Abstract
Archival paraffin-embedded tumor specimens offer a wealth of information for both cancer research and for routine clinical applications. However, the use of formalin-fixed, paraffin-embedded specimens for quantitative real-time PCR is not yet a standard diagnostic method in many laboratories, in particular for the quantification of human papillomavirus (HPV). Particularly high-risk HPV types are involved in almost 100% of the carcinogenesis of cervical cancer. We compared the diagnostic applicability and sensitivity of real-time PCR to that of chromogenic tyramide-signal-amplified in situ hybridization and conventional PCR for the detection of HPV from archival tissue in 164 cases of carcinoma in situ and cervical cancer. Furthermore, we examined whether the viral load of HPV is of prognostic relevance. Our findings indicate that patients in tumor stage I with a lower viral load of HPV type 16 (HPV16; up to 1,000 copies/ng of DNA) had a significantly better survival than HPV 16-negative patients (P = 0.037). We observed a greater sensitivity of both real-time PCR and conventional PCR for the detection of HPV16 and -18 compared to signal amplified in situ hybridization. We found a considerable concordance between HPV16 (κ = 0.661) and HPV18 (κ = 0.781) status as measured by real-time PCR and conventional PCR, indicating similar sensitivities. We recognized an inhibitory effect of formalin fixation and paraffin embedding on the evaluation of real-time PCR quantification.
Formalin-fixed, paraffin-embedded (FFPE) tissues retained in pathology archives are an important resource for molecular pathology studies and for diagnostic applications. The use of in situ hybridization (ISH) and PCR assays have both been reported for DNA analysis in formalin-fixed archival tissues (6, 7, 25, 40, 42, 48). Both assays have the advantage that only small amounts of tissue are required, and both tolerate the degradation of target nucleic acids due to the formalin fixation and paraffin-embedding procedures. Each assay per se has its own advantages and disadvantages.
For a pathologist, the retention of morphology and the histological context of a tumor is very important. Especially in the case of cervical carcinoma, the phenomenon of koilocytosis indicates an infection with human papillomavirus (HPV) (4) that can only be detected by microscopic analysis of tissue sections. Moreover, episomal and integrated viral DNA can only be differentiated by ISH due to the staining pattern (8). With the use of chromogenic tyramide-signal-amplified ISH (CISH) it is even possible to detect and localize single or very few HPV copies within infected nuclei (15, 36, 49). However, this technique is rather cost-intensive and quite involved for routine applications.
Conventional PCR is held to be the most sensitive detection method available (9, 16, 20, 27, 37, 41). Although this is probably true, it has some limitations, as does ISH, in that it cannot quantify viral load in a prognostic-diagnostic significant manner. This restriction can be circumvented by the use of real-time quantitative PCR. Real-time PCR allows the detection and quantification of products generated during PCR process. The reagents most commonly used for real-time PCR include 5′ nuclease assay with TaqMan probes, molecular beacons, and SYBRGreen I intercalating dyes. We used SYBRGreen technology because it is easier to handle in routine applications and to optimize. The use of FFPE specimens for real-time PCR is not yet frequently a standard method in laboratories and therefore is not often reported (28, 29, 33). It is also rarely used for quantification of HPV (46).
HPV is known to be a major risk factor in the development of genito-anal neoplasia (19, 51). Low-risk HPV types (e.g., HPV type 6 [HPV6] and HPV11) are distinguished from intermediate-risk (e.g., HPV31, -33, and -51) and high-risk (e.g., HPV16 and -18) HPV types (3, 32). In particular, high-risk HPV types are involved in almost 100% of the instances of cervical cancer. Cervical squamous cell carcinomas develop from progenitor lesions, so-called cervical intraepithelial neoplasias (CINs; I to III). Viral load of human papillomavirus might be of prognostic-diagnostic significance (21, 31, 45). With increasing amounts of high-risk HPV, the risk for the development of squamous intraepithelial lesions and CINs and further transformation to invasive cervical carcinoma, as well as the severity of cervical disease, is likely to increase (22, 44, 45). Although high-risk HPV is known to be a major risk factor for the development of cervical cancer from CIN, its role in established cervical cancer concerning progression and clinical outcome is still controversial (11, 12, 14, 21, 26, 34, 35, 39). The amount of viral DNA present in the tumor could represent an important aspect for course and strength of disease. Since methods for viral load determination are available and becoming more reliable, that question gains in importance (5, 30, 38).
Ideally, a quantitative assay for viral nucleic acids should possess high sensitivity, specificity, and reproducibility, yield an accurate determination of copy number, and be fast and easy to use in routine work. We compared the applicability, handling, and sensitivity of real-time PCR versus CISH and conventional PCR for the detection of HPV in FFPE cervical cancer tissues.
MATERIALS AND METHODS
Human cervical cancer tissue specimens and cell lines.
A total of 160 cases of invasive cervical carcinomas and 4 cases of carcinoma in situ (CIS) diagnosed between 1989 and 2000 were obtained from the Institute of Pathology, General Hospital Salzburg, Salzburg, Austria. All examinations were carried out upon FFPE tissue blocks. Histological classification was performed according to World Health Organization guidelines, and tumor staging was according to the TNM System of the International Union against Cancer. The patient cohort has been described previously, and numerous clinical and histological parameters were available from previous studies (47). The mean age of the patients was 46.16 (±11.9). The 164 cases examined comprised 4 CIS (2.4%), 82 stage I carcinomas (50%), 49 stage II carcinomas (29.9%), 2 stage III carcinomas (1.2%), and 1 stage IV carcinoma (0.6%). In 26 cases (15.9%), the stage was not defined. Of the specimens, 121 (73.8%) were squamous cell carcinomas and 16 (9.8%) were adenocarcinomas. In 27 cases (16.4%), histological tumor type was not available.
Cervical cancer cell lines (SiHa, HeLa, and CasKi) were obtained from the American Type Culture Collection (Rockville, Md.). They are infected with ascending copy numbers of HPV. SiHa cells contain 1 to 2 copies of HPV16, and CasKi cells contain about 600 copies of HPV16; HeLa cells harbor 40 to 50 copies of HPV18. These cell lines were used for the establishment of an external standard curve for real-time PCR and served as positive controls.
