Abstract
Nineteen isolates of leptospires recovered from patients during three epidemics that occurred at different places and different times in the Andaman Islands and eight isolates from sporadic cases were characterized using serological and molecular genetic techniques. Group sera and monoclonal antibodies were used for antigenic characterization, whereas fluorescent amplified fragment length polymorphism (FAFLP) was used for genotyping. Of the 27 isolates, 19 were identified as belonging to serogroup Grippotyphosa, 3 belonged to serogroup Australis, 2 belonged to serogroup Icterohaemorrhagiae, and 1 each belonged to serogroups Hebdomadis, Canicola, and Sejroe. Analysis of FAFLP data grouped these 27 isolates into two main clusters of genotypes. One of the clusters, populated by 19 isolates, included 16 outbreak isolates. Seven of these 19 isolates belonged to serovar Ratnapura, 10 belonged to serovar Valbuzzi, and 1 each belonged to serovar Grippotyphosa and serovar Saxkoebing. Of the 27 patients from whom isolates were obtained, 9 had severe illness, and 6 of these 9 patients had pulmonary involvement, 1 had pulmonary and hepatorenal involvement, and the remaining 2 had hepatorenal involvement alone. Two patients out of the nine severe cases died subsequently. The isolates from sporadic cases showed great genetic diversity and were also diverse antigenically. Perhaps the strains belonging to a dominant genotype (the outbreak-associated cluster) possessed epidemic potential and higher virulence with a greater predilection to cause pulmonary complications than strains belonging to other genetic backgrounds.
Leptospirosis has been recognized as an important global public health problem because of its increasing incidence in many countries and the occurrence of several large outbreaks in recent years. Leptospirosis had been suspected in India since the early part of the 20th century (5, 9). The first report of bacteriologically confirmed leptospirosis in India and the first common-source outbreak of leptospirosis were reported from the Andaman Islands (16). This outbreak occurred among bund construction workers in a village on South Andaman in 1929 and was caused by leptospires belonging to the groups Akiamy-A (serogroup Grippotyphosa) and Andamans-A (serogroup Andamana). These patients presented with signs and symptoms typical of Weil's syndrome with predominant hepatorenal involvement. There was no report of the status of the disease in these islands between the 1940s and the 1980s.
Seasonal outbreak of febrile illness known locally as Andaman hemorrhagic fever (AHF) was first noticed on South and North Andaman in 1988 (11). Until 1993, when it was identified as a pulmonary form of leptospirosis, AHF was a mysterious disease (11). This was also the first report of severe pulmonary hemorrhage as a complication of leptospirosis in India. Subsequently, several epidemics have occurred on North Andaman and South Andaman. Sporadic cases during the interepidemic period have also been reported from South Andaman, but rarely from North Andaman. Pulmonary involvement has been the predominant complication during the epidemics on North Andaman and was associated with a high case/fatality ratio. A hospital-based surveillance on South Andaman detected few outbreaks and many sporadic cases. Most patients on South Andaman had mild illness, and only a few of them developed complications. It is not clearly understood whether the variation in the clinical presentation during epidemics and sporadic occurrences is due to genotypic differences in the infecting agents, host and ecological factors, or the interplay of both. Study of the genetic makeup of the isolates obtained from patients with different clinical severities of the disease and from different areas and times will be helpful in understanding the roles of specific genetic variants in causing severe disease and also the temporal and spatial distributions of different clones in the islands.
Several PCR-based DNA-fingerprinting methods have been described for genetic characterization of leptospires, for example, random amplified polymorphic DNA and arbitrarily primed PCR. The major disadvantage of these techniques is the lack of reproducibility, as the techniques are very sensitive to the quality of the DNA and to PCR temperature profiles. Recently, fluorescent amplified fragment length polymorphism (FAFLP) (1-4, 8, 10, 14, 18) has been used as a powerful genotyping technique that combines the power of restriction fragment length polymorphism with the flexibility of PCR (18, 10). This technique has been effectively used to study the epidemiologies of some infectious diseases and to understand the evolution of pathogens (1-4, 8, 14). We analyzed the phylogenic relatedness among leptospiral isolates recovered during investigation of three major outbreaks and from several sporadic cases that occurred in the Andaman Islands. This was done with the objective of understanding the distribution and evolution of these pathogens and to study any association between the genetic natures of infecting strains and the clinical presentation of cases.
