Abstract
Carrion's disease is caused by infection with the α-proteobacterium Bartonella bacilliformis. Distribution of the disease is considered coincident with the distribution of its known vector, the sand fly Lutzomyia verrucarum. Recent epidemics of B. bacilliformis infections associated with atypical symptomatology in nonendemic regions have raised questions regarding the historic and present distribution of this bacterium and the scope of disease that infection causes. Phylogenetic relationships and genomic diversity of 18 B. bacilliformis isolates (10 isolates from a region where Carrion's disease is epidemic, Cuzco, Peru, and 8 isolates from a region where Carrion's disease is endemic, Caraz, Peru) were assessed using genomic data generated by infrequent restriction site PCR and gene sequence analysis of the flagellin gltA and ialB genes. A population genetic analysis of the genomic diversity suggests that what was once considered an epidemic region of Peru did not result from the recent introduction of B. bacilliformis.
Bartonella bacilliformis, the causative agent of Carrion's disease (also referred to as bartonellosis, Oroya fever, and verruga peruana), has one of the highest reported human case-fatality rates among bacterial diseases (e.g., 40 to 88% [12, 24]). The classic disease is considered biphasic: the first phase involves a hemolytic anemia and fever, which occurs soon after infection, and this is followed by a cutaneous or verruga phase, which involves skin eruptions and can occur months after infection. Another distinctive feature of Carrion's disease is that the distribution of reported human disease has historically been considered to be restricted to a relatively well-defined altitudinal zone (600 to 3,200 m) of the Andean mountain valleys of South America (3, 17). It has been widely presumed that the limited geographic distribution was a function of the limited range of the sand fly vector, Lutzomyia verrucarum (3, 18). However, recent reports have documented human B. bacilliformis infections over a much larger area than previously reported, including the Andean highlands and the Amazona region east of the Andes mountains, as well as in historically recognized areas where B. bacilliformis is endemic (1, 9, 16, 17). These studies have indicated that in addition to B. bacilliformis occurring over a broader geographical range than previously recognized, the spectrum of symptoms associated with B. bacilliformis infection is highly variable.
This extended range for human B. bacilliformis-associated disease suggests one or more possible scenarios of significant public health importance, including the following: (i) B. bacilliformis-associated disease has existed for many years over a much wider range than previously recognized; (ii) in addition to its endemic distribution, B. bacilliformis has a large but less stable distribution that results from multiple, overlapping introductions of the bacterium via frequent migration between endemic regions and regions that are only marginally capable of maintaining the bacterium; and (iii) for incompletely understood reasons, B. bacilliformis has recently emerged from a limited geographic distribution to significantly expand the endemic region (limits to further expansion are likewise incompletely understood). In addition, in areas outside the range of L. verrucarum, the presence of B. bacilliformis-associated disease implies disease transmission by previously unrecognized vector species; this suggests either recent changes in vector competence or previously unrecognized infection cycles (9).
Comparison of genetic variation between B. bacilliformis isolates from the regions traditionally endemic for Carrion's disease in the Caraz region of Peru with those from a recent Carrion's disease epidemic in the Cuzco region should assist in differentiating among the possible etiologic scenarios described above. For example, a stable, wide distribution of B. bacilliformis should yield similar levels of genetic diversity within and between populations, whereas a recent outbreak should be characterized by lesser variation among samples from the outbreak population compared with those isolates obtained from the population in areas where the disease is considered endemic.
Recent studies have used limited gene sequence data to compare new isolates with existing reference isolates (4, 9). We analyzed nucleotide sequence diversity at three loci: a fragment of the citrate synthase gene or gltA (commonly used for Bartonella species phylogenetic analysis), the invasion-associated locus B (ialB), and the flagellin locus. The latter two loci are believed to be associated with pathogenicity in Bartonella and other bacteria.
In addition to limited gene sequence analysis of B. bacilliformis isolates from two different regions, we used a second method for high-resolution genotypic analysis, infrequent restriction endonuclease site PCR (IRS-PCR). This method has been shown to be useful for genomic analyses of various Bartonella species (14). The IRS-PCR method offers the potential to collect large, robust, and information-rich data sets representing restriction endonuclease-cleaved fragments that are sampled from throughout entire bacterial genomes. We compared levels and distributions of diversity at these genetic markers within and between B. bacilliformis populations and assessed any associations between genetic marker data and patient symptomatology.
