Abstract
In order to determine if variations in rRNA sequence could be used for discrimination of the members of the Bacillus cereus group, we analyzed 183 16S rRNA and 74 23S rRNA sequences for all species in the B. cereus group. We also analyzed 30 gyrB sequences for B. cereus group strains with published 16S rRNA sequences. Our findings indicated that the three most common species of the B. cereus group, B. cereus, Bacillus thuringiensis, and Bacillus mycoides, were each heterogeneous in all three gene sequences, while all analyzed strains of Bacillus anthracis were found to be homogeneous. Based on analysis of 16S and 23S rRNA sequence variations, the microorganisms within the B. cereus group were divided into seven subgroups, Anthracis, Cereus A and B, Thuringiensis A and B, and Mycoides A and B, and these seven subgroups were further organized into two distinct clusters. This classification of the B. cereus group conflicts with current taxonomic groupings, which are based on phenotypic traits. The presence of B. cereus strains in six of the seven subgroups and the presence of B. thuringiensis strains in three of the subgroups do not support the proposed unification of B. cereus and B. thuringiensis into one species. Analysis of the available phenotypic data for the strains included in this study revealed phenotypic traits that may be characteristic of several of the subgroups. Finally, our results demonstrated that rRNA and gyrB sequences may be used for discriminating B. anthracis from other microorganisms in the B. cereus group.
Analysis of 16S rRNA sequences is a simple, commonly used method for the identification of microorganisms (1, 47, 53). However, early studies performed with a limited number of isolates from the Bacillus cereus group revealed that the 16S rRNA sequences of species in this group had as high as a 99 to 100% similarity and, thus, suggested that rRNA sequences might not be useful for discrimination of members of that group (6). However, that study examined only five isolates from the B. cereus group. The B. cereus group contains seven closely related species: Bacillus anthracis, B. cereus, Bacillus thuringiensis, Bacillus mycoides (16, 45, 51), Bacillus pseudomycoides (38), Bacillus weihenstephanensis (33), and Bacillus medusa (15). To date, identification and discrimination of these species has been based on analysis of morphological, biochemical, and immunological characteristics.
Although conserved in sequence overall, the 16S rRNAs actually exhibit great variation in some regions. These differences in 16S rRNA sequence provide the basis for the design of nucleic acid probes of various specificities, ranging from probes targeting all living organisms to group-specific and species-specific probes. Another advantage of using the rRNAs as a target is the fact that these molecules are naturally amplified within the cell. In general, rRNA represents about 80% of total nucleic acids in microbial cells and, thus, is present in many hundreds of thousands of copies per cell. This natural amplification allows for direct detection of rRNA sequences without the need for intermediate amplification via PCR (1).
16S and 23S rRNA are currently considered the most useful molecules for the determination of prokaryotic phylogeny. Analysis of these rRNA sequences has resulted in a tremendous expansion in our knowledge of prokaryotic diversity and has demonstrated the limitations of the existing prokaryotic taxonomy, which is based primarily on the analysis of phenotypic traits (35). Attempts have been made recently to address conflicts between molecular and phenotypic data, such as the work on the phylogenetically heterogeneous genus Pseudomonas (29). Here we have conducted a similar analysis for the B. cereus group. We have investigated the molecular phylogeny of the B. cereus group by extensively analyzing a set of B. cereus group sequences in order to determine if the rRNA sequences contained enough variation to discriminate B. anthracis from other members of the B. cereus group.
Previous work has demonstrated that gyrB gene sequences may also be useful for discrimination of B. cereus group organisms (57). Therefore, in addition to the rRNA analyses, we have also analyzed gyrB sequences for members of the B. cereus group and compared rRNA-based and gyrB-based phylogenies.
MATERIALS AND METHODS
Bacterial strains.
Twelve strains belonging to the B. cereus group were used for sequencing: B. anthracis strain Ames ANR, B. anthracis strain Delta Ames-1, B. anthracis strain Sterne, B. anthracis strain 1, B. anthracis strain 2, B. thuringiensis strain B8, B. cereus strain NCTC 9620, B. cereus strain T, B. thuringiensis strain 4Q281, B. medusa strain ATCC 25621, B. mycoides strain ATCC 6462m, and B. mycoides strain ATCC 10206 (obtained as a generous gift from John Ezzell, U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Frederick, Md.) (see Table 2). Two of the B. mycoides strains were isolated as an occasional admixture from a culture previously identified as B. mycoides strain ATCC 6462. These strains revealed different colony morphologies and were assigned different strain numbers: B. mycoides ATCC 6462m and B. mycoides ATCC 10206. B. thuringiensis strains 4R1, 4D1, 4F1, 4T1, 4W1, 4J4, 4A1, 4A7, 4Q1, 4Q2, and 4M1 were received as a generous gift from Dan Zeigler of the Bacillus Genetic Stock Center. B. cereus HER 1414 was acquired from the National Collection of Type Cultures (NCTC).
TABLE 2.
Classification of bacteria in the B. cereus group according to 16S rRNA sequences
| Subgroup name | Subgroup-specific signature(s) (position)a | Start and end of sequence | Organismr | GenBank accession no. | Position(s) of strain-specific variations |
|---|---|---|---|---|---|
| Anthracis | Consensusa | 11-1554 | B. anthracis Sterneb | AF176321 | —s |
| 1-1451 | B. anthracis Sternec | X55059 | — | ||
| 18-1499 | B. anthracis Sterne | AF290552 | A/W(1147) | ||
| 11-1554 | B. anthracis Ames ANRb | AF155950 | — | ||
| 1-1554 | B. anthracis Ames (TIGR)d | AE017024-26, AE017039 | A/W(1146) | ||
| 11-1554 | B. anthracis Delta Ames-1b | AF155951 | — | ||
| 33-1517 | B. anthracis (wastewater isolate)e | AY043083 | A/T(1146) | ||
| 18-1499 | B. anthracis Vollum | AF290553 | A/W(1147) | ||
| 12-1424 | B. anthracis W21 | AF390088 | C/A(19), T/A(967), A/T(969), TT/GA(1095,1096), T/G(1100), C/A(1113), ins A(1123-1124), A/T(1146), C/T(1200), A/T(1207), C/A(1209), T/C(1220) | ||
| 61-528, 815-1501 | B. anthracis 1b | J. Jackmanf | — | ||
| 61-528, 815-1501 | B. anthracis 2b | J. Jackmanf | — | ||
| Cereus A | Consensusa | 1-1451 | B. cereus NCTC 11143c | X55063 | — |
| 49-1522 | B. cereus WSBC 10037g | Z84576 | A/G(178), G/T(1518) | ||
| 49-1522 | B. cereus WSBC 10030g | Z84575 | C/T(353), T/C(600), T/C(864), A/C(1146), G/T(1518) | ||
| 49-1489 | B. cereus S-5g | AF390086 | C/T(353), A/C(1146) | ||
| 28-1517 | B. cereus AL1g | AY129651 | A/T(1146), del(1459) | ||
| 28-1513 | B. cereus (bovine serum isolate)g | AF206326 | A/G(181), C/T(285), C/T(467), C/T(480), G/A(482), T/C(600), C/T(995), A/C(1146), T/C(1244), T/C(1345), delT(1459) | ||
| 105-1277 | B. cereus ATCC 10702g | AF363440 | — | ||
| 105-1277 | B. cereus DL5g | AF363441 | C/T(906), C/T(1035), G/A(1046), T/G(1141), T/A(1256), A/G(1263) | ||
| 105-1277 | B. cereus DL115g | AF363442 | A/C(462), ins A(471-472), A/C(623), T/C(746), C/A(771), T/C(872), C/T(906), C/T(1035), T/A(1256) | ||
| 1-1554 | B. cereus ATCC 10987d | AE017264-66AE017280 | — | ||
| B. cereus HFR1414h | ? | ||||
| 31-1462 | Bacillus sp. strain JJ-1g | Y15466 | — | ||
| 28-1534 | Bacillus sp. strain BSID723g | AF027659 | C/A(1232) | ||