MATERIALS AND METHODS
Epidemics. (i) Outbreaks on North Andaman.
Two epidemics were observed on North Andaman during October and November, one in 1996 and the other in 1997. In the former, 32 patients were suspected to have leptospirosis, 14 of whom were confirmed. Fifty-eight clinically suspected cases were reported in the latter outbreak, and of these, 26 showed laboratory evidence of leptospirosis. The predominant clinical features presented during the two outbreaks were fever, headache, generalized body ache, cough, and hemoptysis with respiratory distress. The disease was severe in the majority of the patients, and seven patients died (case fatality ratio, 17.5%).
(ii) Outbreak on South Andaman.
The outbreak on South Andaman occurred during October and November 1999 in two different areas, Tushnabad and Manglutan. Ninety-three patients were suspected to have leptospirosis, and for 42 of them, the diagnosis was confirmed by serology or isolation of leptospires from the blood. The disease was mild in a majority of the patients. The most frequently observed clinical features were fever, headache, and generalized body ache. Four patients had icterus, and three had hemoptysis. Two patients died during this epidemic (case fatality ratio, 4.8%).
(iii) Sporadic cases.
During the period between December 1999 and November 2001, a total of 71 confirmed leptospirosis patients were reported from South Andaman. The disease was mild in a majority of the patients. The clinical spectrum of the disease was similar to that of patients who reported during the epidemic that occurred in 1999 in South Andaman. Five patients showed pulmonary involvement, and three patients presented with hepatorenal involvement. Three deaths were also recorded (case fatality ratio, 4.8%). The final events in fatal cases were either massive alveolar hemorrhages and respiratory distress or renal failure.
Isolates.
Four isolates recovered from patients during the investigation of 1996 and two isolates from 1997 outbreaks on North Andaman were available for this work. Twelve isolates from the 1999 epidemic and eight isolates obtained from sporadic cases on South Andaman and in south India were also included. In addition, one isolate from the 1929 outbreak on South Andaman was also included. The details of the isolates are shown in Table 1.
TABLE 1.
Details of isolates recovered from patients during outbreaks and sporadic cases, organ involvement, and outcomes of cases
| Isolate | Area of isolation | Mo and yr of isolationb | Serogroup | Serovar | Organ involvementc
|
Outcome | ||
|---|---|---|---|---|---|---|---|---|
| Pulmonary | Hepatic | Renal | ||||||
| FIS01a | South Andaman | Oct 1999 | Grippotyphosa | Ratnapura | N | N | N | Recovered |
| FIS02a | South Andaman | Oct 1999 | Icterohaemorrhagiae | Copenhageni | N | N | N | Recovered |
| FIS03a | South Andaman | Oct 1999 | Grippotyphosa | Ratnapura | Y | N | N | Recovered |
| FIS04a | South Andaman | Oct 1999 | Grippotyphosa | Ratnapura | N | N | N | Recovered |
| FIS05a | South Andaman | Oct 1999 | Grippotyphosa | Valbuzzi | Y | Y | Y | Expired |
| FIS06 | South Andaman | Nov 1998 | Icterohaemorrhagiae | Copenhageni | N | Y | Y | Recovered |
| FIS07a | North Andaman | Oct 1996 | Grippotyphosa | Valbuzzi | Y | N | N | Recovered |
| FIS08a | North Andaman | Oct 1997 | Grippotyphosa | Valbuzzi | Y | N | N | Expired |
| FIS09a | South Andaman | Oct 1999 | Grippotyphosa | Ratnapura | N | N | N | Recovered |
| FIS10a | South Andaman | Oct 1999 | Grippotyphosa | Ratnapura | N | N | N | Recovered |
| FIS11a | South Andaman | Oct 1999 | Grippotyphosa | Ratnapura | N | N | N | Recovered |
| FIS12a | South Andaman | Oct 1999 | Hebdomadis | Hebdomadis | N | N | N | Recovered |
| FIS14a | South Andaman | Oct 1999 | Sejroe | Saxkoebing | N | N | N | Recovered |
| FIS16a | South Andaman | Nov 1997 | Grippotyphosa | Ratnapura | Y | N | N | Recovered |
| FIS17a | South Andaman | Oct 1999 | Australis | Ramisi | N | N | N | Recovered |
| FIS19 | South Andaman | Oct 2000 | Grippotyphosa | Valbuzzi | N | N | N | Recovered |
| FIS20 | South Andaman | Dec 2000 | Grippotyphosa | Valbuzzi | N | N | N | Recovered |
| FIS21 | South Andaman | Dec 2000 | Australis | Australis | N | N | N | Recovered |
| FIS22 | South Andaman | Dec 2000 | Australis | Australis | N | N | N | Recovered |
| FIS23 | South Andaman | Apr 2001 | Grippotyphosa | Valbuzzi | N | N | N | Recovered |
| FIS24 | South Andaman | July 2000 | Canicola | Canicola | N | N | N | Recovered |
| FIS25a | North Andaman | Oct 1996 | Grippotyphosa | Valbuzzi | Y | N | N | Recovered |
| FIS26a | North Andaman | Oct 1996 | Grippotyphosa | Valbuzzi | Y | N | N | Recovered |
| FIS27a | North Andaman | Nov 1996 | Grippotyphosa | Valbuzzi | N | N | N | Recovered |
| FIS28a | North Andaman | Oct 1997 | Grippotyphosa | Valbuzzi | N | N | N | Recovered |
| FIS29 | South India | Apr 2000 | Grippotyphosa | Ratnapura | N | Y | Y | Recovered |
| CH 31a | South Andaman | Sep 1929 | Grippotyphosa | Grippotyphosa | N | N | N | Recovered |
Isolate recovered during epidemics.