CISH.
For the CISH method, the Enzo Diagnostics kit was used with different probe mixtures for HPV6 and -11 (HPV6/11), -16/18, and -31/33/51 in combination with the Dako GenPoint kit. This method allows the detection and localization of single or very few copies of HPV-infected nuclei. The tyramide reporter molecule amplifies the number of the biotin molecules, which enhances the binding of streptavidin-peroxidase complex. Then, 4-μm sections were mounted and baked onto silanized slides at 60°C for 1 h, and the protocol was performed according to the manufacturer's recommendations.
Extraction of DNA from FFPE sections and cultured cells.
First, 5 to 20 10-μm sections (dependent on tissue size) were collected in 1.5-ml Eppendorf tubes for DNA isolation. To prevent cross-contamination, each block was cut after thorough cleaning of the microtome with a new blade. DNA extraction was carried out by using the QiaAmp DNA minikit (Qiagen) and the appropriate spin columns according to the manufacturer's protocol. The quality (ratio of 260/280 nm) and quantity (absorbance at 260 nm) of isolated DNA were determined by optical density measurement.
HeLa, CasKi, and SiHa cells were grown for about 5 days in the appropriate medium (HeLa and CasKi cells, Dulbecco modified Eagle medium; SiHa cells, RPMI 1600 [Gibco-BRL, Grand Island, N.Y.]). DNA isolation was performed with the QiaAmp DNA minikit according to the protocol for cultured cells grown in monolayer. An aliquot of the cultured cells was collected before adding proteinase K and fixed in 4% buffered formalin to count the cells in a Bürker-Türk cell counter. The number of copies of viral DNA per microliter was derived from the number of cells estimated to be present in that same volume.
Conventional PCR.
HPV detection was made by PCR with DNA from paraffin-embedded cervical cancer tissue. A universal HPV primer pair (termed “consensus primer”) and primer pairs specific for the high-risk HPV16 and -18 were used. The frequently used MY09-MY11 consensus primer contains several degenerate nucleotides and is thus a mixture of 24 primers capable of amplifying a 450-bp fragment within the conserved L1 region common to numerous HPV types; it therefore recognizes a wide spectrum of HPV types (18, 20). This primer pair is routinely used in our lab. The primer sets for HPV16 (E6/E7 region; product size, 96 bp) and HPV18 (product size, 115 bp) (1) were as follows: primer set HPV16fwd (accession no. AF469197, bases 389 to 408; 5′-GGTCGGTGGACCGGTCGATG-3′) and HPV16rev (accession no. AF469197, bases 465 to 484; 5′-GCAATGTAGGTGTATCTCCA-3) and primer set HPV18fwd (accession no. AY262282, bases 7004 to 7023; 5′-CCTTGG ACGTAAATTTTTGG-3′) and HPV18rev (accession no. AY262282, bases 7100 to 7119; 5′-CACGCACACGCTTGGCAGGT-3′). Amplification reactions contained a final volume of 50 μl and were performed with 1× Taq PCR master mix kit (Qiagen), 0.5 μM concentrations of each primer, and either 5 μl of nondiluted or 5 μl of 1/10-diluted DNA samples. PCR was performed by using the following cycling conditions: first, 3 min at 95°C; then 40 cycles of 95°C for 15 s, 59°C (for HPV18) or 60°C (for HPV16) for 20 s, and 72°C for 25 s; with a final elongation of 7 min at 72°C. The cycling protocol for MY09/MY11 was as follows: 3 min 95°C; followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 90 s; with a final elongation of 7 min at 72°C. The quality of sample DNA was validated by amplification of a 286-bp fragment of the housekeeping gene β-globin. 5 μl of each PCR product was electrophoresed in a 1.5% agarose gel (Agarose Multi-Purpose; Roche). Images were recorded by using the Geldoc 2000-System (Bio-Rad).
Real-time PCR.
The I-Cycler instrument (Bio-Rad) permits the simultaneous amplification and quantification of the PCR-product at each cycle. We used the SYBRGreen system for the determination of absolute amounts of viral DNA by standardizing with DNA from cultured cells containing known copy numbers of HPV, which were included in each run in duplicates in the form of a series consisting of five dilutions with amounts ranging from 102 to 106 copies of HPV to create an external standard curve. The final standard regression straight line was made up of the breakpoint signals of both series. A master mix with the SYBRGreen PCR core reagents (PE Biosystems) was optimized for the I-Cycler instrument and contained the following: 5 μl of each probe (diluted 1/100 in H2O), respectively; 5 μl of the cell line-DNA dilution; 1× SYBRGreen buffer; 5 mM MgCl2; a 0.25 mM concentration of each deoxynucleoside triphosphate; a 0.5 μM concentration of each primer; and 1.25 U of AmpliTaq Gold polymerase in a final volume of 50 μl. The reaction mix and samples were loaded into a 96-well plate (Bio-Rad). The thermal cycler conditions were as follows: 40 cycles of 95°C for 12 min, 95°C for 15 s, 59°C (for HPV18) or 60°C (for HPV16) for 20 s, and 72°C for 25 s. A calibration with an external calibration plate containing 50 μl of 40 μM calibration dye (Bio-Rad)/well was performed prior to each run. The absolute viral copy number in the unknown samples was calculated from the Ct value by using the I-Cycler software. Each sample was measured twice, and the average copy number, rounded to whole decimal powers, was used in further calculations and statistics. Normalization was carried out by extrapolating the concentration of copy numbers present in the samples on the basis of known amounts (determined by optical density measurement) applied in real-time PCR.
Specificity and sensitivity of real-time PCR.
Melting curve analysis was performed after PCR amplification to prove product specificity in real-time PCR. Starting at 60°C, the temperature in the thermal chamber was raised to 95°C at a transition rate of 1°C/s. PCR products were considered to be specific if they had deviations of ≤1°C from their expected melting temperatures. Deviations of >1°C indicated that the starting quantity was too small and that the product has been contaminated by unspecific primer-dimer melting signals, thus limiting the instrument's ability for accurate PCR product specification.