| 18-1499 | Bacillus sp. strain AH526g | AF290562 | — | ||
| 52-1517 | Bacillus sp. strain YSS/2001-3g | AF417847 | A/T(1146), A/T(1155), T/G(1402), G/C(1426), ins G(1454-1455) | ||
| 228-1437 | Bacillus sp. strain FO-011g | AF234842 | ins T (276-277), ins C (280-281), C/T(293), C/A(303), A/T(323), C/T(350), G/T(416), C/T(498), C/A(520), A/T(568), G/A(1190), A/T(1205), ins T(1219-1220) | ||
| 11-1554 | B. thuringiensis B8b | AF155955 | — | ||
| Cereus B | C/A(1015)q | 11-1554 | B. cereus NCTC 9620b | AF155952 | — |
| 11-1554 | B. cereus Tb | AF176322 | — | ||
| 28-1513 | B. cereus IAM 12605i | D16266 | — | ||
| 1-1451 | B. cereus NCDO 1771ci | X55060 | — | ||
| 17-1499 | B. cereus ATCC 1778i | AF290546 | — | ||
| 17-1499 | B. cereus ATCC 4579i | AF290547 | — | ||
| 49-1522 | B. cereus ATCC 27877 | Z84581 | A/T(828), G/T(1518) | ||
| 28-1183 | B. cereus (ocular isolate) | AF076031 | C/T(498), del C(520), del (523), T/A(829), C/G(1167) | ||
| 17-1499 | B. cereus ATCG 31293 | AF290548 | — | ||
| 1-1554 | B. cereus BGSC 6A5d | AY224379-88 | — | ||
| 8-1523 | B. cereus SH 01e | AF522353 | A/T(171) | ||
| 11-1249 | B. cereus Tim-r01 | AB050630 | C/T(182) | ||
| 105-1277 | B. cereus DL137 | AF363444 | A/T(170), G/C(407), A/C(623) | ||
| 105-1277 | B. cereus DL122 | AF363443 | A/T(170), G/C(345), A/T(496), A/C(623) | ||
| 20-1534 | Bacillus sp. strain P16 | AY048782 | G/A(28), T/C(30) | ||
| 11-1515 | Bacillus sp. strain 82344 | AF227848 | — | ||
| 45-1513 | Bacillus sp. strain F26 | AF385082 | T/C(49), G/C(530) | ||
| 30-1484 | Glacial ice bacterium SB-12K-9-4 | AF479367 | — | ||
| 30-1481 | Glacial ice bacterium G500K-2 | AF479333 | — | ||
| 30-1475 | Glacial ice bacterium G50-TS3 | AF479356 | — | ||
| 47-1470 | Bacillaceae bacterium PH27B | AF513473 | ins GT(1379-1380) | ||
| 49-1517 | Unident. HTA484 (Mariana Trench isol.) | AB002640 | G/C(72), GC/CG(297,298), del (946), ins C(1111-1112), del (1279) | ||
| 49-1522 | B. thuringiensis WS2614 | Z84584 | A/C(128), G/T(1518) | ||
| 49-1522 | B. thuringiensis WS2617 | Z84585 | G/A(1153), G/T(1518) | ||
| 49-1522 | B. thuringiensis WS2618 | Z84586 | A/G(725), G/T(1518) | ||
| 49-1522 | B. thuringiensis WS2626 | Z84588 | G/T(1518) | ||
| Thuringiensis A | C/A(1015), C/T(192) | 49-1522 | B. thuringiensis WS2623 | Y18473 | G/T(109), A/G(679), T/C(1228), A/G(1503) |
| 49-1522 | B. thuringiensis WS2625 | Z84587 | C/T(565), G/T(1183), G/T(1518) | ||
| 18-1479 | B. thuringiensis ATCC 33679 | AF290549 | C/Y(192) | ||
| B. thuringiensis 4R1,j 4D1,j 4F1,j 4S2,j 4T1, 4W1,j and 4J4j | ? | ||||
| 22-1502 | Bacillus sp. strain FPI/2002 | AY124766 | — | ||
| 8-1514 | Bacillus sp. strain KPU-0013 | AB067810 | C/G(131), C/T(182), G/A(202), del (298), C/T(444), T/G(781), A/C(796), G/A(952), TC/CT(1036,1037), G/T(1313) | ||
| 55-1510 | Unident. sp6 (bovine rumen isolate) | AB003391 | C/T(63), del(108), C/T(182), C/G(227), GC/CG(297,298), del(565), del(1038) | ||
| 20-1530 | Bacillus sp. strain CMB03 | AF406633 | G/A(28), T/C(30), ins C(730-731), A/T(1146), G/A(1254), T/A(1256), A/G(1263), G/A(1266), A/G(1269), G/A(1271), GG/TA(1282,1283), T/C(1287), T/C(1292), A/T(1299), C/T(1301), T/C(1317), T/C(1319), G/T(1322), C/A(1333), A/G(1336), A/G(1338), G/A(1451), T/A(1463), T/G(1477) | ||
| 18-1499 | B. cereus ATCC 43881 | AF290550 | C/Y (182) | ||
| Thuringiensis B | C/A(1015), C/T(192), A/G(77), T/C(90), T/A(92) | ||||
| 11-1554 | B. thuringiensis 4Q281b | AF155954 | — | ||
| 28-1513 | B. thuringiensis IAM 12077k | D16281 | — | ||
| 1-1451 | B. thuringiensis NCIMB 9134ck | X55062 | — | ||
| 18-1499 | B. thuringiensis ATCC 10792k | AF290545 | — | ||
| 37-1491 | B. thuringiensis 82347 | AF157112 | C/Y(182), del C(303), del C(347), ins T(395-396), G/A(1029), C/T(1111), del G(1148), del G(1240), del C(1246), del G(1262), GG/AS(1321,1322), del G(1492) | ||
| 49-1522 | B. thuringiensis (Pieris brassicae isolate) | AF160221 | A/C(161), A/T(183), no C/T(192), G/T(1518) | ||
| 50-1513 | B. thuringiensis Bactisubtil | AF172711 | G/A(733), G/A(778), C/A(857) | ||
| 11-1052 | B. thuringiensis HMB12389 | AF501348 | G/C(39) | ||
| B. thuringiensis 9308,l 20,l Lb5,l 1230,l and L3l | ? | ||||
| B. thuringiensis 4A1,j 4A7,j 4Q1,j 4Q2,j and 4M1j | ? | ||||
| B. cereus Nagoya 126m and 127m | ? | ||||
| 11-1554 | B. medusa ATCC 25621bn | AF155958 | No C/T (192) | ||
| 7-1440 | B. medusa NCIMB10437cn | X60628 | del C(1038), A/G (1383) | ||
| 18-1499 | Bacillus sp. strain AH540 | AF290557 | A/R(77), T/Y(90), T/W(92), T/A(1462) | ||
| 8-1497 | Bacillus sp. strain Fa7 | AY131217 | C/A(19), del (28), no C/T(192) | ||
| 20-1499 | Bacillus sp. strain SVM | AF503203 | C/Y(192) | ||
| 8-1554 | Bacillus sp. strain Kaza-37 | AF441732 | del (25), G/C(634), A/G(828), G/C(949), ins (955-956), del (963), del (982), G/A(983), ins T(990-991), ins T(1000-1001), ins AG(1048-1049), G/A(1049), G/A(1089) | ||
| 8-1554 | Bacillus sp. strain Kaza-31 | AF441728 | no C/T(192), GGG/CCC(309-311), ins C(496-497), ins T(516-517), ins G(523-524), G/A(526), G/C(712), G/C(949), ins A(973-974), ins C(979-980), ins C(988-989), ins T(1009-1010), del G(1029), ins A(1055-1056), G/A(1098), delA(1269) | ||
| 28-1403 | Bacillus sp. strain A23 | AF397398 | No T/A (92) | ||
| 26-1401 | Bacillus sp. strain A24 | AF397399 | — | ||
| 18-1499 | Bacillus sp. strain AH533 | AF290556 | C/Y(182) | ||
| 7-1551 | Bacillus sp. strain Termite isolate bac | X81132 | C/T(182), TT/GG(186,187), G/C(769), TA/CT(822,823), del A(875), G/C(897), TGG/CTA(1281-1283), del C(1301), ins GG(1428-1429), T/A(1461), del T(1463), CG/GT(1473,1474), del CT(1475,1476) | ||
| 28-1513 | Bromate-reducing bacterium B6e | AF442522 | G/K(61), G/S(66), G/R(93), G/S(100), G/R(141), C/T(182), G/C(255), T/K(264), G/S(297), G/A(307), G/R(334), C/A(1130), T/A(1462) | ||
| 30-1489 | Glacial ice bacterium SB100-8-1 | AF479369 | C/T(182), T/A(1462) | ||
| 33-1485 | Unident. bacterium V | AB004761 | No T/A(92), G/C(667), T/A(1462) | ||
| 18-1499 | B. cereus ATCC 53522 | AF290551 | CG/AM(1423,1424) | ||
| 18-1499 | B. cereus AH527 | AF290555 | C/Y(182) | ||
| 29-1532 | B. cereus ATCC 14893 | AJ310098 | CG/AY(43,44) | ||
| 28-1530 | B. cereus biovar toyoi CNCM I-1012/NCIB 40112 | AJ310100 | — | ||
| 62-1511 | B. cereus Biosubtil-Dalat | AJ277907 | del T(76), G/A(388), G/A(402), del G(407), del G(425), G/T(436), G/A(769), G/C(772), G/A(787), ins C(788-789), C/T(1073) | ||
| 62-1511 | B. cereus Bactisubtil | AJ277908 | del A(70), del T(76), G/A(424), G/C(431), G/T(436), del G(680), G/A(769), G/C(772), C/T(1037), T/C(1039), C/T(1041), del AGCA(1045-1048), del A(1048), T/A(1052), AC/GA(1054,1055), del T(1072), C/T(1073), ins A(1103-1104), C/G(1113), ins A(1121-1122) | ||
| B. thuringiensis BT3,ko BT13,o BT15,o BT16,o BTT6,o and BTT8o | ? | ||||
| Mycoides A | C/A(1015), C/T(192), G/A(133), C/T(182), G/A(197), A/G(286), C/T(1029), G/A(1030), T/A(1464) | 1-1451 | B. mycoides DSM2048Tp | X55061 | — |
| 49-1551 | B. mycoides MWS5303-1-4 | Z84591 | T/C(1454), G/T(1518) | ||
| 49-1523 | B. mycoides DRC1 | AF144645 | C/G(63), G/A(1279), TAG/GTA(1319-1321), C/G(1398), A/C(1437), G/T(1441), G/T(1471), T/A(1477), G/A(1492) | ||
| 49-1522 | B. mycoides MWS5303-2-51 | Z84583 | A/G(180), G/T(1518) | ||
| 32-1544 | B. mycoides ATCC 6462p | AB021192 | — | ||
| 228-1415 | B. mycoides FO-080 | AF234860 | G/T(289), G/C(297), del G(311), C/G(387), G/T(392), C/T(520), ins A(532-533), ins G(827-828), A/T(1112), C/T(1117), G/A(1134), G/C(1148), del G(1162), del G(1202) | ||
| 32-574 | B. mycoides B10 | BMY491827 | ins G(111) | ||
| 16-340 | B. mycoides SFLB6 | BMY344516 | A/T(50), A/G(69) | ||