Apr, April; Sep, September; Oct, October; Nov, November; Dec, December.
Y, yes; N, no.
Laboratory diagnosis and criteria for confirmation.
The criteria for laboratory confirmation were based on either successful isolation of leptospires from clinical specimens or positive serology by a microscopic agglutination test (MAT). Fourteen live leptospiral strains belonging to serogroups that have been reported in India were used as antigens. The strains belonged to serogroups Australis (serovar Australis, strain Ballico), Autumnalis (serovar Rachmati, strain Rachmat), Ballum (serovar Ballum, strain Mus 127), Bataviae (serovar Bataviae, strain Swart), Canicola (serovar Canicola, strain Hond Uterecht IV), Cynopteri (serovar Cynopteri, strain 3522C), Grippotyphosa (serovar Grippotyphosa, strain Moskva V), Hebdomadis (serovar Hebdomadis, strain Hebdomadis), Icterohaemorrhagiae (serovar Icterohaemorrhagiae, strain RGA), Javanica (serovar Poi, strain Poi), Pyrogenes (serovar Pyrogenes, strain Salinem), Pomona (serovar Pomona, strain Pomona), Sejroe (serovar Hardjo, strain Hardjoprajitno), and Tarassovi (serovar Tarassovi, strain Peripelitsin). MAT was performed at doubling dilutions from 1 in 25 up to 1 in 400. Those specimens found positive at 1 in 400 were titrated up to the end titers. The criteria for confirmation of a diagnosis by using MAT were the same as those reported earlier (12).
Serological typing: MAT with group sera and monoclonal antibodies.
The serological-typing tests were performed as reported earlier (17). Thirty-seven group-specific rabbit antisera representing 23 pathogenic serogroups were used for serogroup determination (Table 2). A panel of mouse monoclonal antibodies (WHO/FAO Collaborating Centre for Reference and Research, KIT-Biomedical Research, Amsterdam, The Netherlands) capable of differentiating the serovars of serogroups Grippotyphosa (F71C3, F71C9, F165C3, and F165C8), Icterohaemorrhagiae (F52C1, F70C14, F70C24, F89C12, and F82C1), Australis (F81C1, F81C8, F90C5, and F90C6), Sejroe (F13C193, F16C28, F21C21, and F106C53), Canicola (F152C11, F152C14, F152C17, and F152C18), and Hebdomadis (F50C3, F106C1, and F106C5) were used for the identification of serovars.
TABLE 2.