Differences in sensitivity between a gel-free (real-time PCR) and a gel-required (conventional PCR) system should become obvious in comparisons of the sensitivity of product visualization. Therefore, we used starting amounts of HPV16 ranging from 10−1 to 106 copies of HPV DNA from Caski cells. One dilution series was performed in DNA from HPV-negative paraffin-embedded cervical cancer material, and another one was performed in non-paraffin-embedded DNA from HeLa cells. Both samples contained approximately the same amounts of “background DNA” (for the paraffin-embedded sample, 66 ng of DNA/μl; for the HeLa cells, 80 ng of DNA/μl).
The influence of paraffin embedding on the efficacy of amplification and real-time PCR evaluation was tested as follows. The same amount of HPV16 target DNA (104 copies [from CasKi cells]) was added to dilution series made from different HPV-negative paraffin samples and three non-paraffin-embedded samples (HeLa cells). The DNA concentration of the six paraffin-embedded samples ranged from 62.5 to 86 ng/μl, and the DNA concentrations of the HeLa cells were 72, 80, and 91.3 ng/μl. This assay was used to compare nondiluted and diluted paraffin-embedded DNA samples with each other and with DNA from fresh cell material (HeLa cells). We used three 10-fold dilutions to determine at which point of the PCR the inhibiting effects of paraffin (if any) disappeared. Again, this assay was performed by using both real-time PCR and conventional PCR. Visualization of conventional PCR on gel was compared to the results obtained from I-Cycler instrument.
Controls.
For ISH, HPV-positive cases for HPV6/11, -16/18, and -31/33/51 and FFPE cell lines were used. DNA isolated from the cell lines mentioned above (SiHa and CaSki cells for HPV16 and HeLa cells for HPV18) were used as positive controls for conventional PCR and real-time PCR. In conventional PCR and real-time PCR, a negative control containing reagents only was used, and in ISH HPV-negative cases served as negative controls.
Statistical analysis.
Statistical analyses were performed by using the SPSS statistical package (SPSS, Inc., Chicago, Ill.). Kappa statistics were used to assess concordance between pairwise grouped methods. The kappa statistics evaluates the level of agreement adjusted for agreement expected to occur by chance alone. Kappa statistics less than 0.4 represents fair too poor agreement, values of 0.4 to 0.8 represents moderate to good agreement, and values of more than 0.8 represent excellent agreement. Survival analysis was performed by Kaplan-Meyer-statistics (log-rank test).
RESULTS
CISH, conventional PCR, and real-time PCR.
Table 1 gives an overview of the incidence of HPV-positive and -negative cases obtained by CISH, conventional PCR, and real-time PCR. The overall incidence of HPV-positive cases as determined by CISH, independent of the type or multiplicity of infection, reached 47.1%. By conventional PCR, 53 of 142 cases (37.3%) were HPV consensus positive. The overall positive rate for HPV16 and/or HPV18 as determined by conventional PCR reached 74.1%. By real-time PCR, 77.6% were found to be positive for either HPV16 or HPV18.
TABLE 1.
Incidence of HPV-positive cases as determined by CISH, conventional PCR, and real-time PCR
| Method and HPV type(s) | No. of HPV-positive cases/total no. of cases | % |
|---|---|---|
| CISH | ||
| HPV6/11 | 18/144 | 12.5 |
| HPV16/18 | 64/148 | 43.2 |
| HPV31/33/51 | 24/143 | 16.8 |
| HPV total | 66/140 | 47.1 |
| Conventional PCR | ||
| HPV16 | 102/158 | 64.6 |
| HPV18 | 36/161 | 22.4 |
| HPV16/18 | 117/158 | 74.1 |
| HPV consensus | 53/142 | 37.3 |
| Real-time PCR | ||
| HPV16 | 99/161 | 61.5 |
| HPV18 | 34/161 | 21.1 |
| HPV16/18 | 125/161 | 77.6 |
Results of HPV quantitation by real-time are shown in Table 2. HPV16 and -18 have unimodal viral copy numbers of 102 and <100, respectively (HPV16 range, <1 to 105; HPV18 range, <1 to 105). No significant correlation between HPV16 and -18 viral load and tumor stage has been found.
TABLE 2.
Quantification of HPV by real-time PCR
| No. of HPV copiesa | No. of cases
|
|
|---|---|---|
| HPV16 | HPV18 | |
| ≤1 | 9 | 13 |
| 10 | 19 | 9 |
| 102 | 26 | 7 |
| 103 | 23 | 2 |
| 104 | 11 | 0 |
| 105 | 1 | 1 |
That is, The number of copies per nanogram of DNA; values are rounded to whole decimal powers.
Comparison of CISH, conventional PCR, and real-time PCR. (i) PCR versus ISH.
Of 66 CISH-positive cases, only 36 (54.5%) were also determined to be positive by the conventional consensus PCR. Conversely, of 64 HPV-negative cases with ISH, 50 (78.1%) were determined to be negative by conventional consensus PCR (κ = 0.313, 95% confidence interval [CI] = 0.134 to 0.492 [indicating a concordance of 66.2%]).
There was also a fair concordance between the HPV16/18 status as measured by ISH and PCR (κ = 0.275, 95% CI = 0.124 to 0.425). Of 63 ISH-positive cases, 58 (92.1%) were also found to be positive by conventional type-specific PCR. Of 80 ISH-negative cases, only 30 (37.5%) were determined to be negative with conventional type-specific PCR.
A fair concordance was also found between ISH and real-time PCR for HPV16/18 status (κ = 0.259, 95% CI = 0.111 to 0.407), with a similar trend: of 64 HPV16/18-positive cases as determined by ISH, 63 (95.3%) were also found to be positive by real-time PCR, but of 82 ISH-negative cases, only 27 (32.9%) were found to be negative by real-time PCR.
(ii) Real-time PCR versus conventional PCR.
We found a substantial degree of agreement between the HPV16 status revealed by real-time PCR and that determined by conventional PCR (κ = 0.661, 95% CI = 0.539 to 0.783). Of 97 HPV16-positive cases as determined by real-time PCR, 87 (89.7%) were also found to be positive by conventional PCR. Of the 61 HPV16-negative samples as determined by real-time PCR, 46 (75.4%) were also found to be negative by conventional PCR.