| 14-1544 | B. weihenstephanensis DSM11821 | AB021199 | — | ||
| 49-1522 | B. cereus WSBC10201 | Z84577 | A/G(203), no A/G(286), A/G(1513), G/T(1518) | ||
| 49-1522 | B. cereus WSBC10204 | Z84578 | A/G(128), G/T(1518) | ||
| 49-1554 | B. cereus WSBC10206 | Z84579 | G/C(225), AG/GT(1517,1518) | ||
| 49-1522 | B. cereus WSBC10210 | Z84580 | A/G(60), T/C(375), A/G(1298), G/T(1518) | ||
| 18-1499 | B. cereus AH521 | AF290554 | — | ||
| 18-1499 | Bacillus sp. strain AH628 | AF290558 | — | ||
| 18-1499 | Bacillus sp. strain AH648 | AF290559 | — | ||
| 18-1499 | Bacillus sp. strain AH665 | AF290560 | — | ||
| 18-1499 | Bacillus sp. strain AH678 | AF290561 | — | ||
| 28-1517 | Bacillus sp. strain Fa25 | AY131220 | — | ||
| Mycoides B | A/C(189), T/G(200), G/C(208), T/C(1036), A/G(1045) | 11-1554 | B. mycoides ATCC 6462mb | AF155956 | — |
| 11-1554 | B. mycoides ATCC 10206b | AF155957 | — | ||
| 117-423 | B. mycoides jshs5 | AY039819 | T/G(132), T/A(185), ins C(185-186), ins G(202-203), T/G(206), C/G(411) | ||
| 7-1520 | B. pseudomycoides sp. nov. | AF013121 | A/T(55), C/A(341), T/C(495), C/T(516), G/C(566), A/T(929), C/A(958), A/C(1017), T/C(1034), T/G(1040), G/C(1104), C/A(1110), A/C(1121), A/T(1128), C/G(1138), C/A(1232), C/A(1276), T/A(1281), T/A(1390), G/A(1441), G/A(1485), T/A(1508) | ||
| 34-1373 | B. cereus Ki21 | AJ288157 | A/T(95), no T/G(200), del G(202), A/G(329), T/G(752), G/C(778), A/G(793), no T/C(1036), T/A(1350), T/A(1357) |
For more details, see Fig. 1.
Sequenced in this work.
Sequences need to be reexamined; see also Results.
From sequences of whole genome; data represent average sequence of all available 16S rRNA genes.
Sequence submitted in 3′ to 5′ form.
J. Jackman, unpublished.
Final discrimination from Anthracis subgroup will be done after sequencing of 23S rRNA gene (see Results).
Not sequenced; identified through hybridization analysis (see Results).
These four strains are identical and correspond to B. cereus DSM 31; see 23S rRNA sequence of this strain in Table 3.
Not sequenced; assignment to subgroups Thuringiensis A or B was based on hybridization with subgroup-specific probes (S. Bavykin and J. Jackman, unpublished).
These four strains are identical and correspond to B. thuringiensis DSM2046; see the 23S rRNA sequence of this strain in Table 3.
Not sequenced; subgroup-specific signatures A/G(77), T/C(90), and T/A(92) were identified by hybridization with subgroup-specific probes (18).
Not sequenced; subgroup-specific signatures A/G(77), T/C(90), and T/A(92) were identified by hybridization with subgroup-specific probes (55).
According to Bergey's Manual (45), these two strains of B. medusa should be identical.
Not sequenced; subgroup-specific signatures A/G(77), T/C(90), and T/A(92) were identified by hybridization with subgroup-specific probes (13).
According to Bergey's Manual (45), these two strains of B. mycoides should be identical.
Subgroup-specific mutations, which are highlighted in bold, were identical for two or more subgroups and are shown on separate lines to demonstrate connections between different subgroups.
Selected abbreviations of collections and institutions appearing in titles of listed strains: ATCC, American Type Culture Collection, Rockville, Md; NCTC, National Collection of Type Cultures and Pathogenic Fungi, London, United Kingdom; DSM or DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; HER, Center of Reference bacteria and viruses, Laval University, Department of Microbiology, Quebec, Canada; IAM, Institute of Applied Microbiology, University of Tokyo, Tokyo, Japan; NSDO or NSFB, National Collection of Food Bacteria, c/o NCIMB Ltd., Aberdeen, Scotland, United Kingdom; NCIMB or NCIB, National Collections of Industrial and Marine Bacteria Ltd., Aberdeen, Scotland, United Kingdom; CNCM, National Collection of Microorganisms and Cell Cultures, Pasteur Institute, Paris, France; TIGR, The Institute for Genomic Research, Rockville, Md. W = A or T; Y = C or T; S = C or G; M = A or C. Del, deletion; ins, insertion.
—, no strain-specific variations in this strain.
Sequencing of 16S and 23S rRNA genes.
Total DNA was isolated from frozen cell pellets using the guanidine extraction method as described previously (11). 16S rDNA was amplified from total genomic DNA for 12 strains. The 23S rDNA was amplified for 10 of the 12 strains (B. anthracis strains 1 and 2 were excluded). For each amplification reaction mixture, 0.1 μg of bacterial DNA was subjected to PCR in a total volume of 100 μl, with 2.5 U of Taq polymerase (Perkin-Elmer, Boston, Mass.), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.01% (wt/vol) gelatin, a 200 μM concentration of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), and a 6 μM concentration of each of two primers. The primers used for 16S rDNA and 23S rDNA amplification are listed in Table 1. The thermal profile included denaturation at 94°C for 2 min, primer annealing at 45°C for 2 min, extension at 72°C for 2 min, and then 35 cycles of denaturation at 94°C for 15 s, primer annealing at 45°C for 15 s, and extension at 72°C for 4 min. DNA was purified using a QIAquick PCR purification kit (QIAGEN Inc., Valencia, Calif.), and purified PCR products were directly sequenced by the cycle sequencing method using AmpliTaq DNA polymerase FS (Perkin-Elmer), fluorescently labeled dye terminators, and 373A fluorescent sequencer (ABI, Perkin-Elmer). Sequencing primers are shown in Table 1. Both strands of studied DNA fragments were sequenced twice for each strand.
TABLE 1.
Primers used for PCR and for sequencing of 16S and 23S rRNA genes of B. cereus group bacteriaa
| Name | Sequence | Location |
|---|---|---|
| P1 | 5′-GTT TGA TCC TGG CTC AG | 11-27 (16S rRNA) |
| P10 | 5′-CCA GTC TTA TGG GCA GGT TAC | 136-116 (16S rRNA) |
| P11 | 5′-TCC ATA AGT GAC AGC CGA AGC | 226-206 (16S rRNA) |
| P5 | 5′-CTA CGG GAG GCA GCA GTG GG | 340-360 (16S rRNA) |
| P3 | 5′-GWA TTA CCG CGG CKG CTG | 535-517 (16S rRNA) |
| P2 | 5′-GGA TTA GAT ACC CTG GTA GT | 784-803 (16S rRNA) |
| P6 | 5′-CCG TCA ATT CCT TTR AGT TT | 926-907 (16S rRNA) |
| P8 | 5′-TTC GGG AGC AGA GTG ACA GGT | 1029-1049 (16S rRNA) |
| P9 | 5′-TAC ACA CCG CCC GTC ACA CCA | 1392-1412 (16S rRNA) |
| P4 | 5′-RGT GAG CTR TTA CGC | 1513-1492 (16S rRNA) |
| Pr1 | 5′-CCG AAT GGG GVA ACC C | 114-129 (23S rRNA) |
| Pr13 | 5′-CCG TTT CGC TCG CCG CTA CTC | 262-242 (23S rRNA) |
| PB1 | 5′-TAG TGA TCG ATA GTG AAC CAG | 485-505 (23S rRNA) |
| Pr2 | 5′-CAT TMT ACA AAA GGY ACG C | 621-603 (23S rRNA) |
| Pr3 | 5′-GCG TRC CTT TTG TAK AAT G | 603-621 (23S rRNA) |
| PB2 | 5′-TAG TGA TCG ATA GTG AAC CAG | 755-736 (23S rRNA) |
| PB3 | 5′-TAG TGA TCG ATA GTG AAC CAG | 969-990 (23S rRNA) |
| Pr4 | 5′-RGT GAG CTR TTA CGC | 1151-1137 (23S rRNA) |
| Pr5 | 5′-WGC GTA AYA GCT CAC | 1136-1150 (23S rRNA) |
| PB4 | 5′-CAT ACC GGC ATT CTC ACT TC | 1308-1289 (23S rRNA) |
| PB5 | 5′-ACA GGC GTA GGC GAT GGA C | 1408-1426 (23S rRNA) |
| PB8 | 5′-AAC CTT TGG GCG CCT CC | 1679-1661 (23S rRNA) |
| Pr6 | 5′-CYA CCT GTG WCG GTT T | 1673-1659 (23S rRNA) |
| Pr7 | 5′-AAA CCG WCA CAG GTR G | 1659-1673 (23S rRNA) |
| Pr8 | 5′-CAY GGG GTC TTT RCG TC | 2092-2076 (23S rRNA) |
| Pr9 | 5′-GAC GYA AAG ACC CCR TG | 2076-2092 (23S rRNA) |
| Pr10 | 5′-GAG YCG ACA TCG AGG | 2535-2521 (23S rRNA) |
| Pr11 | 5′-CCT CGA TGT CGR CTC | 2521-2535 (23S rRNA) |
| Pr12 | 5′-GYT TAG ATG CYT TC | 2783-2770 (23S rRNA) |
| R1 | 5′-GGC GGC GTC CTA CTC TCA C | 112-95 (5S rRNA) |
Primers P1 to P4, Pr1 to Pr7, and R1 were used for DNA amplification. All other primers were utilized for sequencing. Primers P8, P9, P10, and P11 were selected de novo; other primers were described earlier (for details, see reference 32).