Details of 37 group-specific representative rabbit antisera from 23 serogroups
| Serogroup | Serovar | Strain |
|---|---|---|
| Australis | Australis | Ballico |
| Australis | Bratislava | Jez bratislava |
| Autumnalis | Bangkinang | Bangkinang I |
| Autumnalis | Butembo | Butembo |
| Autumnalis | Carlos | C3 |
| Autumnalis | Rachmati | Rachmat |
| Ballum | Ballum | Mus 127 |
| Ballum | Kenya | Njenga |
| Bataviae | Bataviae | Swart |
| Canicola | Canicola | Hond Utrecht IV |
| Canicola | Schueffneri | VI.90 C |
| Celledoni | Celledoni | Celledoni |
| Cynopteri | Cynopteri | 3522 C |
| Djasiman | Djasiman | Djasiman |
| Grippotyphosa | Grippotyphosa | Moskva V |
| Grippotyphosa | Huanuco | M 4 |
| Hebdomadis | Hebdomadis | Hebdomadis |
| Hebdomadis | Worsfoldi | Worsfoldi |
| Icterohaemorrhagiae | Copenhageni | M 20 |
| Icterohaemorrhagiae | Icterohaemorrhagiae | RGA |
| Javanica | Poi | Poi |
| Louisiana | Louisiana | LSU 1945 |
| Manhao | Manhao | L 60 |
| Mini | Mini | Sari |
| Panama | Panama | CZ 214 K |
| Pomona | Pomona | Pomona |
| Pyrogenes | Pyrogenes | Salinem |
| Sarmin | Rio | Rr 5 |
| Sarmin | Weaveri | CZ 390 |
| Sejroe | Hardjo | Hardjoprajitno |
| Sejroe | Saxkoebing | Mus 24 |
| Shermani | Shermani | 1342 K |
| Tarassovi | Bakeri | LT 79 |
| Tarassovi | Mogdeni | Compton 746 |
| Tarassovi | Rama | 316 |
| Tarassovi | Tarassovi | Perepelitsin |
| Ranarum | Ranarum | ICF |
DNA fingerprinting.
The isolates were characterized by FAFLP analysis as described previously (1-4, 8, 14).
(i) Isolation of DNA.
The isolates were grown at 30°C in Ellinghausen McCullough Johnson and Harris medium and harvested by centrifugation during the late logarithmic phase. DNA was isolated according to the procedure described by Boom et al. (6).
(ii) Enzymes, adapters, and primers.
The sequences of the EcoRI adapters were 5′ CTCGTAGACTGCGTACC 3′ and 3′ CATCTGACGCATGGTTAA 5′, while those of the MseI adapters were 5′ GACGATGAGTCCTGAG 3′ and 3′ TACTCAGGACTCAT 5′ (18). The nonselective forward primer for the MseI adapter site was unlabeled. The reverse primers for the EcoRI adapter site contained a selective base at their 3′ ends (A, G, C, or T) and were labeled with a fluorophore (FAM [6-carboxyfluorescein], JOE [2,7-dimethoxy-4,5-dichloro-6-carboxyfluorescein], NED [2,7′,8′-benzo-5′-fluoro-2′,4,7-trichloro-5-carboxyfluorescein], or TAMRA [6-carboxytetramethylrhodamine]). These primers were obtained commercially (AFLP Microbial Fingerprinting kit; Applied Biosystems, Foster City, Calif.). In each case, genomic DNA was digested with both endonucleases, EcoRI and MseI. The resulting restriction fragments were ligated with the double-stranded adapters, consisting of a core sequence and an enzyme-specific sequence. The restriction ligation reaction was carried out in a single step (1-4, 8, 14).
(iii) Preselective and selective amplifications.
Preselective PCR was carried out using an unlabeled primer set, one for the EcoRI adapter and another for the MseI adapter, whereas selective PCR was performed by using six sets of primers. Four of the forward primers used in the selective PCR contained the EcoRI adapter complementary sequence and an additional nucleotide (A, G, C, or T) at the 3′ ends. The other two forward primers contained an additional AG or AC at the 3′ ends.
(iv) Detection of fragments: Genescan.
Selective PCR products, along with formamide loading dye and the red-colored internal lane standard GS 500 Rox (Applied Biosystems), were loaded onto an ABI Prism 3100 DNA sequencer (Applied Biosystems). Fragment separation was continued for 2.5 h through a performance-optimized polymer 4 (Applied Biosystems). Fragments were detected and compiled by the ABI Data Collection (Perkin-Elmer, Applied Biosystems) software. Gel images were generated, and all lanes were extracted to make individual electropherograms. Fragment analysis was performed with the Genescan Analysis package version 3.1 (Applied Biosystems). Individual sample files were then exported to Genotyper version 2.5 (Applied Biosystems) software for computer-assisted genotyping.
(v) Analysis of data generated on an ABI Prism 3100 sequencer.