As in the case of HPV16, there was also considerable concordance (κ = 0.781, 95% CI = 0.662 to 0.900) between HPV18 status measured by real-time PCR and that measured by conventional PCR. Of 34 HPV18-positive results determined by real-time PCR, 29 (85.3%) were also found to be positive by conventional PCR, and of 127 HPV18-negative results determined by real-time PCR, 120 (94.5%) were also found to be negative by conventional PCR.
Influence of formalin fixation and paraffin embedding on real-time PCR and optimization of SYBRGreen assay conditions.
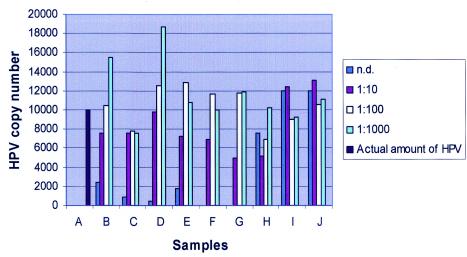
When DNA from paraffin-embedded material was used in real-time PCR, the I-Cycler instrument displayed strongly increased relative fluorescence units right from the start up, which was in contrast to that of the control (non-paraffin-embedded material). This resulted in an underestimation of the starting target DNA quantity after baseline subtraction. This can be seen in the results of the test assay, which show that a more accurate absolute quantitative measurement was reached by a dilution of the paraffin-embedded samples (Fig. 1). In paraffin-embedded samples, viral DNA was measured in proportion at higher dilutions of paraffin-embedded DNA, approaching the actual amount of HPV16 present in the samples. Using nondiluted paraffin samples, there was again a clear underestimation of the actual amount of HPV16 DNA. This effect was not seen with DNA from HeLa cells. A 1 in 100 dilution was used for the subsequent real-time PCR assays because at this dilution nonspecific interference was minimal.
FIG. 1.
Positive effect of dilution of FFPE tissue on copy number determination. A, actual amount (copy numbers) of HPV in each sample; B to G, paraffin-embedded samples; I and J, non-paraffin-embedded samples (HeLa cells); n.d., nondiluted samples; 1:10 to 1:1,000, dilution series of samples.
An attempt was made to differentiate specific and nonspecific product during the quantification process by performing quantifications at a higher temperature (2°C below the melting temperature of the specific product). However, this alteration significantly reduced the sensitivity of the assay. Therefore, we retained the measurement temperature at the extension temperature of 72°C.
Sensitivity of real-time PCR.
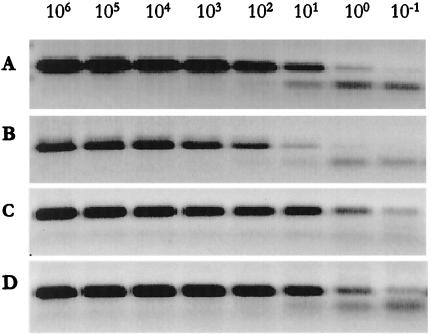
We used a dilution series of HPV16 from 10−1 to 106 copies, diluted with either DNA from paraffin-embedded tissue or cultured cells. There was slight variation in the detection limits at 100 and l0−1 copies of HPV16 between conventional PCR and real-time PCR (Fig. 2). This result was probably due to slight handling variations during gel preparation, to pipetting variations, or to the use of different cycling instruments. When we compared the results with paraffin-embedded material versus non-paraffin-embedded material obtained by real-time PCR on gels, no difference in sensitivity was seen (Fig. 2C and D). A difference in the detection limit between paraffin-embedded and non-paraffin-embedded material was evident when we evaluated the results from the I-Cycler software. The results of HPV16 diluted in paraffin-embedded DNA were continually underestimated, and at a starting amount of ≤10 copies a dramatic reduction in detection was apparent. Only 7% of the actual amount was measured when we used 1 or 10 copies of HPV16 diluted in paraffin-embedded DNA, and a starting amount of 10−1 copies gave a negative value. The melting temperature was specific for all sample dilutions (83°C), except when 10−1 copies were diluted in HeLa cell DNA, in which case the melting temperature was reduced to 82°C. We interpret this as being the result of the formation of primer-dimers, which also explains the overestimation of copy number.
FIG. 2.
Dilution series of HPV16 (106 to 10−1 copies). Comparison of sensitivity between paraffin- and non-paraffin-embedded material performed by conventional PCR and real-time PCR visualized on gel. Rows: A, conventional PCR, paraffin-embedded material (DNA from archival HPV-negative cervical cancer tissue sample); B, conventional PCR, non-paraffin-embedded material (DNA from HeLa-cells); C, real-time PCR, paraffin-embedded material (DNA from archival HPV-negative cervical cancer tissue sample); D, real-time PCR, non-paraffin-embedded material (DNA from HeLa cells).
Problem of long-term storage of FFPE tissue.
Because of the well-known problem of the long-term storage of paraffin-embedded tissue and its influence on DNA quality, we analyzed HPV positivity over time. We grouped the samples into two 6-year periods (1989 to 1994 and 1995 to 2000). We found a significant difference between these two groups (as determined by chi-square test and Fisher exact test) (Table 3).
TABLE 3.
Incidence of HPV-positive cases as determined by ISH, conventional PCR, and real-time PCR
| Method and HPV type(s) | No. of HPV-positive samples/ total no. of samples (%) in:
|
P | |
|---|---|---|---|
| 1989-1994 | 1995-2000 | ||
| ISH | |||
| HPV6/11 | 3/65 (4.6) | 15/79 (19) | 0.011 |
| HPV16/18 | 19/66 (28.8) | 45/82 (54.9) | 0.002 |
| HPV31/33/51 | 5/64 (7.8) | 19/79 (24.1) | 0.013 |
| All HPV types | 19/63 (24.7) | 47/77 (61.0) | <0.0001 |
| Conventional PCR | |||
| HPV16 | 33/71 (46.5) | 69/87 (79.3) | <0.0001 |
| HPV18 | 10/74 (13.5) | 26/87 (29.9) | 0.014 |
| HPV16/18 | 41/71 (57.7) | 76/87 (87.4) | <0.0001 |
| HPV consensus | 8/42 (19) | 45/81 (55.6) | <0.0001 |
| Real-time PCR | |||
| HPV16 | 36/74 (48.6) | 63/87 (72.4) | 0.003 |
| HPV18 | 11/74 (14.9) | 23/87 (26.4) | 0.083 |
| HPV16/18 | 47/74 (63.5) | 78/87 (89.7) | <0.0001 |
Correlation of HPV viral load with survival.