Development of expanded sequence databases.
We retrieved 16S rRNA, 23S rRNA, and gyrB sequences for members of the B. cereus group from GenBank (www.ncbi.nlm.nih.gov). Twelve 16S rRNA and 10 23SrRNA sequences were determined in this study, including one B. anthracis strain (Sterne) resequenced by our laboratory (Table 1). In this article, numbers are shown in all sequences in accordance with numbering in the B. anthracis Ames genome.
Creation of phylogenetic tree.
The 16S, 23S rRNA, and gyrB sequence databases were used to create phylogenetic trees. Sequences of isolates with different names but representing the same strain, incomplete sequences, sequences containing a large number of mistakes, strain-specific variations, or undetermined nucleotides (particularly at the sites of subgroup-specific signatures) were excluded from consideration. For an accurate determination of species similarities, all 5′ and 3′ ends were cut to identical positions along the 16S rRNA, 23S rRNA, and gyrB genes at B. anthracis Ames bp 49 to 1462, 24 to 2779, and 381 to 1499, respectively. The sequences were aligned using the multiprocessor (version 1.81) of CLUSTAL W (http:www.cmbi.kun.nl/bioinf/tools/clustalw.shtml). Aligned sequences were analyzed using the Molecular Evolutionary Genetics Analysis package version 2.1 (31; http://www.megasoftware.net). Rooted and unrooted phylogenetic trees were built with minimum evolution, neighbor-joining, unweighted pair group method with averages (UPGMA), and maximum parsimony methods. During analysis of alignments with the minimum evolution method, gaps were considered missing data points, genetic distances were estimated using nucleotide/Jukes-Cantor (for rRNA) or nucleotide/p-distance (for gyrB) models, where all substitutions were included in pairwise distance calculations. A close-neighbor-interchange search was performed to examine the neighborhood of the neighbor-joining tree to find a potential minimum evolution tree. Bootstrap confidence values were generated using 1,000 permutations of the data set for 16S rRNA and gyrB and 100 permutations for 23S rRNA to derive the nucleotide sequence similarities. In the maximum parsimony method for 16S rRNA and gyrB, gaps in the analysis were treated as missing data points; for 23S rRNA, where insertion G(1218-1219) was considered as a subgroup-specific signature, all sites were included for consideration. We utilized the close-neighbor-interchange algorithm to find the maximum parsimony tree. The random addition option with 10 replicates was used to produce the initial trees. In calculations of tree length, relative weights for different types of changes were specified uniformly using the standard parsimony method. In the both the neighbor-joining and UPGMA methods for gyrB, gaps were treated with the “complete deletion” option, genetic distances were estimated using the nucleotide/p-distance method in neighbor-joining analysis and nucleotide/Jukes-Cantor method for UPGMA, and bootstrap confidence values were generated using 1,000 permutations for both the neighbor-joining and UPGMA methods.
RESULTS
Sequencing of 16S and 23S rRNA genes of B. cereus group microorganisms.
In this study we sequenced 12 16S rRNA and 10 23S rRNA genes (Tables 2 and 3). There are published sequences available (6, 7) for two of the microorganisms that we sequenced, B. medusa NCIMB10437 (ATCC 25621) and B. anthracis Sterne. We found a number of discrepancies between our sequences and these previously published sequences. The published 16S rRNA sequences of B. anthracis Sterne (6; GenBank accession no. X55059) and B. medusa NCIMB10437 (7; GenBank accession no. X60628) have a deletion of a C (position 942) in comparison with other strains of B. anthracis and B. medusa that were later sequenced (Table 2). We did not find this deletion in our resequencing of the 16S rRNA genes of B. anthracis Sterne (GenBank accession no. AF176321) and B. medusa ATCC 25621 (GenBank accession no. AF155958), or in the recent resequencing of this gene in B. anthracis Sterne by Ticknor et al. (50; GenBank accession no. AF290552). We also did not find this deletion in ours or The Institute for Genomic Research's sequencing of B. anthracis Ames (GenBank accession numbers AF267734, AE017024 to AE017026, and AE017039). We believe that the reported deletion was a compression artifact of sequencing this GC-rich region (56), i.e., GGGGCCG instead of GGGGCCCG. We suggest that the same compression artifact at the same site may also have compromised the 16S rRNA sequences of B. cereus NCDO1771, B. cereus NCTC 11143, B. mycoides DSM2048T, and B. thuringiensis NCIMB9134 (6, 7; GenBank accession numbers X55060 to X55063). For this reason, these deletions were not included in Table 2.
TABLE 3.
Classification of bacteria in the B. cereus group according to 23S rRNA sequences
| Subgroup name | Subgroup-specific signature(s) (position)a | Start and end of sequence | Organism | GenBank accession no. | Position(s) of strain-specific variation(s) |
|---|---|---|---|---|---|
| Anthracis | Consensusa | 1-2922 | B. anthracis Sterneb | AF267877 | —k |
| 1-2922 | B. anthracis Delta Ames-1b | AF267876 | — | ||
| 1-2922 | B. anthracis Ames ANRb | AF267734 | — | ||
| 1-2922 | B. anthracis Ames (TIGR)c | AE017024 to -26, AE017039 | — | ||
| 15-2943 | B. anthracis Sterned | S43426 | T/C(491), del CG(1413, 1414), T/C(2651) | ||
| Cereus A | Y/C(594)j | 1-2923 | B. cereus NCTC11143d | S43429 | T/C(2651) |
| G/A(1559) | 1-2923 | B. cereus ATCC 10987c | AE017264 to -66 | — | |
| Insertion G(1218-1219) | AE017280 | ||||
| 1-527 | B. cereus WSBC10030e | Z84589 | — | ||
| B. cereus HER1414f | ? | ||||
| 1-2923 | B. thuringiensis B8b | AF267880 | — | ||
| Cereus B | Y/C(594) | 1-2922 | B. cereus NCTC9620b | AF267878 | — |
| G/A(1559) | 1-2922 | B. cereus Tb | AF267879 | G/R(1559) | |
| T/A(2153) | 1-2787 | B. cereus DSM31g | X94448 | T/C(1275) | |
| 24-2789 | B. cereus LMG6923g | AJ310096 | — | ||
| 1-2922 | B. cereus ATCC 14579c,g | AF016998 to AF017000, AF017013 | — | ||
| 1-2922 | B. cereus BGSC 6A5c | AY224379 to AY224388 | — | ||
| 18-2897 | B. cereus Tim-r01 | AB050631 | ins G(1218-1219), G/T(1268), G/A(1557), ins T(1781-1782), T/A(1938) | ||
| 1-527 | B. thuringiensis WS2617e | Z84594 | — | ||
| 1-527 | B. thuringiensis WS2614e | Z84593 | — | ||
| Thuringiensis A | T/A(2153)Insertion G(1218-1219) | B. thuringiensis strs. 4R1,h 4D1,h 4F1,h 4T1,h 4W1,h 4S2,h and 4J4h | ? | ||
| Thuringiensis B | Y/T(594) | 1-2922 | B. thuringiensis 4Q281b | AF267881 | G/R(1559) |
| T/C(157) | 24-2789 | B. thuringiensis LMG7138i | AJ310738 | G/R(546) | |
| G/A(921), A/G(1020), C/T(1037), G/A(1209), A/G(1251), T/C(1283)C/T(132), A/T(174), G/T(1250) T/A(2153) | 1-2784 | B. thuringiensis DSM2046i | X89895 | C/T(57), T/G(413), ins AATA(479-480), del GG(541-542), G/A(646), C/G(670), G/A(1953), G/A(2055), ins AGT(2556-2557), del G(2573) | |
| B. thuringiensis 4A1,h 4A7,h 4Q1,h 4Q2,h and 4M1h | ? | ||||
| 1-2922 | B. medusa ATCC 25621b | AF267885 | — | ||
| 20-2790 | B. cereus ATCC 14893 | AJ310099 | CA/TC(265,266), T/C(358), G/A(646), C/T(655), G/A(663), C/G(1816), G/C(1849) | ||
| 24-2799 | B. cereus biovar toyoi CNCM 1-1012/NCIB 40112 | AJ310101 | CA/TC(265,266), G/A(646), C/T(655), G/A(663), C/G(1816), G/C(1849) | ||
| Mycoides A | Y/T(594) | 1-527 | B. mycoides DSM2048T | Z84592 | — |
| T/C(157) | 1-527 | B. mycoides MWS5303-1-4 | Z84591 | — | |
| G/A(921), A/G(1020), C/T(1037), G/A(1209), A/G(1251), T/C(1283) | 23-2789 | B. mycoides DSM2048 | AJ310097 | CA/AY(375, 376), T/C(1112), T/C(2651) | |
| CA/TC(265,266), GT/AC(364,365), C/G(1816), G/C(1849) | 1-527 | B. cereus WSBC10206 | Z84590 | — | |
| C/T(132), A/T(174), G/T(1250) T/A(2153) | |||||
| C/T(375) | |||||
| Mycoides B | Y/T(594) | 1-2922 | B. mycoides ATCC 6462mb | AF267884 | — |
| T/C(157) | 1-2922 | B. mycoides ATCC 10206b | AF267883 | — | |
| G/A(921), A/G(1020), C/T(1037), G/A(1209), A/G(1251), T/C(1283) | |||||
| CA/TC(265,266), GT/AC(364,365), C/G(1816), G/C(1849) | |||||
| GA/AG(346,347), TC/CT(358,359), C/A(482), C/T(672), A/T(1219), G/T(1268), A/G(2159) |
For more details, see Fig. 2.