Genescan data of all reference strains and the isolates were imported to Genotyper. The Genotyper software detected the fragment length corresponding to each peak. The amplified products were sized in base pairs within the user-defined categories of marker sizes. The presence or absence of amplicons within the categories was scored by a user-defined Genotyper macro. Allele scores (the presence or absence of amplicons) were converted into binary format (1 or 0). This binary format was converted to a nucleotide sequence (1 = G and 0 = A); therefore, fingerprint profiles could be aligned, and a neighbor-joining tree could be constructed based on these profiles by using the ClustalX and Treeview packages.
RESULTS
Nineteen isolates recovered from patients during epidemics and eight isolates from sporadic cases (Table 1) were analyzed for phylogenic relatedness. Of 27 isolates, 19 belonged to serogroup Grippotyphosa, 3 belonged to serogroup Australis, 2 belonged to serogroup Icterohaemorrhagiae, and 1 each belonged to serogroups Hebdomadis, Canicola, and Sejroe (Table 1).
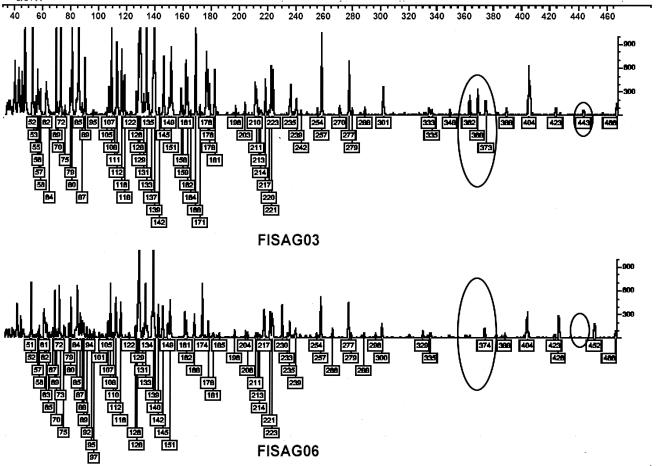
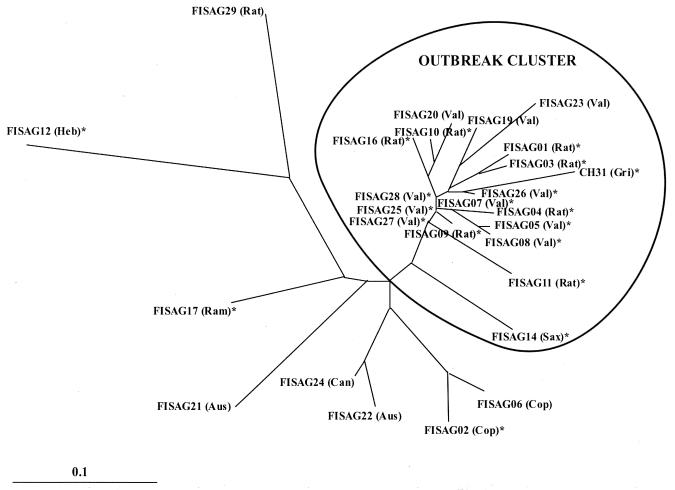
Selective FAFLP PCR with six sets of primers generated a total of 76 polymorphic fragments ranging in size from 50 to 500 bp (Fig. 1). Analysis of a phylogenic tree constructed using the neighbor-joining method revealed that all the isolates grouped into two main clusters (Fig. 2), with the outbreak-associated isolates forming a tight cluster. All of the isolates in the outbreak-associated cluster except one belonged to serogroup Grippotyphosa. Seven of them belonged to serovar Ratnapura, 10 belonged to serovar Valbuzzi, and 1 each belonged to serovars Grippotyphosa and Saxkoebing. Of the 19 isolates in the outbreak-associated cluster, 16 were recovered during various outbreaks, and the remaining 3 were from sporadic cases. However, these epidemics occurred at different times and in different places. The 16 isolates recovered during outbreaks included the strain CH31, isolated during the historical outbreak in 1929. However, isolate CH31 and the non-Grippotyphosa isolate (FISAG14) in this cluster were more distant than the other members of the cluster. Four isolates (FISAG06, FISAG02, FISAG22, and FISAG24) that were outside the outbreak-associated cluster showed close relationship. These isolates belonged to serovars Copenhageni, Australis, and Canicola. The remaining four isolates (FISAG21, FISAG17, FISAG12, and FISAG 29), belonging to serovars Australis, Ramisi, Hebdomadis, and Ratnapura, respectively, were genetically diverse.