Patients positive for HPV16 had a significant better clinical outcome than HPV16-negative patients in the following clinicopathological subgroups: stage I (P = 0.011), no parametrial infiltration (P = 0.048), no vaginal infiltration (P = 0.047), age <35 (P = 0.011), and no lymph node involvement (P = 0.061).
Patients in tumor stage I with lower viral load of HPV16 (up to 1,000 copies/ng DNA) showed significantly better survival than HPV16-negative patients (P = 0.037). This tendency was also seen in the subgroup of patients without parametrial (P = 0.094), vaginal (P = 0.084), and parametrial lymph node (P = 0.098) infiltration.
Patients with a high viral load of HPV16 (>1,000 copies/ng) DNA tended to have a better prognosis than HPV-negative patients. This was statistically not significant, but in the subgroup of patients with lymphatic space invasion (P = 0.086) and lymph node infiltration (P = 0.063), this trend was maintained.
DISCUSSION
In the present study we compared three techniques (CISH, conventional PCR, and real-time PCR) for the detection of human papillomavirus DNA in 164 cases of FFPE cervical CIS and cancer specimens. We were particularly interested in the performance of real-time PCR since this is a new procedure with hitherto limited use in the detection and quantification of DNA in archival tissue.
Our findings indicate a higher sensitivity of both real-time PCR and conventional PCR for the detection of HPV16/18 compared to CISH. We found a considerable concordance between HPV16 and -18 status measured by real-time PCR and versus that determined by conventional PCR. We recognized an inhibitory effect of formalin fixation and paraffin embedding on real-time PCR, resulting in underestimation of the viral copy number.
When we compared CISH with PCR, we found that almost all of the HPV16/18-positive cases identified by ISH were also positive by both PCR and real-time PCR, but only a few of the HPV16/18 ISH-negative cases were also found to be negative by PCR and real-time PCR. These findings indicate a greater sensitivity of HPV16/18 PCR compared to CISH for HPV16/18.
From the cases found to be positive by ISH and negative by consensus PCR, some conclusions can be drawn. For example, the fact that some PCR-negative tissues were strongly positive by ISH indicates that the problem of consensus PCR negativity was not due to low sensitivity. This is most likely due to the length of the template of the consensus primer pair MY09/MY11, which may be fragmented as a consequence of formalin fixation and paraffin embedding (13), which makes it difficult to amplify sequences longer than 200 bp (24). The type-specific primer pairs for HPV16 and -18 amplify products by 96 and 115 bp, respectively, thus explaining the higher prevalence of positive cases (74.1%). These results agree with those of Baay et al. (1), whose primers in HPV16 and -18 were used for the present study. Another reason for the different detection rates could be due to a more efficient amplification with the type-specific primers as a result of diluting the template 1 in 10. This was done due to a paucity of material and because template dilution had a positive effect on the efficiency of previously performed real-time PCR.
The reason for ISH-negative cases that are determined to be positive by consensus PCR could be the specificity. The MY09-MY11 primer pair detects more than 25 different types of human papillomaviruses (20), whereas the ISH hybridization reaction has only probes for seven types of HPV. Another aspect to be considered is that the presence of the virus may not be homogeneously distributed throughout the tissue block and the viral DNA may not be present in equal amounts in the sections processed for ISH and PCR. Nevertheless, when we compared the detection rates of the different methods, consensus PCR detected far fewer HPV-positive cases (37.3%). It has been reported that the use of the MY/GP-nested PCR significantly increases the positivity rate of HPV DNA detection and should be used for samples with a low viral load of HPV (10, 20).
The detection rate with MY09/MY11 was lower compared to those of some other studies (2, 20, 48). This could be due to the fact that most experiments were performed with non-paraffin-embedded material. The lower incidence of HPV-positive cases compared to other studies could also be due to the well-known problem of long-term storage of paraffin-embedded tissue and its influence on DNA quality. Thus, we also did an analysis of positivity by time. The “newer” cases were significantly more often HPV positive than the “older” cases. This finding again reflects the problems associated with the archiving of FFPE tissue.
Other studies have reported essentially the same frequency of HPV-positive cases in archival tissue, and Baay et al. reported that the use of one general primer pair in paraffin-embedded tissue grossly underestimates the prevalence of HPV in the studied population (1, 37, 46).
Upon comparing conventional PCR and real-time PCR, we found a considerable concordance between HPV type-specific (HPV16 and -18) PCRs. The prevalence for high-risk HPV infections detected by these two methods also coincides with those reported in other studies (1, 21, 37, 46). Nevertheless, 37 discrepant results between type-specific conventional PCR and real-time PCR indicates problems with intersystem variation or possibly reporting of false-negative cases by real-time PCR due to deviations in melting temperatures or reporting of false-positive cases by real-time PCR due to unspecific signals. SYBRGreen has the disadvantage of binding double-stranded DNA of any sequence, thus making accurate product determination more difficult. A problem with discerning specific from unspecific peaks in the melting curves was also reported by Szuhai et al. (46). The use of real-time PCR methods based on specific probes (TaqMan probes) might overcome the problem of nonspecificity and thus permit more accurate quantification.
The higher false-negative rate obtained by real-time PCR for HPV16/18 (17.1%) compared to that obtained by conventional PCR for HPV16/18 (9.8%) could reflect the overdilution of several samples. This may have been avoided by repeating the real-time PCR procedure with lower sample dilutions.
Whereas real-time PCR works well with fresh samples, additional problems are encountered in studies with archival tissues. Archival tissue contributes to the complexity of the assay because of the need to dilute the samples to reduce nonspecific paraffin interference that results in a reduction in assay sensitivity. Another aspect to be considered is that PCR-based assays in general suffer from dilution artifacts due to variations in tumor cell content within the given specimen (43). In terms of real-time PCR, an improvement in quantitative assays can be attained by using microdissected tumor material.