23S rDNA sequenced in this work.
From sequences of whole genome; data represent average sequence of all available 23S rRNA genes.
Need to be reexamined (see Results).
Assigned to this subgroup in accordance with the 16S rRNA sequence (see Table 2).
Not sequenced; identified through hybridization analysis (see Results).
These three strains are identical and correspond to B. cereus IAM12605, B. cereus NCDO1771, B. cereus ATCC 11778, and B. cereus ATCC 14579; see 16S rRNA sequences of these strains in Table 2.
Not sequenced; subgroup-specific signatures A/G(77), T/C(90), and T/A(92) were identified by hybridization with subgroup-specific probes (S. Bavykin and J. Jackman, unpublished).
These two strains are identical and correspond to B. thuringiensis IAM12077, B. thuringiensis NCIMB9134, B. thuringiensis ATCC 10792, and B. thuringiensis BT3; see 16S rRNA sequences of these strains in Table 2. Y = C or T; R = G or A.
See footnote q of Table 2.
See footnote s of Table 2.
In addition, our resequencing of the 16S rRNA gene for B. medusa ATCC 25621 did not reveal the C-to-T transition at position 192 (presence of T instead of C found in B. anthracis) or the A-to-G transversion at position 1383 previously reported for B. medusa NCIMB10437 16S rRNA (Table 2).
We did not include in Table 2 the 108 inconsistencies found in the six 16S rRNA sequences mentioned above, which had been deposited in GenBank by Ash et al. (6, 7). These were not supported by the resequencing of B. anthracis Sterne and B. medusa or in any other 16S rRNA sequences (Table 2) and probably represented systematic errors in sequencing which occurred when the base following T was incorrectly read as A, generally, instead of T or G.
We also found differences between the previously published 23S rRNA sequence of B. anthracis Sterne (5; GenBank accession no. S43426) and our resequencing of this isolate (GenBank accession no. AT267877). The differences that we found were the following: T instead of C at position 491, deletion of CG (1413, 1414), and T instead of C at position 2651. We also did not find these changes in any other 23S rRNA sequence in the B. cereus group, including B. anthracis Ames and B. anthracis Delta Ames (Table 3). Therefore, we suggest that these differences in B. anthracis Sterne and also the same differences in B. cereus NCTC 11143 (GenBank accession no. X64646) are due to errors in the previously reported sequences (5).
We did not include in Table 3 the substitutions of A for G, T, and C reported earlier for the 23S rRNA sequences of B. anthracis Sterne and B. cereus NCTC 11143 (5) that were not confirmed by our resequencing of the same gene in B. anthracis Sterne. These inconsistencies with our results, C/A(1037), T/A(1127), G/A(1411), G/A(1827), G/A(1834), C/A(2079), G/A(2182), G/A(2278), and C/A(2391), previously reported in combination with several Ns were not observed in any other isolates (Table 3).
Comparison of 16S and 23S rRNA sequences in the B. cereus group.
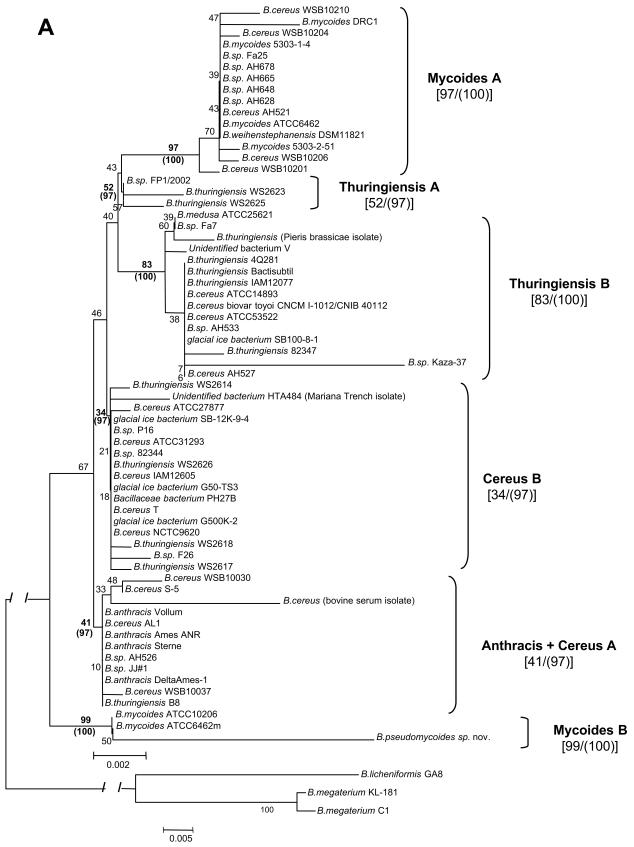
Our analyses indicated that in terms of known 16S and 23S rRNA sequences, B. anthracis was the most homogeneous species within the B. cereus group (Tables 2 and 3). This finding confirms PCR fingerprinting studies that demonstrated almost complete homogeneity of bulk DNA recovered from different strains of B. anthracis (2, 9, 14, 20, 21, 24-27, 30, 34, 39, 40, 43, 44, 48, 54). Because of this homogeneity, we have used the B. anthracis 16S and 23S rRNA sequences as a reference for reporting differences among closely related bacteria within the B. cereus group (Fig. 1 and 2; Tables 2 and 3).
FIG. 1.
Positions of subgroup-specific sequence differences in the 16S rRNA of B. cereus subgroups. The sequence of B. anthracis Sterne (GenBank accession no. AF 176321) has been used as the consensus sequence. Vertical lines indicate nucleotides identical to the consensus sequence. Arrows indicate regions containing subgroup-specific differences. For more details, see Table 2.
FIG.2.
Positions of subgroup-specific differences in the 23S rRNA sequences of B. cereus subgroups. The sequence of B. anthracis Sterne (GenBank accession no. AF176321) has been used as the consensus sequence. Arrows indicate regions containing subgroup-specific differences. Vertical lines indicate nucleotides identical to consensus sequences. Dots note nonsequenced regions. Superscript letters: a, 16S rRNA sequences are available (Table 2); b, whole-genome sequences are available (Table 3); c, 5 of 11 23S rRNA genes contain T(594); d, 4 of 12 23S RNA genes contain the insertion G(1218-129); e, 6 of 12 23S rRNA genes have A(1559); f, 7 of 11 23S rRNA genes in B. cereus ATCC 14579 reveal A(1559); g, 8 of 11 23S rRNA genes contain A(1559).
Analysis of our 16S rRNA sequences and sequences found in GenBank for the other B. cereus group organisms identified six characteristic regions which contained the majority of the positional sequence differences: position(s) 77 to 92, 133, 182 to 208, 286, 1015 to 1045, and 1462 (Fig. 1 and Table 2). Because sequence variation in these regions can be used to divide the B. cereus group organisms into several large subgroups, we have termed the differences located within these regions subgroup-specific differences. The most common were C/A (1015) and C/T (192). A set of subgroup-specific differences that characterized a subgroup were called subgroup-specific signatures (Table 2). In addition, a number of other differences were observed, which we have termed strain-specific differences (Tables 2 and 3). Most of the strain-specific differences were unique to each strain and were located randomly along the 16S rRNA molecule, i.e., they did not occur within the same sites as the subgroup-specific differences. Based on our resequencing experience, it is possible that some of the strain-specific variants represented mistakes in sequencing.
