from identification of a species by PCR that the commercial kits could not identify. For example, the multiplex PCR identified five E. mundtii isolates, whereas those isolates were identified as E. faecium, E. casseliflavus, or E. gallinarum by the commercial kits. All four species share similar biochemical traits and belong in group II of the classical phenotypic characterization table, suggesting a close relationship (3). Standard biochemical testing results concurred with results from PCR, as those five isolates were also identified as E. mundtii (Table 2). For PCR, there was only one isolate (B3) for which results did not agree with results from any other test. Since outcomes of the traditional phenotypic tests can be variable, some misidentification of the isolates can occur using those methods. However, no distinct pattern could be discerned from the isolates for which two groups gave matching identifications but the remaining three groups did not.
When all five identification methods were compared against each other, discrepancies were apparent (Table 3). The multiplex PCR agreed with BBL Crystal for 82% of the total samples. Although percent agreement increased when results from PCR were compared with those from ID 32 Strep (85%), the highest percent agreements were observed between PCR and VITEK (90%) and PCR and standard testing (93%). With the exception of the BBL-VITEK comparison (83%) and the standard testing-VITEK comparison (85%), the percent agreement between PCR and all other methods was higher than for any other combination of identification methods. The lowest percent agreements in identification were 79% for standard testing and ID 32 Strep, 78% for standard testing and BBL Crystal, and only 77% between BBL Crystal and ID 32 Strep (Table 3). When source of isolates was examined, percent agreement between PCR and VITEK was higher than any other combination for swine, whereas percent agreement between PCR and standard testing was higher for poultry (96%), environmental (96%), and retail food samples (100%) (Table 3). Ninety-six percent agreement between BBL Crystal and VITEK was also observed for environmental samples. This discrepancy in agreement between PCR and VITEK can be explained by the identification of E. mundtii by PCR but not by VITEK. Two E. mundtii isolates from environmental samples and three from retail food samples were identified by PCR.
TABLE 3.
Comparison of enterococci identification methods
| Source | No. tested | No. in agreement (%) for the two methods
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| STD/PCR | STD/BBL | STD/VITEK | STD/ID 32 Strep | PCR/BBL | PCR/Vitek | PCR/ID 32 Strep | BBL/VITEK | BBL/ID 32 Strep | VITEK/ID 32 Strep | ||
| Swine | 25 | 20 (80) | 15 (60) | 20 (20) | 14 (56) | 18 (72) | 24 (96) | 18 (72) | 17 (68) | 12 (48) | 18 (72) |
| Poultry | 25 | 24 (96) | 19 (76) | 23 (92) | 21 (84) | 19 (76) | 23 (92) | 22 (88) | 19 (76) | 19 (76) | 21 (84) |
| Park | 25 | 24 (96) | 22 (88) | 21 (84) | 22 (88) | 23 (92) | 22 (88) | 23 (92) | 24 (96) | 23 (92) | 22 (88) |
| Retail food | 25 | 25 (100) | 22 (88) | 21 (84) | 22 (88) | 22 (88) | 21 (84) | 22 (88) | 23 (92) | 23 (92) | 21 (84) |
| Total | 100 | 93 (93) | 78 (78) | 85 (85) | 79 (79) | 82 (82) | 90 (90) | 85 (85) | 83 (83) | 77 (77) | 82 (82) |
DISCUSSION
Characterization and identification of enterococci by using the traditional phenotypic differentiation can be a tedious process requiring numerous tests. Strains are classified based upon growth in various media, biochemical reactions in those media, motility, and pigmentation. Although more than 20 species can be identified using these methods, tests are typically performed in test tubes and often require long periods of incubation before results can be interpreted (3, 4). In addition, grouping and identification of strains with phenotypic tests have been determined using type strains and strains isolated from human sources (17). Strains isolated from nonhuman sources may be atypical and may not conform to the criteria used for standard phenotypic characterization. Problems such as time constraints and number of samples to be processed can be overcome to some degree by using commercial identification kits. However, their accuracy has also been assessed using type strains and strains from clinical sources. Moreover, they usually only identify a maximum of 10 enterococcal species, which must also conform to the testing scheme. In addition, auto-reading of results should be utilized with commercial kits, as manual interpretation of results can lead to erroneous identification, and aberrant reactions will result in no species identification of the strain. An auto-reader was used for the commercial identification kits in this study and, therefore, factors other than manual reading of results must have accounted for the low overall agreement in identification.