FIG. 1.
Genotyper plots representing outbreak-associated (FISAG03) and sporadic (FISAG06) isolates. The traces representing allelic variation between the two profiles are circled.
FIG. 2.
Neighbor-joining network depicted using the PHYLIP package. The tree reveals genetic affinities among outbreak-associated and sporadic isolates of different serovars recovered in India. The asterisks indicate outbreak-associated strains. Aus, Australis; Can, Canicola; Cop, Copenhageni; Gri, Grippotyphosa; Heb, Hebdomadis; Ram, Ramisi; Rat, Ratnapura; Sax, Saxkoebing; Val, Valbuzzi.
Of the 27 patients from whom the isolates were obtained, 9 had severe illness with complications, such as renal failure, abnormal liver function, or pulmonary involvement. Seven of the nine isolates recovered from patients with complications were in the outbreak-associated cluster. Of these seven isolates, five belonged to serovar Valbuzzi and the remaining two belonged to serovar Ratnapura. Of the other two isolates from patients with complications, one belonged to serogroup Icterohaemorrhagiae, serovar Cophenhageni, and the other belonged to serogroup Grippotyphosa, serovar Ratnapura. However, these two were found to be genetically distant and therefore did not cluster together. The isolates obtained from sporadic cases on South Andaman and in south India showed great genetic diversity (Fig. 2). Their serogroups were also diverse (Table 1).
As far as the composition of the outbreak cluster is concerned, we are sure that all the isolates falling in this cluster were Leptospira interrogans. Given that the reference strain of serovar Ratnapura (Wumalasena) is L. kirschneri, our Ratnapura isolates did not appear to be L. kirschneri, as species-specific (17) PCR amplification, based on serovar Icterohaemorrhagiae-derived primers, did amplify amplicons characteristic of L. interrogans. Also, the serovar Bim-derived primers (17) could not amplify L. kirschneri-specific amplicons. Sequencing of the PCR products of four serovar Valbuzzi isolates, generated by serovar Icterohaemorrhagiae-derived primers, indicated that these isolates were indeed L. interrogans. In addition, the historical outbreak strain CH31 was also found to be L. interrogans, though the reference strain and other strains belonging to serovar Grippotyphosa were L. kirschneri.
DISCUSSION
In the Andaman Islands, leptospirosis occurs as outbreaks during the postmonsoon period or occasionally as sporadic cases. Involvement of the respiratory system is a common complication, and this occurs frequently during epidemics on North Andaman. Pulmonary involvement manifesting as hemoptysis is associated with high morbidity and mortality. The manifestation of the disease in patients affected during the 1999 epidemic and in the sporadic cases on South Andaman was different. The disease was mild to moderate among a majority of the patients. Hepatorenal involvement was also a common complication, in addition to pulmonary hemorrhages, among the severe cases.
FAFLP analysis of leptospiral strains recovered from North and South Andaman and south India revealed two underlying genetic groups. The outbreak-associated cluster (Fig. 2) comprised isolates recovered on different occasions and in different places. The strain CH31 (serovar Grippotyphosa, serogroup Grippotyphosa) was recovered from a patient with mild nonicteric leptospirosis during an outbreak in 1929 on South Andaman. FAFLP data showed that CH31 was more distant than the other members of serogroup Grippotyphosa in the outbreak-associated cluster. Although the reference strain, Moskva V, and the other three strains, DF, GG, and STB, of serovar Grippotyphosa belong to L. kirschneri (7), strain CH31 belonged to L. interrogans.