MATERIALS AND METHODS
Source of isolates.
Isolation of B. bacilliformis isolates from blood cultures of Carrion's disease patients or asymptomatic individuals has been previously described in detail (7, 9). In brief, 10 isolates from Cuzco, Peru, were obtained from a case-control study conducted during an outbreak of bartonellosis in the Urubamba area in May 1998 (Fig. 1) (9). Eight isolates from the Caraz area were obtained during ongoing studies conducted by the Uniformed Services University of Health Sciences and the Instituto Nacional de Salud. Isolates were stored in liquid nitrogen and further propagated at the Centers for Disease Control and Prevention. DNA was isolated according to established protocols (14). Isolates used in this study were passed two times or less.
FIG. 1.
Map of Peru indicating the sampled regions.
Genetic sequencing and fragment analysis.
Purified DNA was resuspended to a concentration of 50 μg ml−1. Primers for the citrate synthase gene (gltA) were used to amplify a 379-bp product by PCR (19). The DNA sequence from the citrate synthase gene product (338 bp) helped to confirm the Bartonella isolate identity. Primers used to PCR amplify a 1,067-bp product of the flagellin gene and a 688-bp product of the ialB gene were published by Sander et al. (21) and Coleman and Minnick (8), respectively. PCR-amplified DNA products were sequenced in both directions by use of dideoxy sequencing Big Dye kits (Applied Biosystems Inc., Foster City, Calif.) using an ABI model 310 automated sequencer (Applied Biosystems Inc.).
Isolates identified as B. bacilliformis were subjected to IRS-PCR analysis using all four primer pair combinations (A, T, G, and C) as previously described (14). Fluorescently labeled, amplified products were separated on an ABI 310 automated sequencer (Applied Biosystems Inc.), and patterns were subsequently analyzed with the BioNumerics software package version 2.0 (Applied Maths, Kortijk, Belgium). Subsets of the IRS-PCR data were generated independently by two of the authors (T. M. Hambuch and S. A. Handley) with no significant differences in the results.
Data analysis.
Measurements of sequence diversity from the three genetic loci and the IRS-PCR patterns were calculated using the ARLEQUIN program version 2.0 (23). Only nonindependent fragments from the IRS-PCR data were analyzed. Fragments were identified as nonindependent from each other if they were polymorphic among the isolates but showed nonrandom association with other fragments within isolates, as implemented by testing for linkage disequilibrium in ARLEQUIN version 2.0. The pairwise comparisons of diversity within and between sites were performed using the method of Kimura (15) and reported as gene diversity (fragment data) or nucleotide diversity (sequence data).
Several population parameter estimates were calculated from the IRS-PCR and sequence data. These included Θ, Fst, and the mismatch distribution test. These measures are designed to detect if the distribution of genetic variation within a population is different from that of the entire sample, as would be expected in an epidemic; however, they employ different assumptions about numbers and distributions of polymorphism under various circumstances. Multiple tests allowed us to consider several permutations of how much variation existed and how it was distributed. Θ is a population parameter representing the effective population size and the mutation rate (Θ = Neμ for a haploid population) and is based on the number of segregating sites and the sample size (27). Measures of the inbreeding coefficient, Fst, were estimated using this same program. The measure Fst compares the amount of variation within one population to the amount of variation present among all samples (27a) and provides a measure of how genetically distinct populations are from each other. The mismatch distribution test was calculated to ascertain if the B. bacilliformis population had undergone a recent demographic expansion. The mismatch distribution test estimates how well the distribution of the observed numbers of differences between pairs of haplotypes matches Θ. P values associated with the mismatch distribution test use a parametric bootstrap to assess how the data match expectations (23, 27).
Phylogenetic relationships among the B. bacilliformis isolates were inferred from the IRS-PCR data using neighbor-joining methods after applying the Jaccard correlation coefficient to estimate similarities, as instituted in the BioNumerics program version 2.0 (Applied Maths). Bootstrap analysis was performed using 1,000 rounds of resampling with replacement. IRS-PCR data from a Bartonella clarridgeiae isolate were used as an outgroup to root the phylogenetic tree.