Real-time PCR is becoming more and more available due to its possible application for time-saving high-throughput analysis. However, it is still a very expensive test. In particular, the use of this technology in FFPE specimens requires dilution of the extracted DNA by 1:10 to 1:1,000 (Fig. 2), suggesting that every specimen needs to be tested three times (at dilutions 1:10, 1:100, and 1:1000), multiplying the cost. Adding to the “teething problem” of this technology in FFPE specimens is the difficulty in interpreting certain curves because of nonspecific amplifications and the eventual need to run a gel to resolve the problem. The inability to type HPV, rather than having to run even more reactions, and the need to microdissect tumor tissue to perform real time-PCR also add to the cost.
With respect to the clinical relevance of HPV viral load, our findings indicate that HPV16-positive patients with stage I carcinoma had a significantly better prognosis than HPV-negative patients. In several other studies a similar trend has been observed. Riou et al. found that stage I cervical cancer patients with no detectable HPV DNA showed a higher risk of overall relapse and of distant metastases than comparable HPV-positive patients (39). Higgins et al. also reported that HPV-negative carcinomas were associated with elevated mortality (17). In the study of Ikenberg et al., patients with HPV-negative tumors tended to have a higher risk of death (21). Compared to other studies (44, 45) we did not find a statistically significant increase of HPV viral load with increasing severity of disease. However, in contrast to these studies we did not compare different stages of CINs but different stages of invasive carcinomas.
In performing further subgroup analysis, we found out that patients with a lower viral load of HPV16 in early stage carcinoma had a significantly better survival rate than HPV16-negative patients. We suppose that an infection with HPV16 in early stage cervical carcinoma may favor prognosis due to an altered immune status shifted from a silent to an active status. Tumor cells are well known for their ability to escape from immune surveillance by inducing an immunosuppressed state. Thus, any infection could be helpful and supportive for activating the immune system. It is even suggested that an infection with HPV benefits the prognosis of cancer patients due to an activated immune status (31).
In the subgroup of women with a higher-stage carcinoma with lymph node infiltration and lymphatic space invasion, a high viral load of HPV16 tended to favor prognosis compared to HPV16-negative patients. However, poorly differentiated tumor cells show an extremely altered metabolism, including loss of cell cycle control mechanisms, with p53 as an often mutated tumor suppressor gene. In both HPV-positive and HPV-negative tumors, there is loss of function of wild-type p53—in the former because the protein product of the viral E6 gene complexes with that of the p53 gene, leading to its inactivation, and in the latter because the protein itself is altered (23). Ikenberg et al. also found a weak trend for better survival in patients with a higher copy number but independent of the FIGO stage (21). The fact that lower viral load of HPV16 in higher-stage carcinoma is associated with a poorer outcome compared to higher viral load suggests that patients with lower viral load have more “potent” HPV E6/E7 proteins or other oncogenic factors. This might also explain the observation that high-risk HPVs of all types are able to induce malignant transformation even when present at low levels (50).
However, in many cases of CIN I, the cell is cured of the virus and reverts to a morphologically normal state. Thus, the immune system seems to play an important role in viral clearance, regression of dysplasia, and tumor development and progression in general; maybe it is even the decisive factor, if a HPV infection appears in a latent form without any consequences for the organism or if cervical cancer can develop. It is possible that some of the HPV-negative tumors could represent a form of carcinoma in which HPV has already been cleared and the immune status has returned to a dormant state. However, the absence of HPV in some tumors could also indicate that (i) they originated through different oncogenic mechanisms and that they may represent a biologically distinct subset of tumors that carry a poorer prognosis (39) or (ii) that there may be some “nastier” HPV types that were not detected, causing the poor outcome.
In summary, we observed a greater sensitivity of both real-time PCR and conventional PCR compared to CISH. However, there are still some problems concerning performance and evaluation of real-time PCR with FFPE material. It appears that cervical cancer patients can be divided into groups differing in their clinical outcome: an HPV16-negative subgroup revealing a poor prognosis and, in contrast, a lower-stage subgroup with a lower viral load and a higher-stage subgroup with a higher viral load, both showing a better survival. These findings are crucial in order to offer each patient an individualized therapy, and it highlights the importance of quantifying viral load, thus emphasizing the necessity of real-time PCR.
Acknowledgments
K.B. received a scholarship from the Medizinische Forschungsgesellschaft Salzburg, General Hospital Salzburg, Salzburg, Austria.
REFERENCES
- 1.Baay, M. F., W. G. Quint, J. Koudstaal, H. Hollema, J. M. Duk, M. P. Burger, E. Stolz, and P. Herbrink. 1996. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J. Clin. Microbiol. 34:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birner, P., B. Bachtiary, B. Dreier, M. Schindl, E. A. Joura, G. Breitenecker, and G. Oberhuber. 2001. Signal-amplified colorimetric in situ hybridization for assessment of human papillomavirus infection in cervical lesions. Mod Pathol. 14:702-709. [DOI] [PubMed] [Google Scholar]
