Abstract
We report a rapid and accurate real-time PCR-based method to quantify wild-type and lamivudine-resistant hepatitis B virus by using a common forward primer paired with different reverse primers. Excellent concordance was demonstrated between sequencing and the discriminatory real-time assay; however, a mixture of quasispecies was more frequently detected by discriminatory real-time PCR.
The most widely used antiviral treatment for hepatitis B virus (HBV) is lamivudine. Reduced virologic response to lamivudine therapy can occur due to the emergence of drug-resistant quasispecies (6). Specific mutations which result in replacement of methionine (M) in the tyrosine-methionine-aspartate-aspartate (YMDD) catalytic site motif of the C domain of the HBV reverse transcriptase (rt) by valine (V), isoleucine (I), or serine (S) confer resistance to lamivudine (1). The resultant changes in the amino acid sequence within the polymerase protein are designated rtM204V, rtM204I, and rtM204S, respectively, according to the new genotype-independent nomenclature (3, 7, 13, 14).
Mutations leading to lamivudine resistance are generally detected by conventional DNA sequencing after PCR amplification of a selected portion of the viral polymerase gene. The advantage of sequencing includes obtaining information over a span of several nucleotides. However, sequencing is expensive and laborious and has a low level of sensitivity for minority quasispecies, usually only detecting to a level of 20% of the total virus population (8). Other molecular techniques developed to detect the changes associated with lamivudine resistance include restriction fragment length polymorphism, a 5′ nuclease assay, a real-time PCR assay using the LightCycler (4, 16), and line probe assay technology (15) (INNO-LiPA HBV DR; Innogenetics, Ghent, Belgium). Using the principle of primer selection, we have developed a quantitative sensitive real-time PCR assay able to detect minority drug-resistant quasispecies at a level of 1 in 1,000. This assay is useful in the early detection of drug resistance and will assist in the study of viral fitness in vivo.
Serum samples were collected from 38 individuals chronically infected with HBV and were either naïve to antiviral therapy or had commenced treatment with lamivudine (n = 21). A total of 75 samples were analyzed, including 38 samples from individuals naïve to lamivudine and 37 samples from individuals on lamivudine.
HBV DNA was extracted from 200 μl of serum by using the QIAamp DNA Mini kit (QIAGEN, Chatsworth, Calif.). Amplification of the HBV polymerase gene has been described previously (5). The INNO-LiPA HBV DR (Innogenetics) testing was performed in accordance with the methods previously published (15). HBV viral load was determined by using real-time PCR as previously described (10).
Discrimination between the wild type and two mutants (rtM204I and rtM204V) was possible by using primers in which the relevant base pair mutation was at or near the terminal 3′ base of the primer. A primer-template mismatch at the 3′ terminus significantly compromises polymerase efficiency and allows for discrimination between mutants and the wild type even with a single base pair mismatch (9). The efficiency of discrimination by the primer between wild-type and mutant templates is dependent on the specific 3′-terminal base, with maximal reduction of PCR product yield of 100-fold with a A:G, G:A, or C:C mismatch (9).
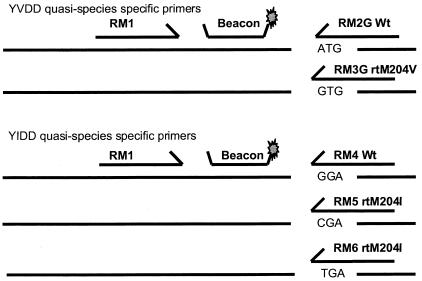
We designed a common forward primer to a highly conserved sequence within the polymerase open reading frame encoding the C domain of the viral enzyme (RM1, 5′-CACCTGTATTCCCATCCCAT-3′) and different reverse linear primers that would allow selective amplification of YVDD (rtM204V), YIDD (rtM204I), and YMDD (wild type) quasispecies (Fig. 1). The amplicon was detected and quantified by using real-time PCR and a molecular beacon (5′-FAM-GGTCCCATTTGTTCAGTGGTTCGTAGGGCTTTGGGACC-DABCYL-3′) that annealed to a highly conserved region between the forward and reverse primers. The total viral load was quantified by using a separate PCR with primers and beacon in the precore region (10). Real-time PCR was performed with conditions identical to those previously described (10).