RESULTS
Locations, dates, and symptomatology associated with the isolates are presented in Table 1. Five of the 10 isolates from Cuzco were obtained from individuals without a history of acute illness consistent with bartonellosis. As described by Ellis et al. (9), cases were identified through clinical records, whereas three groups of control or asymptomatic individuals were identified as members of the same household, age-matched neighbors, and age-matched residents living at least 5 km from the case by interviews and questionnaires. Isolates were recovered in exactly the same manner from the blood of both case (symptomatic) and control (asymptomatic) individuals, and the surprising number of infected, asymptomatic cases was discussed by Ellis et al. (9). In addition, verruga was not observed in any of the bartonellosis patients from Cuzco, and no evidence of verruga in the community was found up to 6 months after infection (9).
TABLE 1.
Summary of patient characteristics corresponding to the isolates used in this studya
| Isolate | Collection location | Collection date | Reported symptomatology | Age (yrs) | Sex |
|---|---|---|---|---|---|
| Caraz 1 | Choquechaca | June 1997 | Biphasic | 9 | Male |
| Caraz 2 | Culashpampa | January 1997 | History of acute | 21 | Female |
| Caraz 3 | Culashpampa | January 1997 | Acute | 7 | Male |
| Caraz 4 | Paty | June 1997 | Acute | 4 | Female |
| Caraz 5 | Care-Remonta | June 1997 | Acute | 18 | Male |
| Caraz 6 | San Pedro | June 1997 | Biphasic | 8 | Male |
| Caraz 7 | Conchup | January 1997 | History of acute | 8 | Female |
| Caraz 8 | Caraz | June 1997 | Verruga | 14 | Female |
| Cuzco 1 | Ollantaytambo | 14 May 1998 | Acute | 7 | Female |
| Cuzco 2 | Urubamba | 19 May 1998 | Acute | 11 | Female |
| Cuzco 3 | Calca | 18 May 1998 | Asymptomatic | 11 | Female |
| Cuzco 4 | Urubamba | 16 May 1998 | Asymptomatic | 12 | Male |
| Cuzco 5 | Ollantaytambo | 19 May 1998 | Acute | 11 | Female |
| Cuzco 6 | Urubamba | 13 May 1998 | Asymptomatic | 12 | Male |
| Cuzco 7 | Ollantaytambo | 15 May 1998 | Acute | 5 | Male |
| Cuzco 8 | Urubamba | 22 May 1998 | Asymptomatic | 18 | Female |
| Cuzco 9 | Ollantaytambo | 15 May 1998 | Asymptomatic | 4 | Male |
| Cuzco 10 | Calca | April 1998 | Acute | 11 | Female |
These data were collected and reported by Ellis et al. (9) and Chamberlin et al. (7). For Caraz samples, villages lie within a semiarid river valley at an elevation of approximately 2,300 to 2,400 m, within 30 mi of latitude 9° 7′ 96"S, longitude. 77° 48′ 633"W. For Cuzco samples, elevations varied from 2,825 to 3,250 m, within 35 mi of latitude 13° 17′ 59"S, longitude 71° 86′ 24"W.
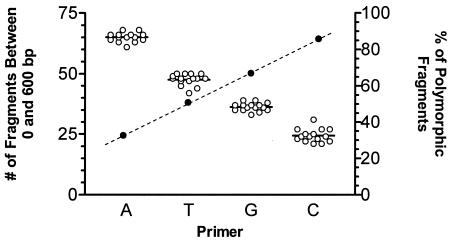
The combination of all four possible primer sets produced a large set of data on which to perform further data analysis (Fig. 2). Independently, each primer combination produced a consistent number of fragments for each B. bacilliformis isolate. The number of fragments produced varied, dependent on which primer was used, with the XbaI-A primer producing the largest number of fragments and the XbaI-C primer producing the fewest. Conversely, the XbaI-C primer produced the data set with the highest average percentage of polymorphic fragments, while many of the fragments produced using the XbaI-A primer were nonpolymorphic.