- 3.Burd, E. M. 2003. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne, P., C. Woodman, C. Meanwell, K. Kelley, and J. Jordan. 1986. Koilocytes and cervical human papillomavirus infection. Lancet i:205-206. [DOI] [PubMed] [Google Scholar]
- 5.Chang, D. Y., C. Y. Hsieh, R. J. Chen, S. C. Lee, and S. C. Huang. 1995. Comparison of detection of human papillomavirus 16 DNA in cervical carcinoma tissues by Southern blot hybridisation and nested polymerase chain reaction. J. Med. Microbiol. 43:430-435. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L., A. H. Graf, C. Hauser-Kronberger, O. Dietze, R. R. Tubbs, and G. W. Hacker. 1999. Detection of human papillomavirus in cervical carcinoma: comparison of peroxidase, Nanogold, and catalyzed reporter deposition (CARD)-Nanogold in situ hybridization. Mod Pathol. 12:689-696. [PubMed] [Google Scholar]
- 7.Clementi, M. 2000. Quantitative molecular analysis of virus expression and replication. J. Clin. Microbiol. 38:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, K., C. S. Herrington, J. E. Stickland, M. F. Evans, and J. O. McGee. 1991. Episomal and integrated human papillomavirus in cervical neoplasia shown by non-isotopic in situ hybridisation. J. Clin. Pathol. 44:990-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delvenne, P., M. A. Fontaine, C. Delvenne, A. Nikkels, and J. Boniver. 1994. Detection of human papillomaviruses in paraffin-embedded biopsies of cervical intraepithelial lesions: analysis by immunohistochemistry, in situ hybridization, and the polymerase chain reaction. Mod Pathol. 7:113-119. [PubMed] [Google Scholar]
- 10.Evander, M., K. Edlund, E. Boden, A. Gustafsson, M. Jonsson, R. Karlsson, E. Rylander, and G. Wadell. 1992. Comparison of a one-step and a two-step polymerase chain reaction with degenerate general primers in a population-based study of human papillomavirus infection in young Swedish women. J. Clin. Microbiol. 30:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco, E., J. Bergeron, L. Villa, M. Arella, L. Richardson, J. Arseneau, and G. Stanimir. 1996. Human papillomavirus DNA in invasive cervical carcinomas and its association with patient survival: a nested case-control study. Cancer Epidemiol. Biomarkers Prev. 5:271-275. [PubMed] [Google Scholar]
- 12.Franco, E. L. 1992. Prognostic value of human papillomavirus in the survival of cervical cancer patients: an overview of the evidence. Cancer Epidemiol. Biomarkers Prev. 1:499-504. [PubMed] [Google Scholar]
- 13.Frank, T. S., S. M. Svoboda-Newman, and E. D. Hsi. 1996. Comparison of methods for extracting DNA from formalin-fixed paraffin sections for nonisotopic PCR. Diagn. Mol. Pathol. 5:220-224. [DOI] [PubMed] [Google Scholar]
- 14.Girardi, F., P. Fuchs, and J. Haas. 1992. Prognostic importance of human papillomavirus type 16 DNA in cervical cancer. Cancer 69:2502-2504. [DOI] [PubMed] [Google Scholar]
- 15.Graf, A. H., A. L. Cheung, C. Hauser-Kornberger, N. Dandachi, R. R. Tubbs, O. Dietze, and G. W. Hacker. 2000. Clinical relevance of HPV16/18 testing methods in cervical squamous cell carcinoma. Appl. Immunohistochem. Mol. Morphol. 8:300-309. [PubMed] [Google Scholar]
- 16.Guerrero, E., R. W. Daniel, F. X. Bosch, X. Castellsague, N. Munoz, M. Gili, P. Viladiu, C. Navarro, M. L. Zubiri, N. Ascunce, et al. 1992. Comparison of ViraPap, Southern hybridization, and polymerase chain reaction methods for human papillomavirus identification in an epidemiological investigation of cervical cancer. J. Clin. Microbiol. 30:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins, G. D., M. Davy, D. Roder, D. M. Uzelin, G. E. Phillips, and C. J. Burrell. 1991. Increased age and mortality associated with cervical carcinomas negative for human papillomavirus RNA. Lancet 338:910-913. [DOI] [PubMed] [Google Scholar]
- 18.Hildesheim, A., M. H. Schiffman, P. E. Gravitt, A. G. Glass, C. E. Greer, T. Zhang, D. R. Scott, B. B. Rush, P. Lawler, M. E. Sherman, et al. 1994. Persistence of type-specific human papillomavirus infection among cytologically normal women. J. Infect. Dis. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 19.Howley, P. M. 1991. Role of the human papillomaviruses in human cancer. Cancer Res. 51:5019s-5022s. [PubMed] [Google Scholar]
- 20.Husnjak, K., M. Grce, L. Magdic, and K. Pavelic. 2000. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J. Virol. Methods 88:125-134. [DOI] [PubMed] [Google Scholar]
- 21.Ikenberg, H., W. Sauerbrei, U. Schottmuller, C. Spitz, and A. Pfleiderer. 1994. Human papillomavirus DNA in cervical carcinoma: correlation with clinical data and influence on prognosis. Int. J. Cancer 59:322-326. [DOI] [PubMed] [Google Scholar]
- 22.Josefsson, A. M., P. K. Magnusson, N. Ylitalo, P. Sorensen, P. Qwarforth-Tubbin, P. K. Andersen, M. Melbye, H. O. Adami, and U. B. Gyllensten. 2000. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 355:2189-2193. [DOI] [PubMed] [Google Scholar]
- 23.Kaelbling, M., R. D. Burk, N. B. Atkin, A. B. Johnson, and H. P. Klinger. 1992. Loss of heterozygosity on chromosome 17p and mutant p53 in HPV-negative cervical carcinomas. Lancet 340:140-142. [DOI] [PubMed] [Google Scholar]
- 24.Karlsen, F., M. Kalantari, M. Chitemerere, B. Johansson, and B. Hagmar. 1994. Modifications of human and viral deoxyribonucleic acid by formaldehyde fixation. Lab. Investig. 71:604-611. [PubMed] [Google Scholar]
- 25.Kiene, P., K. Milde-Langosch, M. Runkel, K. Schulz, and T. Loning. 1992. A simple and rapid technique to process formalin-fixed, paraffin-embedded tissues for the detection of viruses by the polymerase chain reaction. Virchows Arch. A Pathol. Anat. Histopathol. 420:269-273. [DOI] [PubMed] [Google Scholar]
- 26.Kristensen, G. B., F. Karlsen, A. Jenkins, J. Kaern, V. M. Abeler, and C. G. Trope. 1996. Human papilloma virus has no prognostic significance in cervical carcinoma. Eur. J. Cancer 32A:1349-1353. [DOI] [PubMed] [Google Scholar]