FIG. 1.
Location and sequence of the common forward primer (RM1), common beacon, and specific reverse primers. The target sequence of each mutation is shown.
For detection of the rtM204I variants, a primer specific for the mutation G/C (RM5, 5′-CCCCAATACCACATCATCG-3′) was used. Later modifications were made (described in detail below), leading to inclusion of an additional primer that also detected the G/T substitution (RM6, 5′-CCCCCAATACCACATCATCA-3′) (final concentration, 0.8 pmol/μl). In initial experiments, only the RM5 primer was used. For detection of rtM204V variants, a primer specific for the A/G substitution at nucleotide 206 was used (5′-CCCCAATACCACATCATCCGC-3′). For validation of these assays and in order to determine the degree of cross-priming, a reverse primer specific for wild-type HBV at nucleotide 206 was used to detect non-rtM204I wild-type HBV (RM4, 5′-CCCCCAATACCACATCATCC-3′) and non-rtM204V (5′-CCCCAATACCACATCATCCGT-3′) within the samples. For detection of drug-resistant quasispecies in clinical samples only two separate PCRs are needed.
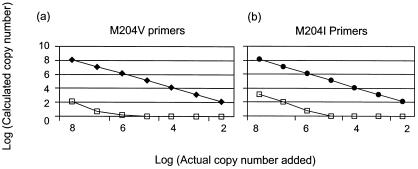
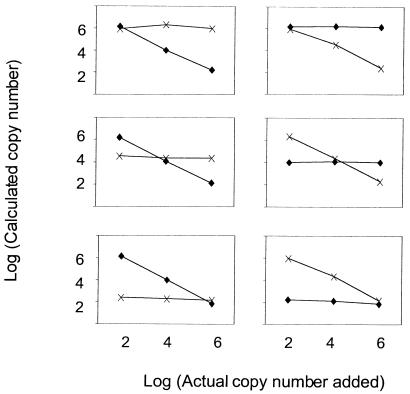
As expected, nonspecific priming to the alternate template (i.e., mutant primers to wild-type template and vice versa) was observed. The degree of cross-priming was first quantified by using a dilution series of template ranging from 108 to 102 copies, and PCR was performed with the mismatched primers (Fig. 2). In most cases, nonspecific priming occurred when the mismatched target was present at >106 copies per PCR. The calculated copy number (of the mismatched target) was at least 4 logs less than the actual copy number present, representing a trivial degree of cross-priming. Mixing experiments were then performed over a 4-log range (wild-type and mutant species were mixed, with concentrations ranging from 102 to 106 copies per PCR; Fig. 3). The actual and calculated copy numbers were identical for the wild-type and mutant templates. The intra-assay variability of each assay was similar to that of the precore quantitative assay (11) and was calculated to be 0.3 log (data not shown).
FIG. 2.
Dilutions of known concentrations of plasmids containing the wild-type (Wt, open squares) or rtM204V and rtM204I mutations (closed diamonds and circles, respectively) were tested with primer sets designed to detect the rtM204V and rtM204I mutations. The actual copy number per PCR is demonstrated on the x axis. The calculated copy number per PCR is shown on the y axis. (a) M204V primers accurately quantify the M204V template. When the M204V primers are used to detect the wild-type template, a false-positive signal is detected when the concentration of the wild-type template is >106 copies; however, the calculated value (or false-positive detection) is reduced to 100 copies. (b) The discrimination between the wild type and mutants is less stringent for the wild type when the M204I primers are used, but the reduction in signal remains at least 4 logs.
FIG. 3.