FIG. 2.
Summary of IRS-PCR fingerprinting data. The left y axis indicates the number of fragments (open circles) identified for each primer set used for the IRS-PCR analysis (A, T, G, or C extension). The horizontal bar indicates the mean number of fragments. The right y axis indicates the average number of polymorphic bands in each IRS-PCR set of data (darkened circles connected with a dashed line). The analysis in Table 2 was carried out on a combined data set of polymorphic bands only.
From a combined set of data consisting of fragments from all four possible primer combinations, 118 polymorphic loci were identified. One shared haplotype was found within the Cuzco population, and one was found within the Caraz population (Table 2); in each case, these haplotypes were shared by relatives living in the same household. No haplotypes were shared between the two geographically distinct study populations. Pairwise measures of gene diversity, estimates of the population parameter Θ, the inbreeding coefficient Fst, and the mismatch distribution test probabilities for the IRS-PCR data are all presented in Table 2. For each measure reported, the levels of diversity and degree of genetic differentiation within each population were comparable (by gene diversity and Θ). No evidence of genetic differentiation between populations was detected by the Fst or mismatch distribution test.
TABLE 2.
Measures of genetic diversity from IRS-PCR dataa
| Isolate | n | No. of shared haplotypes | Gene diversity | θ | Fst | Probability | Probability of mismatch distribution |
|---|---|---|---|---|---|---|---|
| All isolates | 18 | 2 | 0.967 ± 0.03 | 31.97 ± 10.7 | NA | NA | 0.190 |
| Cuzco | 10 | 1 | 0.933 ± 0.08 | 33.96 ± 12.8 | 0.00 | 1 | 0.280 |
| Caraz | 8 | 1 | 0.927 ± 0.08 | 33.48 ± 14.2 | 0.00 | 1 | 0.230 |
| Between populations | NA | 0 | NA | NA | 0.00 | 0.9997 | NA |
Numbers of shared haplotypes reflect the number of identical haplotypes collected within a population. Mean pairwise differences and θs are pairwise comparisons of diversity across all loci (± standard deviation). Tests of specific patterns of diversity (i.e., Fst and mismatch distribution) employed a parametric bootstrap and indicated no statistically significant levels of genetic differentiation or population expansion. NA, not applicable.
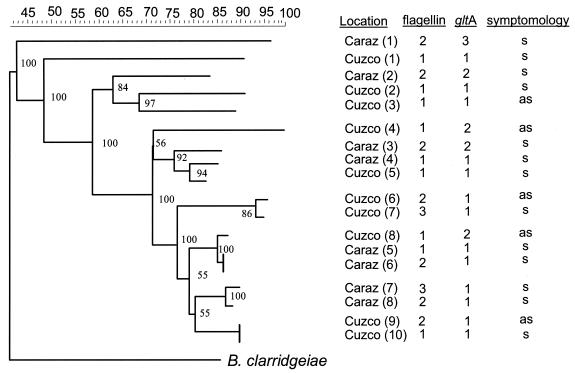
The neighbor-joining tree, based on IRS-PCR data, produced a diverse topology with several well-supported basal clades as well as several peripheral tips (Fig. 3). The tree was rooted using B. clarridgeiae, the closest recognized relative of B. bacilliformis. There is a tendency for pairs of isolates found in the same region to cluster together but only in the peripheral tips of the tree (e.g., Cuzco 6 and 7). More striking was the overall trend for isolates from the Cuzco population to be distributed throughout the tree, suggesting relatively deep, well-supported divergences among samples from the Cuzco region.
FIG. 3.
Neighbor-joining tree of B. bacilliformis isolates based on IRS-PCR data. Numbers at nodes reflect bootstrap values after 1,000 resampling events (1:10). Ruler across top reflects percent similarity among fragment patterns. Isolate designations correspond to those listed in Table 1. Allelic designations of nucleotide sequences from the flagellin and gltA alleles are mapped onto the IRS-PCR tree. Symptomatology (s/as) refers to the symptoms (s) or lack thereof (a) of the patient from whom the isolate was obtained.