The serovars Ratnapura and Valbuzzi appeared to be the dominant infecting serovars during the recent outbreaks in the Andaman Islands, and some of these isolates were recovered from patients with pulmonary hemorrhage. Sequencing of the PCR products of four isolates belonging to serovar Valbuzzi (FISAG07, FISAG25, FISAG26, and FISAG27) using species-specific primers showed that they belonged to L. interrogans (17). The reference strain Valbuzzi, serovar Valbuzzi, belongs to L. interrogans, whereas the strain Dyster of serovar Valbuzzi belongs to L. kirschneri (7). Strong antigenic similarities among some serovars of serogroup Grippotyphosa are well documented (17), and these similarities sometimes interfere with the identification of serovars. The monoclonal antibody patterns of serovars Ratnapura and Valbuzzi were almost similar, and hence, these antibodies failed to differentiate the two serovars (17). Genetic similarity between the two serovars with respect to a high-resolution genotyping technique such as FAFLP further substantiated this relatedness among the members of serovars Valbuzzi and Ratnapura. The reference strain of serovar Ratnapura (strain Wumulsena) belongs to L. kirschneri (7). However, since the isolates belonging to serovars Valbuzzi, Ratnapura, and Saxkoebing formed a tight cluster, along with strain CH31 of serovar Grippotyphosa of L. interrogans, it is possible that all the isolates in the outbreak-associated cluster were L. interrogans. This was subsequently proved by species-specific PCR. Grippotyphosa is frequently identified as the infecting serogroup, based on MAT titers in patients' sera during several upsurges, outbreaks, and sporadic cases of leptospirosis (11, 15, 17). Serogroup Grippotyphosa was also frequently encountered among domestic and free-living animals (13). Although genotypic data for animal isolates are not available, it can be assumed that a genetically similar group of strains might be circulating in different animal hosts in the islands.
Some of the isolates in the outbreak-associated cluster were obtained in 1996 and 1997 on North Andaman from patients with AHF (the local name for leptospirosis with pulmonary involvement). We assume that these AHF isolates spread to South Andaman because of increased transport of people and domestic animals between the two areas and perhaps via rodents infesting cargo and passenger ships that sail between North and South Andaman frequently.
Of the nine severe cases in which isolation was successful, isolates belonging to the outbreak cluster were the causative agents in seven patients. Two of the patients with severe illness who were infected by strains in the outbreak-associated cluster died (Table 1).
There are eight isolates outside the outbreak-associated cluster. Of these eight isolates, five were recovered from sporadic cases. Seven of these eight isolates were obtained from patients on South Andaman, and the remaining isolate was from south India. Four isolates (FISAG06, FISAG02, FISAG22, and FISAG24) outside the outbreak-associated cluster were closely related to each other. These isolates were identified as belonging to serovars Copenhageni, Australis, and Canicola, and the strains of these serovars studied so far belong to L. interrogans. Although these serovars belong to the species L. interrogans, they were well separated from the strains in the outbreak-associated cluster, indicating the discriminatory power of FAFLP for resolving genetic differences that are important in molecular epidemiology. The remaining four isolates (FISAG29, FISAG12, FISAG21, and FISAG17) in this group were genetically diverse. These isolates belong to serovars Ratnapura, Hebdomadis, and Ramisi. The reference strains of serovar Hebdomadis belong to L. interrogans, whereas the reference strains of serovar Ratnapura and serovar Ramisi belong to L. kirschneri. The antigenic nature and genetic makeup of leptospires are complex and do not often coincide with each other. Leptospires belonging to different serogroups and serovars may be of the same genomospecies. For example, several serovars of 15 different serogroups were identified as belonging to L. interrogans sensu stricto (7). Conversely, strains belonging to different genomospecies may be serologically indistinguishable. This is seen in different strains of serovar Hardjo, which belong to several different genomospecies, such as L. interrogans, L. borgpetersenii, and L. meyeri (19).
The present study has demonstrated that FAFLP is a powerful technique for resolving genetic differences that are important in studying the molecular epidemiology and dissemination dynamics of leptospires. Whole-genome fingerprinting profiles generated in our study may be useful for electronic deposition and downloading for interlaboratory comparisons and are suitable for storage in clinicoepidemiological databases. Future developments involving genome sequence-based modeling of FAFLP, in the context of the four spirochetal genomes available in the public domain, should facilitate the identification of novel marker loci helpful in addressing important questions related to the evolution of leptospires in different geographical regions and niches.
Acknowledgments
This study was undertaken as part of the AmpliBASE pathogen barcode program of CDFD, supported by funds from the Department of Biotechnology, Government of India, to N.A. and S.E.H. through core grants. P.V. is the In-Charge of the Leptospirosis Reference Laboratory at RMRC, Port Blair, India. N.A. is a staff scientist and leads the pathogen evolution program at CDFD.
We are thankful to Mahfooz Alam, Farhana Kauser, and Abid Hussain for help during FAFLP analysis. We are thankful to Rudy Hartskeerl for helpful discussions and suggestions.