Mixing experiments of mutant (M204V) and wild-type DNA were performed and analyzed by using primers containing the wild-type or mutant sequence. Discriminatory real-time PCR accurately quantified wild-type and mutant input DNA over a 4-log range independent of whether 106 (upper panels), 104 (middle panels), or 102 (lower panels) copies of mutant (left panels) or wild type (right panels) were added. Both the wild type (diamonds) and mutants (crosses) were accurately detected over a 4-log range regardless of their input ratio.
Initially the assay was evaluated by using sera from individuals never treated with lamivudine (n = 38). Real-time selective PCR identified the wild type only at position 204 (rtM204). This was concordant with sequencing in all 38 individuals. Sequencing and real-time selective PCR were then performed on sera (n = 47) from 21 individuals following the introduction of lamivudine (Table 1). There was full concordance between sequencing and real-time PCR results for 19 samples (10 were wild type and 9 had the rtM204V change). Real-time PCR detected a mixed quasispecies in 13 samples, whereas sequencing only detected a single species. However, in all cases the dominant quasispecies was concordant with the results of sequencing.
TABLE 1.
Comparison of results obtained by DNA sequencing with those obtained by discriminatory real-time PCR
| Time pointa | Variantb | No. of concordant samples | No. of samples showing mix with PCR but not sequencing | No. of discordant samples | VLc mean (range) log copies per ml |
|---|---|---|---|---|---|
| A | Wild type | 38 | 0 | 0 | 7.95 (3.86-8.79) |
| B | M204V | 9 | 9 | 3 | 7.34 (4.59-8.88) |
| M204I | 0 | 2 | 12 | 7.92 (3.67-7.81) | |
| Wild type | 10 | 2 | 0 | 6.63 (4.08-8.28) | |
| Total | 19 | 13 | 15 | ||
| C | M204I | 9 | 1 | 0 | 8.08 (4.79-9.00) |
Time points are the following: A, pretherapy; B, posttherapy, prior to addition of the second M204I primer; C, posttherapy, following addition of the second M204I primer.
As defined by direct sequencing.
VL, HBV viral load.
In 15 samples, results from sequencing and real-time PCR were discordant. For the three discordant cases where sequencing identified the rtM204V mutation, close examination of the sequences showed that instead of the most common mutation associated with the rtM204V change (i.e., GTG) to which the primer was designed, other mutations were responsible for the rtM204V modification. In one case the patient isolate sequence was GTG, and in the two other samples the sequence was GTT.
The remaining 12 samples having a discordant result were all identified by sequencing as rtM204I, and in each case the viral isolate contained the sequence ATT. The primer used to detect rtM204I (primer RM5) was designed to detect sequence ATC. Given that stringency of primer binding is most efficient when there is complementarity at the 3′ end of the primer, we presume that primer RM5 was inefficient in binding to mutant virus containing ATT. Therefore, for detection of rtM204I variants, a mix of primers specific for both the G/C and the G/T substitution were subsequently used.
Three commonly described sequences can code for rtM204I: ATC, ATT, and ATA. The most common sequence coding for rtM204I in all HBV genotypes is ATT. We designed primers that specifically recognized each of these three sequences (primers RM5, RM6, and RM7, respectively). Data for cross-priming of RM5 to wild-type template has been shown in Fig. 2. RM6 behaved in a fashion similar to that of RM5, with minimal cross-priming to the wild type. The specificity of RM7 for the wild type (i.e., the mismatched template) was 1 log less than that of RM5 and RM6. Due to this lack of specificity, RM7 was not used in the primer mix. When the sensitivity of detection of RM5 and RM6 was evaluated to detect the nonmatched mutant quasispecies, we determined that RM5 detected ATC only (i.e., the complementary sequence) and did not detect ATT or ATA. RM6, however, was able to prime to ATA as well as the complementary sequence ATT. Therefore, the combination of RM5 and RM6 will reliably detect the three major mutants that code for rtM204I.