As expected for a reasonably conserved gene, minimal variation in gltA nucleotide sequencing data was observed, with only three alleles being detected (Fig. 3). Allele 1 matched GenBank accession number U28076 (B. bacilliformis strain KC584) exactly, while allele 2 had only one silent base pair change, C to A at position 190 (GenBank accession number AF478356). Allele 3, found only once (Caraz 1), had one silent base pair change, T to A at position 191 (GenBank accession AF478356). The distribution of these alleles was superimposed on the IRS-PCR neighbor-joining tree (Fig. 3), where it became evident that all alleles except allele 3 were approximately equally shared between populations. At the gltA locus, there was more variation in the Caraz group than in the Cuzco group (Table 3); however, the differences were not significant (P > 0.05) and were the result of the bias introduced into a pairwise comparison by the addition of the third, rare allele. Tests of allelic distributions of diversity (i.e., Fst and mismatch distribution) employed a parametric bootstrap and detected no significant levels of genetic differentiation or population expansion (Table 3).
TABLE 3.
Diversity and distribution of diversity of sequence dataa
| Isolate source | Nucleotide diversity
|
θ
|
Fst (probability)
|
|||
|---|---|---|---|---|---|---|
| gltAb | Flagellin | gltA | Flagellin | gltA | Flagellin | |
| Caraz | 0.0043 ± 0.0033 | 0.0050 ± 0.0032 | 1.54 ± 0.96 | 4.24 ± 2.15 | 0 (1) | 0 (1) |
| Cuzco | 0.0021 ± 0.0019 | 0.0048 ± 0.0031 | 0.71 ± 0.54 | 4.24 ± 2.03 | 0 (1) | 0 (1) |
| Both | 0.0030 ± 0.0023 | 0.0052 ± 0.0031 | 1.16 ± 0.68 | 3.48 ± 1.53 | 0 (1) | 0 (1) |
Values are means ± standard deviations. Fst values indicate differences in distribution within and between populations (0 = no differences, 1 = complete differentiation); corresponding probabilities are reported in parentheses. As there was no variation detected in the ialB locus, it has been excluded.
Differences at the gltA locus between the Caraz and Cuzco samples are explained by the third allele, found once in the Cuzco population. Since this measure is one of pairwise diversity, it reflects differences among alleles over frequencies of those alleles. These differences are not significant (P > 0.05).
Three distinct flagellin gene sequences were detected (Fig. 3). Allele number 1 was identical to nucleotide sites 180 to 1329 (which comprise the protein coding region of GenBank accession number L20677). Allele 2 had two silent changes (A to G at nucleotide position 642 and C to T at nucleotide position 654) and was found in both populations (GenBank accession number AF478358). Finally, relative to allele 1, flagellin allele 3 possessed nine base pair changes. Flagellin allele 3 shared with flagellin allele 2 the changes at positions 642 and 654 and, in addition, contained the following changes: T to C at 807, G to A at 819, G to A at 822, T to C at 831, T to C at 838, A to G at 927, and T to C at 969. Only one of these base pair changes results in a predicted amino acid change (nucleotide site 822, amino acid site 274, methionine to isoleucine; GenBank accession number AF478359). Strikingly, all three alleles were found in both populations. The distribution of flagellin alleles can be seen superimposed on the IRS-PCR tree (Fig. 3). Both nucleotide diversity measures and Fst values suggested that variation was comparable in and between both populations (Table 3). No variation was found at the ialB locus, as all sequences matched exactly with those published by Coleman and Minnick (8) (data not shown).
DISCUSSION
Population-based studies that contrast B. bacilliformis genotypic diversity in two different regions have the potential to provide important clues for better understanding the apparent recent emergence of B. bacilliformis-associated human disease. During an outbreak, a pathogen is typically introduced to a novel set of susceptible hosts and vectors. Such a pathogen is expected to experience a population “bottleneck” upon introduction to the new population, followed by rapid population expansion (2). This should result in a “founder effect,” such that very little genetic polymorphism from the source population survives the bottleneck and all bacterial isolates within the outbreak population would be expected to be more closely related to each other than to isolates from areas where the bacterium is endemic. In other words, genetic diversity among all outbreak isolates should be lower when compared to isolates sampled from a more stable, endemic population of bacteria. Phylogenetic analyses of human immunodeficiency virus (28), influenza virus (10, 26), Norwalk-like virus (11), Mycobacterium tuberculosis (13), and echoviruses (22) have produced data suggesting that all samples from a specific disease outbreak group could be traced to a single founder and were more closely related to each other than to isolates outside the outbreak.