It is necessary to stress that all of the different subgroup-specific differences were not equally important for subgroup identification. For example, 16S rRNA subgroup-specific differences C/T (182) and T/A (1462) were found in all microorganisms from the Cereus A subgroup. However, both of these alterations, as well as C/Y (192), sometimes appeared in Thuringiensis B isolates (B. thuringiensis 82347, Bacillus sp. strain AH540, Bacillus sp. strain AH533, Bacillus sp. strain Termite isolate bac, bromate-reducing bacterium B6, glacial ice bacterium SB100-8-1, unidentified bacterium V, and B. cereus AH527) (Table 2). Alterations C/T (182) and C/Y (182) also were found in some Thuringiensis A microorganisms (Bacillus sp. strain KPU-0013, B. cereus ATCC 43881) (Table 2). These findings suggested close relationships for the subgroups Thuringiensis A, Thuringiensis B, and Mycoides A.
Analysis of the 23S rRNA sequences for the B. cereus group organisms revealed 12 regions within which the majority of the sequence variation occurred (Fig. 2 and Table 3). The differences within these regions were analogous to the subgroup-specific variants found in the 16S rRNA.
However, due to the limited number of 23S rRNA sequences in the database, it may be that not all of these differences are subgroup specific. Some of the regions which appear to contain subgroup-specific differences may actually contain strain-specific variations. For example, the Mycoides B subgroup signature contained five subgroup-specific differences in the 16S rRNA and nine subgroup-specific differences in 23S rRNA sequences that were not found in other subgroups (Fig. 2). However, available rRNA sequences for the Mycoides B subgroup currently include only five strains for which 16S rRNA sequences have been determined and two strains for 23S rRNA sequences (Tables 2 and 3). Among them, B. mycoides ATCC 6462m and B. mycoides ATCC 10206 had identical 16S and 23S rRNA sequences (Fig. 2; Tables 2 and 3) as well as 16S-23S rRNA spacers (GenBank accession numbers AF267905 and AF267906). However, they differed in colony morphology (see Materials and Methods). If additional members of the Mycoides B subgroup will be sequenced, we may find that some of the subgroup-specific differences are actually strain specific. Further work is needed to address this issue.
The most common subgroup-specific differences in 23S rRNA sequence occurred at positions 157 and 594 (Table 3; Fig. 2). The presence of these common variants among the subgroups supports a phylogenetic relationship among them.
Grouping of microorganisms in the B. cereus group according to 16S rRNA sequences.
The B. cereus group can be divided into seven subgroups based on 16S rRNA sequence differences (Table 2). We have labeled each of these subgroups according to the name of the most common member of the subgroup: Anthracis, Cereus A and B, Thuringiensis A and B, and Mycoides A and B.
The following subgroups reflect the 16S rRNA sequence relationships (Table 2). Subgroup Anthracis includes eight strains of B. anthracis. Most of the published 16S rRNA sequences in this subgroup contain a polymorphic site at position 1146 or 1147.
Subgroup Cereus A includes 17 members which do not contain any subgroup-specific sequence differences from the B. anthracis consensus but which were not identified as B. anthracis by conventional taxonomic methods. Ten of these 17 isolates do contain strain-specific sequence differences. However, at least six of the other seven members, B. cereus NCTC 11143, B. cereus ATCC 10702, B. thuringiensis B8, Bacillus sp. strain JJ-1, and Bacillus sp. strain AH526, have sequences identical to subgroup Anthracis in the region of the 16S rRNA compared. Two isolates of the subgroup Cereus A, B. cereus WSBC10037 and B. cereus WSBC10030, have been previously characterized as mesophilic (33). B. thuringiensis B8 apparently represents a misclassification, because it does not contain any cry genes (J. Jackman, personal communication).
Subgroup Cereus B includes 23 strains of B. cereus and B. thuringiensis that differ from B. anthracis by a C-to-A change at position 1015. B. cereus NCTC 9620, B. cereus T, B. cereus IAM12605, B. cereus NCDO1771, B. cereus ATCC 11778, B. cereus ATCC 14579, B. cereus ATCC 31293, B. cereus BGSC 6A5, Bacillus sp. strain 82344, and glacial ice bacterium strains SB-12K-9-4, G500K-2, and G50-TS3 do not differ from one another in 16S rRNA sequence and, thus, they would be indistinguishable based on 16S rRNA hybridization. Strains B. cereus IAM12605, B. cereus NSDO1771, B. cereus ATCC 11778, and B. cereus ATCC 14579 represent the same strain and correspond to B. cereus DSM31 and B. cereus LMG6923, whose 23S rRNA sequences are considered below (Tables 2 and 3).
Subgroups Thuringiensis A and Thuringiensis B include, respectively, 15 and 42 strains of microorganisms which contain two and five subgroup-specific sequence differences, respectively, C/A (1015) and C/T (192) being shared among the two subgroups. These two subgroups include mainly B. thuringiensis strains. Six strains in the subgroup Thuringiensis B (B. thuringiensis 4Q281, B. thuringiensis IAM12077, B. thuringiensis NCIM9134, B. thuringiensis ATCC 10792, Bacillus sp. strain A24, and B. cereus biovar toyoi CNSMI-1012/NCIB40112) have identical 16S rRNA sequences. Strains B. thuringiensis IAM12077, B. thuringiensis NCIMB9134, B. thuringiensis ATCC 10792, and B. thuringiensis BT3 comprise the same strain and correspond to B. thuringiensis DSM2046, the 23S rRNA sequence of which is considered below (Tables 2 and 3). Two other members of this subgroup, B. medusa ATCC 25621 and B. medusa NCIMB10437, should be identical according to those in Bergey's Manual (45). However, according to our sequencing and hybridization studies (data not shown), strain B. medusa ATCC 25621 does not contain the subgroup-specific alteration C/T (192), whereas according to published sequences (7), B. medusa NCIMB10437 does contain this sequence variant.
In the last two subgroups, Mycoides A and Mycoides B, 18 B. mycoides strains group in subgroup Mycoides A and 5 fall under subgroup Mycoides B. Subgroups contain, respectively, nine and five subgroup-specific differences in their signatures. Strains B. mycoides DSM2048 and B. mycoides ATCC 6462 according to Bergey's Manual (45) represent the same strain. Psychrotolerant isolates B. weihenstephanensis DSM11821 and B. cereus strains WSBC 10201, 10204, 10206, and 10210 were also included in subgroup Mycoides A. Nine isolates, B. mycoides strain DSM2048T, B. mycoides ATCC 6462, B. weihenstephanensis DSM11821, B. cereus AH521, and Bacillus sp. strains AH628, AH648, AH665, AH678, and Fa25 have identical 16S rRNA sequences. Subgroup Mycoides B contains B. cereus Ki21 and B. pseudomycoides sp. nov., which may have split off from the other three isolates in this subgroup rather early in their evolution, as they have a large number of strain-specific sequence differences (Table 2).
The same groups containing the same microorganisms were identified in a rooted phylogenetic tree that was generated with the minimum evolution method using 16S rRNA sequences of the B. cereus group with Bacillus licheniformis strain GA8 (GenBank accession no. AY162136), Bacillus megaterium strain C1 (GenBank accession no. AJ491841), and B. megaterium strain KL-181 (GenBank accession no. AY030336) 16S rRNA sequences as an outgroup. This grouping was also completely and independently confirmed by reconstructing an unrooted 16S rRNA tree with the maximum parsimony method (Fig. 3A).
FIG. 3.
Genetic relationship among B. cereus group strains. 16S rRNA (A) and 23S rRNA (B) phylogenetic trees obtained by minimum evolution method demonstrate the division of the B. cereus group into subgroups. Subgroup names are marked with bold on the right side of the brackets. Bootstrap volumes are reported on the branches. Numerals indicated in the quadrant parentheses denote the bootstrap volumes for each subgroup. During calculation of consensus parsimonious trees 7615 and 219 most-parsimonious trees were obtained for 16S rRNA and 23S rRNA, respectively. The percentages of most-parsimonious trees that support each subgroup in consensus parsimonious trees are presented in round parentheses. Bars indicate the scales of genetic distances.
Although the affiliations in the tree are consistent with those we defined earlier by signature analysis (Table 2 and Fig. 1), these groupings do not correspond exactly to the current taxonomy, which divides the B. cereus group into seven species: B. anthracis, B. cereus, B. thuringiensis, B. medusa, B. mycoides, B. pseudomycoides sp. nov., and B. weihenstephanensis (15, 16, 33, 38, 45, 51).
Grouping of microorganisms in the B. cereus group according to 23S rRNA sequences.