Other methods for identification of enterococci have utilized molecular techniques such as PCR and sequencing (1, 6, 9, 12, 22). A previous report identified the manganese-dependent superoxide dismutase gene sodA as an ideal gene for species identification of enterococci (16). The superoxide dismutase gene has been used to distinguish genera and species of mycobacteria, streptococci, staphylococci, and enterococci (14, 15, 24). For differentiating species of enterococci, sodA gene sequences were used to create a library of sequences. Other unknown isolates were compared to the type strains and subsequently identified by percent homology to those reference strains. Although sequencing is becoming more available, it can be expensive and time-consuming if a number of isolates need to be analyzed. In order to overcome these limitations, PCR primers to unique sodA sequences in each enterococcal species were designed.
When coupled with genus primers, the multiplex PCR provides an accurate and quick method for identification of enterococci, without the need for extensive phenotypic tests. Genus primers designed by Deasy et al. (2) were used in each reaction to confirm the genus enterococci. Previously, these primers were rigorously tested against a number of gram-positive bacteria and only produced product from bacteria belonging to the enterococci. Although those authors acknowledged that the genus primer may also amplify a product from Carnobacterium, we have not encountered this problem and have found these primers to be specific for Enterococcus and negative for other bacterial genera. Moreover, a genus- and species-specific PCR has been developed for Carnobacterium, but the enterococcal primers were not used for that purpose (19).
The inclusion of certain species in the multiplex PCR was based on previous reports and availability of isolates. A single species primer pair was designed for detection of both E. villorum and E. porcinus, because previous studies have shown that these two species are the same (3). Also of interest was the ability of the PCR, for some isolates, to distinguish E. casseliflavus from E. flavescens, even though those two species have been reported to comprise a single species (3). Some isolates were clearly identified as either E. casseliflavus or E. flavescens, while other isolates were positive for both sets of primers. In addition, the species E. haemoperoxidus and E. moraviensis were not included in the multiplex PCR because sodA sequences from multiple isolates were not available for comparison (20). Moreover, although E. seriolicida and E. solitarius may eventually be reclassified into other genera, they are presently classified as enterococci and were included in the analysis (21, 23).
As with every identification system, multiplex PCR has limitations and will not identify every isolate. Difficulty with identification was encountered when pure cultures were not used in the analysis. If isolates were not streaked at least twice onto nonselective medium, then mixed signals resulted. This could be due to either contamination of the selected colony with smaller colonies not visible to the naked eye or clumping of cells. Mixed cultures for enterococci have been observed previously when using the BBL Crystal ID kit for identification (8). Errant bands that did not correspond to bands in control samples were regarded as nonspecific and were not used for identification purposes. These bands were often either of lower or higher molecular weights than control bands and were easily recognized as being different from the expected size. It could be argued that the lower- or higher-molecular-weight bands could have resulted from the addition or loss of DNA between the primers, but when all seven groups of primers were tested a perfect match to the control strains was always made, suggesting that the errant bands were artifacts. Sequencing of errant products also revealed that it was possible for one primer from one species and another primer from another species to form a product, albeit of the incorrect size. Although the multiplex PCR was designed to amplify specific regions of enterococcal species, this property can be both a positive and a negative feature. Species-specific primers amplified only the designated species; however, while major variations in sodA sequences would not be expected, minor sequence variations among strains of the same species could result in the absence of expected species amplicons. Isolates from environmental and retail food samples were found to be more difficult to identify to the species level and sometimes to the genus level. These isolates have not been extensively characterized, and it would not be surprising if their sodA sequences were more variable than those from other sources or if those sources contained previously unidentified species. Presently, approximately 15% of isolates from environmental and retail food samples remain unidentified at the species level when using multiplex PCR. Studies are ongoing in order to determine significant differences, if any, in the sodA gene sequence of such isolates.
Acknowledgments
We thank Lari Hiott, Carolina Hall, Sandra House, and Tiffanie Woodley for technical assistance and Katie Gay and Katie Lewis (CDC) for standard biochemical testing. We also thank Mark Englen for critical review of the manuscript.