While the number of isolates for our two populations of B. bacilliformis was limited, IRS-PCR fragment analysis sampled large numbers (118) of informative genetic sites distributed throughout the B. bacilliformis genome and was appropriate for addressing population-level questions; in fact, theoretical work has shown that a large number of polymorphic loci can be more powerful in the evaluation of population genetic diversity than large numbers of samples (25). Conversely, although specific gene sequence data were available for three B. bacilliformis loci (gltA, flagellin, and ialB), these data provided little information regarding possible genetic factors related to variable pathogenicity. The spectrum of illness caused by B. bacilliformis infection could not be associated with either B. bacilliformis genomic IRS-PCR patterns or sequence data from B. bacilliformis genes, homologs of which are known to be involved in pathogenicity in other bacteria.
All measures of genetic diversity among the samples isolated during the Cuzco outbreak were comparable to those derived from isolates made in the Caraz area, where Carrion's disease was recognized as endemic (Tables 2 and 3). These measures have included considerations of the amounts of diversity (i.e., numbers of haplotypes and alleles), degrees of diversity (pairwise comparisons of the degrees of differentiation among haplotypes and allelic variants), and distributions of diversity (by Fst and the mismatch distribution test). For all measures, using both pan-genomic fragment data and sequence data of three loci, there were no measures that detected significant differences in variation between populations or distinguished one population from the other. Thus, these data clearly do not support the hypothesis that the Cuzco samples represent a single, recent outbreak.
Three possible scenarios could explain the observed results: (i) frequent travel of infected individuals between Cuzco and Caraz; (ii) B. bacilliformis has a larger and more stable distribution than previously recognized and includes a larger spectrum of symptomatic and asymptomatic infections than traditionally recognized; or (iii) B. bacilliformis has invaded neighboring regions in frequent, overlapping “waves” and then has been maintained in those marginal regions, where it has caused occasional periodic epidemics. Temporarily hospitable environmental conditions capable of allowing occasional overlapping invasions into regions where Carrion's disease has not been considered endemic might correspond with changes in weather patterns or possibly with changes in host dynamics (e.g., naive host populations). The several outbreaks of B. bacilliformis-associated disease documented since 1993 might support the latter hypothesis, particularly since in the investigation of at least one of these apparent outbreaks concurrent abnormalities were reported in the weather patterns (16). However, it is difficult to reconcile this hypothesis with the extreme variability in symptoms among infected individuals. Both epidemiological studies in Caraz and Cuzco, in which these isolates were obtained, included surveys of the arthropod vectors. The suspected vector, L. verrucarum, made up 99% of all sand flies collected in the Caraz region, whereas only Lutzomyia peruensis was found in the Cuzco region. The L. peruensis flies were PCR surveyed for the presence of Bartonella, and one example of B. bacilliformis was identified from 104 flies (9). Thus, there is no reason to suspect that sand fly movement might explain our findings. Furthermore, travel of infected individuals directly between Cuzco and Caraz would also be unlikely. Travel from Cuzco or Caraz to the capital city of Lima, for example, happens with some frequency; however, travel between Cuzco and Caraz is unlikely (S. Romero, personal communication).
Variability of symptoms associated with B. bacilliformis isolates exhibiting high genetic diversity and the presence of asymptomatic, infected individuals have been observed many times (1, 5, 6, 9, 16, 17, 20). It remains unclear whether the lack of symptoms in some infected individuals results from infection with less-virulent forms of the bacterium or if host factors might explain these differences. All eight isolates from the Caraz area were from patients that had experienced acute illness (7), while half of the samples from Cuzco were isolated from individuals with atypical or asymptomatic infections (Table 1; Fig. 1) (9). Observations from two epidemics (9, 16) noted that children were primarily affected, and these researchers independently suggested that this might mean that B. bacilliformis was endemic in those regions and that adults had partial immunity.