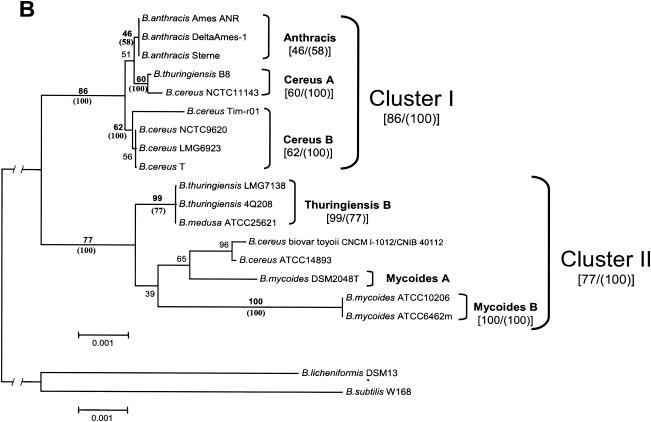
For all microorganisms whose 23S rRNA genes were sequenced (Table 3), 16S rRNA sequences were also available. This finding gave us a unique opportunity to confirm the division of the B. cereus group into seven subgroups that was done according to 16S rRNA sequences (Fig. 1 and 3A; Table 2). Unfortunately, there were no 23S rRNA sequences available for any of the organisms from subgroup Thuringiensis A. Analyzing 23S rRNA signatures obtained for six subgroups did not reveal any serious contradictions with the groupings based on 16S rRNA sequence analysis (compare data in Table 2 and Fig. 3A with Fig. 2 and Table 3 data). However, in contrast to 16S rRNA sequences, 23S rRNA sequences of isolates in the Anthracis subgroup revealed a set of variations that provided the possibility for differentiation of the Anthracis subgroup from all the other six subgroups (Table 3; Fig. 2).
Subgroup Cereus A contains B. cereus ATCC 10987, B. thuringiensis B8, and B. cereus NCTC 11143 (Table 2). The 23S rRNA sequences of these two organisms include subgroup-specific alterations at positions 594 and 1559 and an insertion G(1218-1219) (Fig. 2; Table 3). The 23S rRNA sequence of B. cereus WSBC10030 was sequenced only partially (Fig. 2; Table 3). It does not contain any differences that are specific for any other subgroups, but also it does not cover subgroup-specific sites for the Cereus A subgroup.
Subgroup Cereus B contains seven strains. We observed that 23S rRNA sequences of this subgroup contain three subgroup-specific differences at positions 594, 1559, and 2153 (Fig. 2). Unfortunately, the 23S rRNA sequences available for B. thuringiensis WS2614 and B. thuringiensis WS2617 do not extend beyond position 527 from the 5′ end of the gene. Strains B. cereus DSM31, B. cereus ATCC 14579, and B. cereus LMG6923 represent the same strain that corresponds to B. cereus IAM12605, B. cereus NCDO1771, and B. cereus ATCC 11778 (Table 3), whose 16S rRNA sequences are also available (Table 2).
Subgroup Thuringiensis B consists of 10 microorganisms containing 12 subgroup-specific alterations. B. thuringiensis LMG7138 and B. thuringiensis DSM2046 (Table 3) represent one strain, whose synonyms are B. thuringiensis IAM 12077, B. thuringiensis NCIMB9134, B. thuringiensis ATCC 10792, and B. thuringiensis BT3, whose 16S rRNA sequences were considered above (Table 2).
B. mycoides DSM2048, B. mycoides DSM2048T, B. mycoides MWS5303-1-4, and B. cereus WSBC10206 form subgroup Mycoides A. Although only 527 nucleotides of sequence from the 5′ end of 23S rRNAs are available for the last three organisms, they revealed enough (8) variations to be discriminated from members of all other subgroups (Table 3; Fig. 2).
Subgroup Mycoides B includes only two microorganisms, B. mycoides ATCC 6462m and B. mycoides ATCC 10206. We found 23 specific differences in their 23S rRNA sequences (Table 3; Fig. 2).
The subgroups described above were consistent with the 23S rRNA phylogenetic trees obtained by two independent methods where the minimum evolution tree was rooted using B. licheniformis strain DSM 13 (GenBank accession no. X68433) and Bacillus subtilis strain W168 (GenBank accession no. K00637) as an outgroup (Fig. 3B). Phylogenetic analysis also revealed a division of B. cereus group bacteria into two clades which we called cluster I, which includes subgroups Anthracis, Cereus A, and Cereus B, and cluster II, which consists of subgroups Thuringiensis B, Mycoides A, and Mycoides B (Fig. 3B). Thuringiensis A could not be placed in a cluster due to the lack of 23S rRNA sequences for any microorganisms which were members of this subgroup (based on their 16S rRNA sequences). We also found that microorganisms B. cereus biovar toyoi and B. cereus 14893, which according to their 16S and 23S rRNA sequences unambiguously belonged to subgroup Thuringiensis B, also contained four alterations in their 23S rRNA (CA/TC [265, 266], C/G [1816], and G/C[1849]) that are typical for the Mycoides A and Mycoides B subgroups. The same connection between different subgroups was found for B. cereus Tim-r01 (subgroup Cereus B), which shares its G(1218-1219) insertion with subgroup Cereus A and variation G/T(1268) with subgroup Mycoides B (Fig. 2; Table 3).
Therefore, the division of the B. cereus group members into the specified subgroups was supported by both the 16S and 23S rRNA sequence analyses.
Subgroup identification through hybridization.
Recently, two 16S rRNA probes containing A/G (77), T/C (90), and T/A (92) differences were described as a tool for discrimination of B. thuringiensis from B. cereus (18). In this study, we demonstrated that this signature is sufficient only for identification of bacteria included in the Thuringiensis B group (Fig. 1; Table 2). Using these probes, this signature was found in B. thuringiensis strains 9308, 20, Lb5, 1230, and L3 (18), in B. thuringiensis serotypes Galleriae and Israelensis (55) (Table 4), in B. cereus strains Nagoya 126 and 127 (55), and in B. thuringiensis strains BT3, BT13, BT15, BT16, BTT6, and BTT8 (13). We included all these microorganisms in the Thuringiensis B subgroup (Table 2).
TABLE 4.
Classification of B. cereus group bacteria with sequenced gyrB genes according to their 16S rRNA gene sequences
| Subgroup namea | Organisme | GenBank accession no.
|
|
|---|---|---|---|
| 16S rRNA | gyrB | ||
| Anthracis | B. anthracisAmes | AF155950 | AE017024 |
| B. anthracis Pasteur 2 | —f | AF090333 | |
| Cereus A | B. cereusATCC 10987b | NC003909 | AE017264 |
| Anthracis or Cereus A | Bacillus sp. strain H-01c | AY461742 | AY461763 |
| Bacillus sp. strain H-03c | AY461744 | AY461764 | |
| Bacillus sp. strain H-05c | AY461746 | AY461766 | |
| Bacillus sp. strain H-06c | AY461747 | AF136389 | |
| Bacillus sp. strain H-07c | AY461748 | AY461767 | |
| Bacillus sp. strain H-08c | AY461749 | AY461768 | |
| Bacillus sp. strain H-09c | AY461750 | AY461769 | |
| Bacillus sp. strain H-12c | AY461753 | AY461772 | |
| Bacillus sp. strain H-17c | AY461758 | AY461776 | |
| Cereus B | B. cereusATCC 14579 | AF290547 | AE016998 |
| B. thuringiensis serotype Aizawai | AY461760 | AY461778 | |
| Bacillus sp. strain H-04 | AY461745 | AY461765 | |
| Bacillus sp. strain H-11 | AY461752 | AY461771 | |
| Bacillus sp. strain H-13 | AY461754 | AY461773 | |
| Bacillus sp. strain H-14 | AY461755 | AY461774 | |
| Bacillus sp. strain H-15 | AY461756 | AY461775 | |
| Thuringiensis A | Bacillus sp. strain H-10 | AY461751 | AY461770 |
| Bacillus sp. strain H-18 | AY461759 | AY461777 | |
| Thuringiensis B | B. thuringiensisIAM12077 | D16281 | AF090331 |
| B. thuringiensis serotype Galleriae | AY461761 | AY461779 | |
| B. thuringiensis serotype Israelensis | AY461762 | AY461780 | |
| Bacillus sp. strain ES-027 | AY461789 | AY461783 | |
| Bacillus sp. strain H-16 | AY461757 | AF136387 | |
| Thuringiensis A or B | Bacillus sp. strain SAFN-003d | AY167823 | AY461786 |
| Bacillus sp. strain 83-3Cd | AF526900 | AY461781 | |
| Mycoides A | B. mycoidesATCC 6462 | AB021192 | AF090332 |
| Bacillus sp. strain SAFR-048 | AY167860 | AY461787 | |
| Bacillus sp. strain FO-080 | AY461791 | AY461785 | |
Subgroups were identified in accordance with 16S rRNA sequences of the organisms.
Belongs to subgroup Cereus A according to 16S rRNA and 23S rRNA sequences, which were extracted from whole-genome sequences of this organism (GenBank accession nos. AE017264, AE017265, AE01266, and AE017280).
Because 23S rRNA genes of the organisms are not sequenced yet, the isolates were assigned to Anthracis or Cereus A subgroups.
Because of incomplete 16S rRNA sequences, these two isolates were identified as belonging to Thuringiensis A or Thuringiensis B subgroup.