REFERENCES
- 1.Ahmed, N., A. Bal, A. A. Khan, M. Alam, A. Kagal, V. Arjunwadkar, A. Rajput, A. A. Majeed, S. A. Rahman, S. Banerjee, S. Joshi, and R. Bharadwaj. 2002. Whole genome fingerprinting and genotyping of multiple drug resistant (MDR) isolates of Pseudomonas aeruginosa from endophthalmitis patients in India. Infect. Genet. Evol. 1:237-242. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, N., L. Caviedes, M. Alam, K. R. Rao, V. Sangal, P. Sheen, et al. 2003. Distinctiveness of Mycobacterium tuberculosis genotypes from human immunodeficiency virus type 1-seropositive and -seronegative patients in Lima, Peru. J. Clin. Microbiol. 41:1712-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, N., A. A. Khan, A. Alvi, S. Tiwari, C. S. Jyothirmayee, F. Kauser, M. Ali, and C. M. Habibullah. 2003. Genomic analysis of Helicobacter pylori from Andhra Pradesh, South India: molecular evidence for three major genetic clusters. Curr. Sci. 85:101-108. [Google Scholar]
- 4.Ahmed, N., M. Alam, A. A. Majeed, S. A. Rahman, A. Cataldi, D. Cousins, et al. 2003. Genome sequence based, comparative analysis of the fluorescent amplified fragment length polymorphisms (FAFLP) of tubercle bacilli from seals provides molecular evidence for a new species within the Mycobacterium tuberculosis complex. Infect. Genet. Evol. 2:193-199. [DOI] [PubMed] [Google Scholar]
- 5.Barker, F. A. 1926. Leptospirosis with special reference to the existence of spirochaetosis, icterohaemorrhagica, or Weil's disease in the Andaman Islands. Ind. Med. Gaz. 61:479-488. [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-Van Dillen, and J. Van der Noorda. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49:839-858. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, I. M., N. Ahmed, S. M. Beesley., A. A. Khan, C. A. O Moráin, and C. J. Smyth. 2003. Fine structure molecular typing of Irish Helicobacter pylori isolates and their genetic relatedness to strains from four different continents. J. Clin. Microbiol. 41:5755-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary, A. K. 1903. Jaundice at Port Blair, Andaman Islands. Ind. Med. Gaz. 38:409-412. [PMC free article] [PubMed] [Google Scholar]
- 10.Savelkoul, P. H. M., H. J. M. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, et al. 1995. Amplified fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sehgal, S. C., M. V. Murhekar, and A. P. Sugunan. 1995. Outbreak of leptospirosis with pulmonary involvement in North Andaman. Indian J. Med. Res. 102:9-12. [PubMed] [Google Scholar]
- 12.Sehgal, S. C., P. Vijayachari, and V. Subramaniam. 1997. Evaluation of Leptospira microcapsule agglutination test (MCAT) for serodiagnosis of leptospirosis. Indian J. Med. Res. 106:504-507. [PubMed] [Google Scholar]
- 13.Sharma, S., P. Vijayachari, A. P. Sugunan, and S. C. Sehgal. 1999. Leptospiral carrier state and seroprevalance among animal populations—a cross-sectional sample survey in Andaman and Nicobar Islands. Epidemiol. Infect. 130:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiqi, N., R. Das, N. Pathak, S. Banerjee, N. Ahmed, V. M. Katoch, and S. E. Hasnain. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a TAP like efflux pump. Infection 32:109-111. [DOI] [PubMed] [Google Scholar]
- 15.Singh, S. S., P. Vijayachari, A. Sinha, A. P. Sugunan, M. A. Rasheed, and S. C. Sehgal. 1999. Clinico-epidemiological study of hospitalized cases of severe leptospirosis. Indian J. Med. Res. 109:94-99. [PubMed] [Google Scholar]
- 16.Taylor, J., and A. N. Goyle. 1931. Leptospirosis in Andamans. Indian Med. Res. Memoirs 20. Thacker, Spink and Co., Calcutta, India.
- 17.Vijayachari, P., S. C. Sehgal, M. G. Goris, W. J. Terpstra, and R. Hartskeerl. 2003. Leptospira interrogans serovar valbuzzi: a cause of severe pulmonary haemorrhages in Andaman Islands. J. Med. Microbiol. 52:913-918. [DOI] [PubMed] [Google Scholar]
- 18.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Lee, M. Hornes, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuerner, R., D. Haake, B. Alder, and R. Segers. 2000. Technological advances in the molecular biology of leptospira. J. Mol. Microbiol. 2:455-462. [PubMed] [Google Scholar]