In conclusion, our population-based study of genome diversity in B. bacilliformis isolates offers further insight into the biology and demography of this important pathogen. We found no evidence for significant genetic differentiation between what has previously been considered a stable, endemic, source population of B. bacilliformis and the B. bacilliformis isolates recovered during an apparent outbreak in an area where the bacterium was not thought to be endemic; both populations were equally diverse. Conversely, these data suggest that the Cuzco epidemic probably represented infections related to a diverse, relatively stable B. bacilliformis population and did not represent a single-source outbreak. These data support the previous observation that significant, observable genotypic variation exists within populations of B. bacilliformis, which may well have significant implications for understanding the natural histories of the related bacterial entities, their suspected arthropod vectors, and perhaps the human diseases they may cause (5). More-extensive population studies of naturally occurring B. bacilliformis are clearly warranted to better understand the significance of the observed genetic variation within populations of B. bacilliformis. In addition, more-detailed consideration of the possible roles of host-pathogen interactions appears necessary, particularly those related to possible host factors (including host immune status), that may be important in determining the manifestations of human Carrion's disease.
Acknowledgments
We express our appreciation to the health professionals in the Instituto Nacional de Salud and Urubamba Regional Hospital for their collaboration while investigating the Cuzco outbreak in 1998. We also thank James Childs and Gregory Dasch for helpful comments.
REFERENCES
- 1.Amano, Y., J. Rumbea, J. Knobloch, J. Olson, and M. Kron. 1997. Bartonellosis in Ecuador: serosurvey and current status of cutaneous verrucous disease. Am. J. Trop. Med. Hyg. 57:174-179. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. M., and R. M. May. 1991. Infectious diseases of humans: dynamics and control. Oxford University Press, Oxford, England.
- 3.Benenson, A. S. 1995. Bartonellosis in control of infectious diseases manual, 16th ed., p. 63-64. American Public Health Association, Washington, D.C.
- 4.Birtles, R. J., J. Canales, P. Ventosilla, E. Alvarez, H. Guerra, A. Llanos-Cuentas, D. Raoult, N. Doshi, and T. G. Harrison. 1999. Survey of Bartonella species infecting intradomicillary animals in the Huayllacallan Valley, Ancash, Peru, a region endemic for human bartonellosis. Am. J. Trop. Med. Hyg. 60:799-805. [DOI] [PubMed] [Google Scholar]
- 5.Birtles, R. J., N. K. Fry, P. Ventosilla, A. G. Caceres, E. Sanchez, H. Vizcarra, and D. Raoult. 2002. Identification of Bartonella bacilliformis genotypes and their relevance to epidemiological investigations of human bartonellosis. J. Clin. Microbiol. 40:3606-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvajal, H. L., B. G. Paulson, P. F. Zerega, V. M. Loaiza, and C. M. Palacios. 1978. Bartonelosis en el Ecuador. Verruga peruana. Su estudio historico, epidemiologico, immunologico, clinico e histopathologico. Rev. Ecuat. Hig. Med. Trop. 31:37-47. [Google Scholar]
- 7.Chamberlin, J., L. Laughlin, S. Gordon, S. Romero, N. Solorzano, and R. L. Regnery. 2000. Serodiagnosis of Bartonella bacilliformis infection by indirect fluorescence antibody assay: test development and application to a population in an area of bartonellosis endemicity. J. Clin. Microbiol. 38:4269-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, S. A., and M. F. Minnick. 2003. Differential expression of the invasion-associated locus B (ialB) gene of Bartonella bacilliformis in response to environmental cues. Microb. Pathog. 34:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, B. A., L. D. Rotz, J. A. Leake, F. Samalvides, J. Bernable, G. Ventura, C. Padilla, P. Villaseca, L. Beati, R. Regnery, J. E. Childs, J. G. Olson, and C. P. Carrillo. 1999. An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am. J. Trop. Med. Hyg. 61:344-349. [DOI] [PubMed] [Google Scholar]
- 10.Fitch, W. M., R. M. Bush, C. A. Bender, K. Subbarao, and N. J. Cox. 2000. The Wilhelmine E. Key 1999 Invitational lecture. Predicting the evolution of human influenza A. J. Hered. 91:183-185. [DOI] [PubMed] [Google Scholar]
- 11.Foley, B., J. O'Mahony, C. Hill, and J. G. Morgan. 2001. Molecular detection and sequencing of “Norwalk-like viruses” in outbreaks and sporadic cases of gastroenteritis in Ireland. J. Med. Virol. 65:388-394. [DOI] [PubMed] [Google Scholar]
- 12.Gray, G. C., A. A. Johnson, S. A. Thornton, W. A. Smith, J. Knobloch, P. W. Kelley, L. Obregon Escudero, M. Arones Huayda, and F. S. Wignall. 1990. An epidemic of Oroya fever in the Peruvian Andes. Am. J. Trop. Med. Hyg. 42:215-221. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez, M. C., J. C. Galan, J. Blazquez, E. Bouvet, and V. Vincent. 1999. Molecular markers demonstrate that the first described multidrug-resistant Mycobacterium bovis outbreak was due to Mycobacterium tuberculosis. J. Clin. Microbiol. 37:971-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handley, S. A., and R. L. Regnery. 2000. Differentiation of pathogenic Bartonella species by infrequent restriction site PCR. J. Clin. Microbiol. 38:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, England.
