Most antigens encountered by the immune system enter the body through the mucosal surfaces of the respiratory, gastrointestinal (GI), and urogenital tract, and the vast majority of pathogens use these tissues as portals of entry. An additional feature of these tissues is their constitutive and ever evolving relationship with highly diverse and site-specific microbial communities referred to as the microbiota (1). Barrier sites are therefore charged with the formidable task of mediating the host relationship with its microbiota, protecting the host from environmental and pathogenic challenges while preserving vital physiological functions.
At these sites, failure to control responses can lead to severe inflammatory disorders including asthma, inflammatory bowel disease, psoriasis, or food allergies. The incidence of these chronic inflammatory diseases, as with most pathologies targeting barrier tissues, has risen significantly for the past several decades (2). Understanding how immunity is controlled at barrier tissues as well as the key stressors of these environments is not only of major public health interest, but is also the object of intensive investigation. This issue of Immunological Reviews presents articles from investigators involved in the exploration of the mechanisms by which mucosal barrier sites induce and control innate and adaptive immune responses.
The body's epithelial surfaces act as a scaffold to sustain diverse communities of commensals that include bacteria, archaea, fungi, protozoa, and viruses (3-7). With an estimated composition of 100 trillion cells, commensals outnumber host cells by at least a factor of 10 and encode at least 100 times more unique genes than their host's genome (8). These microbes represent a formidable challenge for the immune system, and as such, a large fraction of the mucosal immune network is aimed at limiting reactivity or exposure to the microbiota. To maintain a homeostatic relationship with the microbiota and limit antigenic and pathogen exposure, protection is primarily mediated by an epithelial layer that forms a physical barrier and is supported by both adaptive and innate arms of the immune system. Over the past few years, it has become clear that epithelial cells, via their capacity to produce mucus, antimicrobial peptides, metabolites, or cytokines, are a central determinant of immunity in tissue microenvironments (9). As discussed in this volume by Gunnar Hansson and colleagues (10), the mucus layer, built around the gel-forming mucins, covers all mucosal surfaces therefore limiting pathogenic invasion and contact with epithelial cells. However, this primary shield is not absolute, and a certain degree of controlled reactivity to the microbiota contributes to reinforce barrier immunity and function. Appropriate responses to the microbiota are established soon after birth upon exposure of the immune system to commensals during the passage through the birth canal. Expanding on this concept, Fulde and Hornef (11) discuss how early adaptation of innate cells to microbial stimulation set the tone of subsequent responses to the microbiota. As further discussed by Fung et al. (12), containment of the microbiota thorough life represents a formidable task for the mucosal immune system that requires the continuous and combined action of epithelial cells, adaptive immunity, and innate population of lymphocytes. One of these mechanisms is associated with immunoglobulin A (IgA) responses. About three quarters of total antibody production is composed of IgA, in the order of 3–5 g produced per day in humans, with the vast majority secreted across barrier surfaces. The reviews of Slack et al. (13), Kato et al. (14), and Gutzeit et al. (15) discuss the various and fundamental roles of IgA in regulating mucosal immunity. Notably, these authors discuss how IgA shape the microbiota, mediate pathogen clearance, neutralize toxins, and prevent adhesion of commensal bacteria to the epithelial surfaces by generating steric hindrance.
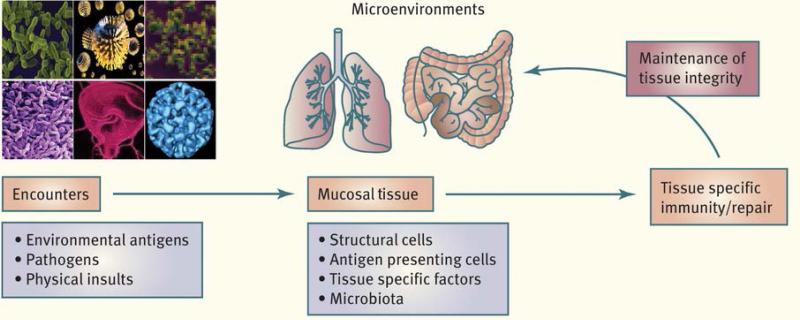
The complex and dynamic control of mucosal immunity requires an arsenal of defined tissue-specific factors and highly specialized innate and adaptive immune cell types, specifically conditioned by the mucosal environment to induce tissue tailored immunity (Figure 1). These networks converge to prevent overt reactivity to commensal and environmental antigens and to favor the induction and maintenance of tolerogenic responses. Antigen-presenting cells (APCs) play an important role in this tissue immune specialization, and each mucosal site is seeded by defined APCs. The gut notably is home to a highly specialized and complex network of APCs. These gut APCs, in addition to monocytes recruited from the blood during inflammation, form the mononuclear phagocyte system of the GI tract. Reviews from Agace and colleagues (16), Mowat and colleagues (17), and Maria Rescigno (18) highlight the complexity and versatility of the intestinal APC network and particularly how the function and development of these cells is conditioned by local cues derived from the diet or epithelial cells. The results of this local tuning are the induction of tolerogenic responses via the induction of various populations of regulatory T cells. Together, these reviews also discuss the contextual role of gut APCs in both maintaining tolerance in steady state and promoting effector responses during infection or chronic inflammation. Indeed, tolerance to the microbiota or environmental antigen is not the only fate of immune responses at mucosal sites. Reactivity of barrier tissue is a vital requirement for the host, as the majority of pathogenic microbes target or are acquired through mucosal surfaces of the GI, respiratory, and genital tracts. To this end, and as discussed by McAleer and Kolls (19) and Cho and Kelsall (20), mucosal tissues are constitutively enriched in cytokines such as interleukin-17 (IL-17), IL-22, or type I interferon (IFN). Despite the specialization of each tissue and microenvironment, the expression of these cytokines is relatively conserved between mucosal sites such as the lung and GI tract. Via their capacity to act both on epithelial cells and to control inflammatory cells, IL-22 and cytokines of the IL-17 family contribute to both mucosal homeostasis and immunity (21, 22). Stephen McSorley (23) also discusses how immunity to a pathogen such as Salmonella develops and is controlled in the GI tract, while Grencis et al. (24) highlight how the mucosal immune system has evolved mechanisms promoting type 2 immunity to allow coexistence with nematodes.