- 27.Kuypers, J. M., C. W. Critchlow, P. E. Gravitt, D. A. Vernon, J. B. Sayer, M. M. Manos, and N. B. Kiviat. 1993. Comparison of dot filter hybridization, Southern transfer hybridization, and polymerase chain reaction amplification for diagnosis of anal human papillomavirus infection. J. Clin. Microbiol. 31:1003-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehmann, U., and H. Kreipe. 2001. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 25:409-418. [DOI] [PubMed] [Google Scholar]
- 29.Lin, M. H., T. C. Chen, T. T. Kuo, C. C. Tseng, and C. P. Tseng. 2000. Real-time PCR for quantitative detection of Toxoplasma gondii. J. Clin. Microbiol. 38:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorincz, A. T. 1996. Hybrid Capture method for detection of human papillomavirus DNA in clinical specimens: a tool for clinical management of equivocal Pap smears and for population screening. J. Obstet. Gynaecol. Res. 22:629-636. [DOI] [PubMed] [Google Scholar]
- 31.Mellin, H., L. Dahlgren, E. Munck-Wikland, J. Lindholm, H. Rabbani, M. Kalantari, and T. Dalianis. 2002. Human papillomavirus type 16 is episomal and a high viral load may be correlated to better prognosis in tonsillar cancer. Int. J. Cancer 102:152-158. [DOI] [PubMed] [Google Scholar]
- 32.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 33.Mygind, T., S. Birkelund, E. Falk, and G. Christiansen. 2001. Evaluation of real-time quantitative PCR for identification and quantification of Chlamydia pneumoniae by comparison with immunohistochemistry. J. Microbiol. Methods 46:241-251. [DOI] [PubMed] [Google Scholar]
- 34.Pilch, H., S. Gunzel, U. Schaffer, B. Tanner, P. Brockerhoff, M. Maeurer, M. Hockel, G. Hommel, and P. G. Knapstein. 2001. Human papillomavirus (HPV) DNA in primary cervical cancer and in cancer free pelvic lymph nodes: correlation with clinico-pathological parameters and prognostic significance. Zentbl. Gynakol. 123:91-101. [DOI] [PubMed] [Google Scholar]
- 35.Pilch, H., S. Gunzel, U. Schaffer, B. Tanner, P. Brockerhoff, M. Maeurer, M. Hockel, G. Hommel, and P. G. Knapstein. 2001. The presence of HPV DNA in cervical cancer: correlation with clinico-pathologic parameters and prognostic significance: 10 years experience at the Department of Obstetrics and Gynecology of the Mainz University. Int. J. Gynecol. Cancer. 11:39-48. [DOI] [PubMed] [Google Scholar]
- 36.Qian, X., R. A. Bauer, H. S. Xu, and R. V. Lloyd. 2001. In situ hybridization detection of calcitonin mRNA in routinely fixed, paraffin-embedded tissue sections: a comparison of different types of probes combined with tyramide signal amplification. Appl. Immunohistochem. Mol. Morphol. 9:61-69. [PubMed] [Google Scholar]
- 37.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 35:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajeevan, M. S., S. D. Vernon, N. Taysavang, and E. R. Unger. 2001. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J. Mol. Diagn. 3:26-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riou, G., M. Favre, D. Jeannel, J. Bourhis, V. Le Doussal, and G. Orth. 1990. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA. Lancet 335:1171-1174. [DOI] [PubMed] [Google Scholar]
- 40.Schadendorf, D., K. H. Tiedemann, N. Haas, and B. M. Czarnetzki. 1991. Detection of human papillomaviruses in paraffin-embedded condylomata acuminata: comparison of immunohistochemistry, in situ hybridization, and polymerase chain reaction. J. Investig. Dermatol. 97:549-554. [DOI] [PubMed] [Google Scholar]
- 41.Sebbelov, A. M., C. Svendsen, H. Jensen, S. K. Kjaer, and B. Norrild. 1994. Prevalence of HPV in premalignant and malignant cervical lesions in Greenland and Denmark: PCR and in situ hybridization analysis on archival material. Res. Virol. 145:83-92. [DOI] [PubMed] [Google Scholar]
- 42.Shibata, D. K., N. Arnheim, and W. J. Martin. 1988. Detection of human papilloma virus in paraffin-embedded tissue using the polymerase chain reaction. J. Exp. Med. 167:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slamon, D. J., W. Godolphin, L. A. Jones, J. A. Holt, S. G. Wong, D. E. Keith, W. J. Levin, S. G. Stuart, J. Udove, A. Ullrich, et al. 1989. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244:707-712. [DOI] [PubMed] [Google Scholar]
- 44.Sun, C. A., J. F. Liu, D. M. Wu, S. Nieh, C. P. Yu, and T. Y. Chu. 2002. Viral load of high-risk human papillomavirus in cervical squamous intraepithelial lesions. Int. J. Gynaecol. Obstet. 76:41-47. [DOI] [PubMed] [Google Scholar]
- 45.Swan, D. C., R. A. Tucker, G. Tortolero-Luna, M. F. Mitchell, L. Wideroff, E. R. Unger, R. A. Nisenbaum, W. C. Reeves, and J. P. Icenogle. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J. Clin. Microbiol. 37:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szuhai, K., E. Sandhaus, S. M. Kolkman-Uljee, M. Lemaitre, J. C. Truffert, R. W. Dirks, H. J. Tanke, G. J. Fleuren, E. Schuuring, and A. K. Raap. 2001. A novel strategy for human papillomavirus detection and genotyping with SYBRGreen and molecular beacon polymerase chain reaction. Am. J. Pathol. 159:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trattner, M., A. H. Graf, S. Lax, R. Forstner, N. Dandachi, J. Haas, H. Pickel, O. Reich, A. Staudach, and R. Winter. 2001. Prognostic factors in surgically treated stage ib-iib cervical carcinomas with special emphasis on the importance of tumor volume. Gynecol. Oncol. 82:11-16. [DOI] [PubMed] [Google Scholar]
- 48.Unger, E. R., S. D. Vernon, D. R. Lee, D. L. Miller, and W. C. Reeves. 1998. Detection of human papillomavirus in archival tissues: comparison of in situ hybridization and polymerase chain reaction. J. Histochem. Cytochem. 46:535-540. [DOI] [PubMed] [Google Scholar]
- 49.Wiedorn, K. H., H. Kuhl, J. Galle, J. Caselitz, and E. Vollmer. 1999. Comparison of in-situ hybridization, direct and indirect in-situ PCR as well as tyramide signal amplification for the detection of HPV. Histochem. Cell Biol. 111:89-95. [DOI] [PubMed] [Google Scholar]
- 50.zur Hausen, H. 1996. Papillomavirus infections: a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]
- 51.zur Hausen, H. 1987. Papillomaviruses in human cancer. Cancer 59:1692-1696. [DOI] [PubMed] [Google Scholar]