- 16.Kosek, M., R. Lavarello, R. H. Gilman, J. Delgado, C. Maguina, M. Verastegui, A. G. Lescano, V. Mallqui, J. C. Kosek, S. Recavarren, and L. Cabrera. 2000. Natural history of infection with Bartonella bacilliformis in a nonendemic population. J. Infect. Dis. 182:865-872. [DOI] [PubMed] [Google Scholar]
- 17.Maguina, C., and E. Gotuzzo. 2000. Bartonellosis. New and old. Infect. Dis. Clin. North Am. 14:1-22. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi, H., R. C. Shannon, E. B. Tilden, and J. R. Tyler. 1929. Etiology of Oroya fever. XIV. The insect vectors of Carrion's disease. J. Exp. Med. 49:993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norman, A. F., R. Regnery, P. Jameson, C. Greene, and D. C. Krause. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ollague, W. 1991. Enfermedad de Carrion y verruga peruana en Ecuador, p. 387-396. In F. Gararza (ed.), In lecciones de medicina tropical 3. University of Guayaquil Press, Guayaquil, Ecuador.
- 21.Sander, A., A. Zagrosek, W. Bredt, E. Schiltz, Y. Piemont, C. Lanz, and C. Dehio. 2000. Characterization of Bartonella clarridgeiae flagellin (FlaA) and detection of antiflagellin antibodies in patients with lymphadenopathy. J. Clin. Microbiol. 38:2943-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savolainen, C., T. Hovi, and M. N. Mulders. 2001. Molecular epidemiology of echovirus 30 in Europe: succession of dominant sublineages within a single major genotype. Arch. Virol. 146:521-537. [DOI] [PubMed] [Google Scholar]
- 23.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin version 2.000: a software for population genetics data analysis, 2.0 ed. Genetics and Biometry Laboratory, University of Geneva, Switzerland, Geneva, Switzerland.
- 24.Schultz, M. G. 1968. A history of bartonellosis (Carrion's disease). Am. J. Trop. Med. Hyg. 17:503-515. [DOI] [PubMed] [Google Scholar]
- 25.Slatkin, M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suarez, D. L. 2000. Evolution of avian influenza viruses. Vet. Microbiol. 74:15-27. [DOI] [PubMed] [Google Scholar]
- 27.Watterson, G. A. 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7:256-276. [DOI] [PubMed] [Google Scholar]
- 27a.Wright, S. 1951. The genetical structure of populations. Ann. Eugen. 15:323-354. [DOI] [PubMed] [Google Scholar]
- 28.Yusim, K., M. Peeters, O. G. Pybus, T. Bhattacharya, E. Delaporte, C. Mulanga, M. Muldoon, J. Theiler, and B. Korber. 2001. Using human immunodeficiency virus type 1 sequences to infer historical features of the acquired immune deficiency syndrome epidemic and human immunodeficiency virus evolution. Philos. Trans. R. Soc. London B 356:855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]