Figure 1. Tailored immunity.
Maintenance of tolerance and restoration of mucosal homeostasis following insults or exposure to pathogens relies on a complex and coordinated set of innate and adaptive responses. Such tissue-specific immune specialization relies on the capacity of defined and site-specific populations of immune cells to integrate local cues such as microbial products, metabolites, or nutrients. These tailored networks are essential to the induction of responses in a way that preserve the physiological and functional requirements of a defined tissue.
As previously discussed, barrier sites are primary sites of inflammatory disorders. When operating optimally, the mucosal immune system network interweaves the innate and adaptive arms of immunity in a dialogue that selects, calibrates, and terminates responses in the most appropriate manner. However, as discussed by Elson (25), Jabri (26), Umetsu (27), and Strober (28) and their colleagues, breakdown in the capacity of the host to maintain regulatory responses to the multitude of antigens encountered by mucosal tissues represents the underlying cause of severe inflammatory disorders such as inflammatory bowel disease, allergy, and asthma that increasingly affect barrier sites in westernized countries. Collectively, this group of reviews highlight the rapidly growing knowledge of the pathways and cell types that regulate immune cell homeostasis at mucosal surfaces and inflammation and discuss some of the future challenges to be addressed in understanding barrier immunity in the context of infection, inflammation, and tissue repair.
Acknowledgement
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- 1.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 3.Foulongne V, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg PB, Lepp PW, Relman DA. Archaea and their potential role in human disease. Infect Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelaseyed T, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulde M, Hornef MW. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev. 2014;260:21–34. doi: 10.1111/imr.12190. [DOI] [PubMed] [Google Scholar]
- 12.Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev. 2014;260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slack E, Balmer ML, Macpherson AJ. B cells as a critical node in the microbiota-host immune system. Immunol Rev. 2014;260:50–66. doi: 10.1111/imr.12179. [DOI] [PubMed] [Google Scholar]
- 14.Kato LM, Kawamoto S, Maruya M, Fagarasan S. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev. 2014;260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 15.Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol Rev. 2014;260:76–85. doi: 10.1111/imr.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekiaris V, Persson EK, Agace WW. Intestinal dendritic cells in the regulation of mucosal immunity. Immunol Rev. 2014;260:86–101. doi: 10.1111/imr.12194. [DOI] [PubMed] [Google Scholar]
- 17.Bain CC, Mowat AM. Macrophages in intestinal homeostasis and inflammation. Immunol Rev. 2014;260:102–117. doi: 10.1111/imr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rescigno M. Dendritic cell-epithelial cell crosstalk in the gut. Immunol Rev. 2014;260:118–128. doi: 10.1111/imr.12181. [DOI] [PubMed] [Google Scholar]
- 19.McAleer JP, Kolls JK. Directing traffic: IL-17 and IL-22 coordinate pulmonary immune defense. Immunol Rev. 2014;260:129–144. doi: 10.1111/imr.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho H, Kelsall BL. The role of type I interferons in intestinal infection, homeostasis, and inflammation. Immunol Rev. 2014;260:145–167. doi: 10.1111/imr.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberl G. Development and evolution of RORgammat+ cells in a microbe's world. Immunol Rev. 2012;245:177–188. doi: 10.1111/j.1600-065X.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- 22.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McSorley SJ. Immunity to instestinal pathogens: lessons learned from Salmonella. Immunol Rev. 2014;260:168–182. doi: 10.1111/imr.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grencis RK, Humphreys NE, Bancroft AJ. Immunity to gastrointestinal nematodes: mechanisms and myths. Immunol Rev. 2014;260:183–205. doi: 10.1111/imr.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander KL, Targan SR, Elson CO. Microbiota activation and regulation of innate and adaptive immunity. Immunol Rev. 2014;260:206–220. doi: 10.1111/imr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abadie V, Jabri B. IL-15: a central regulator of celiac disease immunopathology. Immunol Rev. 2014;260:221–234. doi: 10.1111/imr.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeKruyff RH, Yu S, Kim HY, Umetsu DT. Innate immunity in the lung regulates the development of asthma. Immunol Rev. 2014;260:235–248. doi: 10.1111/imr.12187. [DOI] [PubMed] [Google Scholar]
- 28.Strober W, Asano N, Fuss I, Kitani A, Watanabe T. Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn's disease. Immunol Rev. 2014;260:249–260. doi: 10.1111/imr.12193. [DOI] [PubMed] [Google Scholar]