REFERENCES
- 1.Baele, M., P. Baele, M. Vaneechoutte, V. Storms, P. Butaye, L. A. Devriese, G. Verschraegen, M. Gillis, and F. Haesebrouck. 2000. Application of tRNA intergenic spacer PCR for identification of Enterococcus species. J. Clin. Microbiol. 38:4201-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deasy, B. M., M. C. Rea, G. F. Fitzgerald, T. M. Cogan, and T. P. Beresford. 2000. A rapid PCR based method to distinguish between Lactococcus and Enterococcus. Syst. Appl. Microbiol. 23:510-522. [DOI] [PubMed] [Google Scholar]
- 3.Facklam, R. R., M. G. S. Carvalho, and L. M. Teixeiram. 2002. History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci, p. 1-54. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 4.Facklam, R. R., and M. D. Collins. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Garrote, F., E. Cercenado, and E. Bouza. 2000. Evaluation of a new system, VITEK 2, for identification and antimicrobial susceptibility testing of enterococci. J. Clin. Microbiol. 38:2108-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goh, S. H., R. R. Facklam, M. Chang, J. E. Hill, G. J. Tyrrell, E. C. Burns, D. Chan, C. He, T. Rahim, C. Shaw, and S. M. Hemmingsen. 2000. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J. Clin. Microbiol. 38:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton-Miller, J. M., and S. Shah. 1999. Identification of clinically isolated vancomycin-resistant enterococci: comparison of API and BBL Crystal systems. J. Med. Microbiol. 48:695-696. [DOI] [PubMed] [Google Scholar]
- 8.Hudson, C. R., P. J. Fedorka-Cray, M. C. Jackson-Hall, and L. M. Hiott. 2003. Anomalies in species identification of enterococci from veterinary sources using a commercial biochemical identification system. Lett. Appl. Microbiol. 36:245-250. [DOI] [PubMed] [Google Scholar]
- 9.Ke, D., F. J. Picard, F. Martineau, C. Menard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1999. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 37:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moellering, R. C., Jr. 1992. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 14:1173-1176. [DOI] [PubMed] [Google Scholar]
- 11.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naimi, A., G. Beck, and C. Branlant. 1997. Primary and secondary structures of rRNA spacer regions in enterococci. Microbiology 143:823-834. [DOI] [PubMed] [Google Scholar]
- 13.Ozawa, Y., P. Courvalin, and M. Gaiimand. 2000. Identification of enterococci at the species level by sequencing of the genes for d-alanine:d-alanine ligases. Syst. Appl. Microbiol. 23:230-237. [DOI] [PubMed] [Google Scholar]
- 14.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poyart, C., G. Quesnes, and P. Trieu-Cuot. 2000. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J. Clin. Microbiol. 38:415-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruoff, K. L., L. de la Maza, M. J. Murtagh, J. D. Spargo, and M. J. Ferraro. 1990. Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sader, H. S., D. Biedenbach, and R. N. Jones. 1995. Evaluation of Vitek and API 20S for species identification of enterococci. Diagn. Microbiol. Infect. Dis. 22:315-319. [DOI] [PubMed] [Google Scholar]
- 19.Scarpellini, M., D. Mora, S. Colombo, and L. Franzetti. 2002. Development of genus/species-specific PCR analysis for identification of Carnobacterium strains. Curr. Microbiol. 45:24-29. [DOI] [PubMed] [Google Scholar]
- 20.Svec, P., L. A. Devriese, I. Sedlacek, M. Baele, M. Vancanneyt, F. Haesebrouck, J. Swings, and J. Doskar. 2001. Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov., isolated from water. Int. J. Syst. Evol. Microbiol. 51:1567-1574. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira, L. M., V. L. Merquior, M. C. Vianni, M. G. Carvalho, S. E. Fracalanzza, A. G. Steigerwalt, D. J. Brenner, and R. R. Facklam. 1996. Phenotypic and genotypic characterization of atypical Lactococcus garvieae strains isolated from water buffalos with subclinical mastitis and confirmation of L. garvieae as a senior subjective synonym of Enterococcus seriolicida. Int. J. Syst. Bacteriol. 46:664-668. [DOI] [PubMed] [Google Scholar]
- 22.Tyrrell, G. J., R. N. Bethune, B. Willey, and D. E. Low. 1997. Species identification of enterococci via intergenic ribosomal PCR. J. Clin. Microbiol. 35:1054-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams, A. M., U. M. Rodrigues, and M. D. Collins. 1991. Intrageneric relationships of enterococci as determined by reverse transcriptase sequencing of small-subunit rRNA. Res. Microbiol. 142:67-74. [DOI] [PubMed] [Google Scholar]
- 24.Zolg, J. W., and S. Philippi-Schulz. 1994. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J. Clin. Microbiol. 32:2801-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]