A further four isolates containing the ATT variant of rtM204I (three specimens contained only rtM204I while one contained a mixture of rtM204 and M204I, as determined by sequencing) were studied by using the rtM204I primer mix. There was concordance between sequencing and real-time PCR for the rtM204-M204I mixture and one of the rtM204I mutants. For the other two rtM204I mutants, the dominant quasispecies detected by real-time PCR was rtM204I (but in a mixture with the wild type).
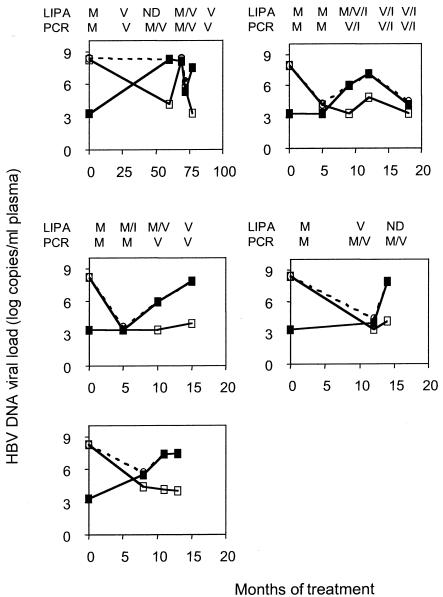
Five individuals were monitored over time following the introduction of lamivudine in order to determine the relative kinetics of wild-type and drug-resistant quasispecies by using discriminatory real-time PCR, sequencing, and LiPA (Fig. 4). In all but one case discriminatory real-time PCR detected the drug-resistant species at the same time it was detected by LiPA.
FIG. 4.
Longitudinal study of HBV viral load and quasispecies analysis following introduction of lamivudine (shown on the x axis in months). Total HBV viral load (dashed lines), M204V (closed squares), and the wild type at M204 (open squares) were quantified over time. Results from LiPA and PCR sequencing are shown above each graph as M (wild type), V (M204V), I (M204I), or ND (not done).
There have been numerous methods published to date that rapidly identify lamivudine-resistant HBV mutants. These methods include the LiPA (15), restriction fragment length polymorphism, and real-time PCR assays using the LightCycler (4, 16) or the 5′ nuclease assay (1). The main advantage of our discriminatory real-time PCR assay is the ability to detect minority quasispecies at a level of 1 in 1,000. Other assays that use common primers and a specific probe to identify mutants and the wild type are limited in the linear range by which quantification is possible due to primer competition for template. For example, the LightCycler real-time assay will only detect a minority population of 25% of the total, which is a level of detection similar to that of direct sequencing (16). Studies examining LiPA (12) have also shown earlier and more frequent detection of minority quasispecies, but this assay is not quantitative and has a limit of detection of 10%.
The most common reason for discrepant results between sequencing and our discriminatory real-time PCR assay could be attributed to polymorphisms in the YMDD region. By the inclusion of two primers for detection of the YIDD change, we were able to detect all three variants (ATC, ATT, and ATA). For the YVDD mutant, the primer only detected GTG and not the minor infrequent double mutation (GTT). The GTT mutation occurs at a significantly lower frequency, accounting for approximately 1% of YVDD variants (A. Bartholomeusz and L. Yuen, personal communication). We designed primers that also detected these YVDD quasispecies, but significant nonspecific cross-priming occurred in the wild type, making this approach impractical. Given the infrequent prevalence of these mutations, we did not think that the ability to detect only the GTG variant would compromise the clinical utility of the assay.
The discriminatory real-time PCR assay for lamivudine-resistant mutations could potentially be applied to the clinical setting to monitor HBV-infected patients receiving antiviral therapy while simultaneously measuring HBV viral load. As newer antiviral agents are introduced, such as adefovir and tenofovir, and as resistance is encountered (2), the application of these assays could allow for an earlier change in antiviral therapy before viral breakthrough occurs.
Acknowledgments
This work was supported in part by Innogenetics, Ghent, Belgium. S.R.L. is supported by an NHMRC Practitioner Fellowship, The Victor Hurley Trust, The Alfred Research Trust, and the NHMRC Centre for Clinical Research Excellence in Infectious Diseases at the University of Melbourne.
REFERENCES
- 1.Allen, M., M. Deslauriers, C. Andrews, G. Tipples, K. Walters, D. Tyrrell, N. Brown, and L. Condreay. 1998. Identification and characterisation of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Angus, P., R. Vaughan, S. Xiong, H. Yang, W. Delaney IV, C. Gibbs, and C. Brosgart. 2003. Resistance to adefovir dipivoxil therapy associated with development of a novel mutation in the HBV polymerase. Gastroenterology 125:292-297. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomeusz, A., R. Schinazi, and S. Locarnini. 1998. Significance of mutations in the hepatitis B virus polymerase selected by nucleoside analogues and implications for controlling chronic disease. Viral Hepatitis Rev. 4:167-187. [Google Scholar]
- 4.Cane, P. A., P. Cook, D. Ratcliffe, D. Mutimer, and D. Pillay. 1999. Use of real-time PCR and fluorimetry to detect lamivudine resistance-associated mutations in hepatitis B virus. Antimicrob. Agents Chemother. 43:1600-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coia, G., A. Ayres, G. G. Lilley, P. J. Hudson, and R. A. Irving. 1997. Use of mutator cells as a means for increasing production levels of a recombinant antibody directed against hepatitis B. Gene 201:203-209. [DOI] [PubMed] [Google Scholar]
- 6.Delaney IV, W., S. Locarnini, and T. Shaw. 2001. Resistance of hepatitis B virus to antiviral drugs: current aspects and directions for future investigation. Antivir. Chem. Chemother. 12:1-35. [DOI] [PubMed] [Google Scholar]
- 7.Gan, R. B., M. J. Chu, L. P. Shen, S. W. Qian, and Z. P. Li. 1987. The complete nucleotide sequence of the cloned DNA of hepatitis B virus subtype adr in pADR-1. Sci. Sin. B 30:507-521. [PubMed] [Google Scholar]
- 8.Gunthard, H. F., S. D. Frost, A. J. Leigh-Brown, C. C. Ignacio, K. Kee, A. S. Perelson, C. A. Spina, D. V. Havlir, M. Hezareh, D. J. Looney, D. D. Richman, and J. K. Wong. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J. Virol. 73:9404-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwok, S., D. E. Kellogg, N. McKinney, D. Spasic, L. Goda, C. Levenson, and J. J. Sninsky. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin, S., R. Ribeiro, T. Walters, G. Lau, S. Bowden, S. Locarnini, and A. Perelson. 2001. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology 34:1012-1020. [DOI] [PubMed] [Google Scholar]
- 11.Lewin, S., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. Ho, and M. Markowitz. 1999. The use of real-time PCR and molecular beacons to detect virus-replication in HIV-1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lok, A. S., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. De Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niesters, H., R. De Man, S. Pas, E. Fries, and A. Osterhaus. Identification of a new variant in the YMDD motif of the hepatitis B virus polymerase gene selected during lamivudine therapy. J. Med. Microbiol. 51:695-699. [DOI] [PubMed]
- 14.Stuyver, L., S. Locarnini, A. Lok, D. Richman, W. Carman, J. Dienstag, and R. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:7751-7757. [DOI] [PubMed] [Google Scholar]
- 15.Stuyver, L., C. Van Geyt, S. De Gendt, G. Van Reybroeck, F. Zoulim, G. Leroux-Roels, and R. Rossau. 2000. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J. Clin. Microbiol. 38:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whalley, S. A., D. Brown, C. G. Teo, G. M. Dusheiko, and N. A. Saunders. 2001. Monitoring the emergence of hepatitis B virus polymerase gene variants during lamivudine therapy using the LightCycler. J. Clin. Microbiol. 39:1456-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]