Microorganisms whose names are marked in bold were also subgroup identified based on their 23S rRNA sequences.
—, not sequenced.
We also included for consideration (Fig. 2; Tables 2 and 3) isolates B. cereus HER1414 and two sets of B. thuringiensis strains, B. thuringiensis strains 4R1, 4D1, 4F1, 4S2, 4T1, 4W1, and 4J4 and B. thuringiensis strains 4A1, 4A7, 4Q1, 4Q2, and 4M1, whose 16S and 23S rRNA sequences have not yet been sequenced. According to hybridization experiments of rRNA isolated from these bacteria with oligonucleotide probes specific to all described 16S rRNA subgroup-specific signatures (Fig. 1; Table 2), these strains were identified as belonging to subgroups Cereus A, Thuringiensis A, and Thuringiensis B, respectively. 23S rRNA signatures identified in these microorganisms after hybridization with probes specific to regions 3, 9, 10, and 12 are shown on Fig. 2 (S. Bavykin and J. Jackman, unpublished data).
Grouping of microorganisms in the B. cereus group according to gyrB gene sequences.
Recently, a set of parallel 16S rRNA and partial (more then 60% of the whole gene length) gyrB sequences for B. cereus group isolates was placed in GenBank. We classified these microorganisms (Table 4) according to subgroup-specific signatures found for 16S rRNA sequences (Fig. 1 and Table 2). Additional information about the 23S rRNA obtained from whole-genome sequences of B. cereus ATCC 10987 made it possible to identify this organism as belonging to the Cereus A subgroup (Fig. 2; Table 3) and, therefore, to differentiate it from bacteria of the Anthracis subgroup. Because 23S rRNA genes for isolates Bacillus sp. strains H-01, H-03, H-05, H-06, H-07, H-08, H-09, H-12, and H-17 are not yet published, we identified these microorganisms as belonging to the Anthracis or Cereus A subgroups. The absence of 98 and 139 nucleotides in the beginning of the 16S rRNA sequences for isolates Bacillus sp. strain SAFN-003 and Bacillus sp. strain 83-3C, respectively, led us to identify these organisms as belonging to the Thuringiensis A or Thuringiensis B subgroups. Finally, among 30 microorganisms for which both 16S RNA and gyrB gene sequences were published, we identified members of six of the seven subgroups, identified using rRNA sequences (Tables 2 and 3 and Fig. 1 to 3). Unfortunately, there were no gyrB sequences available in GenBank for any members of the Mycoides B subgroup.
The same groups containing the same microorganisms were identified in rooted and unrooted phylogenetic trees that were generated with minimum evolution (compare Table 4 and Fig. 4), neighbor-joining, UPGMA, and maximum parsimony methods with the gyrB sequences of B. cereus group organisms and the Bacillus pumilis strain KL-052 gyrB sequence (GenBank accession no. AY167878) as an outgroup. All four trees revealed completely identical branching patterns (data not shown) with high bootstrapping values from all four methods for five of six subgroups (Fig. 4). The subgroups in all four trees were organized into two clusters, as was observed for the 23S rRNA tree (Fig. 3B). Cluster I contained subgroups Anthrax, Cereus A, and Cereus B, and Cluster II included subgroups Thuringiensis B and Mycoides A. Additionally, the Thuringiensis A subgroup was located in cluster I of the gyrB tree, but it was not completely separated from subgroup Cereus B. Unfortunately, as was mentioned above, there are no 23S rRNA sequences for Thuringiensis A subgroup organisms available.
FIG. 4.
Grouping of the B. cereus group strains based on gyrB gene sequences. Names of the subgroups were identified through 16S rRNA sequences of their members (Table 4) and are displayed with bold on the right sides of the brackets. Subgroup identities of the microorganisms, whose names appear in bold, were confirmed by their 23S rRNA sequences. The phylogenetic tree was constructed by the minimum evolution method. Bootstrap volumes are reported on the branches. Numerals indicated in the quadrant parentheses designate the bootstrap volumes for each subgroup. Phylogenetic trees obtained by neighbor-joining, UPGMA, and maximum parsimony methods gave identical branching patterns. Bootstrap numbers for these three methods are presented in round parentheses. Bar indicates scale of genetic distances.
Subgroups Anthracis and Cereus A were identified in the gyrB phylogenetic trees (Fig. 4) in accordance with the presence in these subgroups of B. anthracis AMES and B. cereus ATCC 10987, whose correspondence to subgroups Anthracis and Cereus A, respectively, were confirmed through independent grouping of 16S rRNA and 23S rRNA sequences (Tables 2 to 4). The presence of B. anthracis Pasteur 2 and B. anthracis Ames at the same subgroup (Fig. 4) also confirms this identification. Isolates Bacillus sp. strain SAFN-003 and Bacillus sp. strain 83-3C were found in the gyrB phylogenetic tree in subgroup Thuringiensis B (Fig. 4).
gyrB as a phylogenetic marker for subgroup identification in the B. cereus group.
In accordance with our 16S rRNA analysis, we aligned gyrB sequences for 31 B. cereus group isolates. Analysis of the gyrB alignment resulted in identification of 93 unique subgroup-specific differences (Fig. 5). As shown in Table 5, we defined these differences for each subgroup as subgroup-specific signatures. Because the number of members of each subgroup was not statistically large enough, we defined subgroup-specific signatures according to the rule that all members of the subgroup should contain all of the selected subgroup-specific differences. In total, we identified 15 unique subgroup-specific gyrB differences for the Anthracis subgroup (Table 5) which were absent from the other five subgroups (Fig. 5). For subgroups Cereus A, Thuringiensis B, and Mycoides A, we identified 12, 13, and 32 unique subgroup-specific differences, respectively (Table 5). For subgroups Cereus B and Thuringiensis A, we did not find significant differences in the gyrB sequences. For this reason, we defined one set of signatures for these two subgroups (Table 5).
FIG. 5.
Positions of subgroup-specific differences in the gyrB gene in the B. cereus group. Only differences that are unique for one of the subgroups are shown. (Members of each subgroup are displayed in Fig. 4.) Differences indicated for each subgroup were present in gyrB sequences of all members of that subgroup. The sequence complementary to the B. anthracis Ames gyrB gene (GenBank accession no. AE017024) has been used as the consensus sequence. Vertical lines indicate nucleotides identical to the consensus sequence. Multiple nucleotide substitutions (w, y, m, and r) indicate that more than one subgroup member revealed alternative differences. w, a or t; y, c or t; m, a or c; r, a or g.
TABLE 5.
Subgroup-specific signatures in gyrB in B. cereus group microorganisms
| Subgroup name | Subgroup-specific signatures |
|---|---|
| Anthracis | Consensusa: G(195), C(313), G(315), G(318), A(348), C(399), C(531), G(600), C(616), C(651), C(808), T(891), A(945), G(1008), C(1092) |
| Cereus A | A/C(267), T/C(387), G/A(600), T/C(636), T/G(642), T/C(696), T/C(891), G/A(900), G/A(1047), G/A(1074), A/G(1083), A/C(1149) |
| Cereus B and Thuringiensis A | G/A(51), T/G(69), A/G(225), T/C(234), A/T(255), T/C(363), A/G(432), T/C(435), T/C(495), T/C(528), T/C(594), T/C(684), G/A(723), T/A(810), A/G(813), A/T(843), A/T(861), T/G(1068) |
| Cereus B | A/M(186), A/R(489), A/R(777), A/R(1077), A/R(1028) |
| Thuringiensis A | A/G(489), A/G(777), A/G(1128) |
| Thuringiensis B | C/A(190), G/T(195), C/T(252), A/G(354), T/C(360), C/G(474), G/A(561), T/C(573), T/A(762), A/G(795), A/G(879), A/T(885), A/G(930) |
| Mycoides A | T/A(60), T/A(114), T/C(129), A/C(145), T/G(150), T/C(158), C/A(174), T/A(177), T/A(183), A/T(186), A/G(191), A/T(192), A/C(204), T/A(207), C/T(223), A/C(236), A/T(438), A/C(441), T/G(456), A/C(492), C/T(510), T/C(543), A/G(567), T/C(663), G/A(717), A/G(861), A/G(885), T/C(928), T/C(963), A/G(978), T/C(996), A/C(1157) |
For more details, see Fig. 5.
We also found that isolates B. sp. SAFN-003 and B. sp. 83-3C, which were found in the Thuringiensis B subgroup in the gyrB phylogenetic tree, included all 13 subgroup-specific differences (Fig. 5) that were present in subgroup Thuringiensis B (Table 5). This confirmed that these two isolates belonged in the Thuringiensis B subgroup.
DISCUSSION
Identification of subgroups in the B. cereus group.
Based on analysis of the 16S rRNA sequence alignments, we divided the B. cereus group into the seven above-mentioned subgroups, each containing microorganisms with similar 16S rRNA gene sequences. The strains within each subgroup include all or most of the alterations specific for that subgroup (Fig. 1 and Table 2).
Some of the 16S rRNA subgroup-specific differences, indicated in Table 2, have already been used by other researchers for identification of certain Bacillus sp. (18, 33, 41), but a systematic analysis of all B. cereus group microorganisms in accordance with their 16S and 23S rRNA sequences had not been done before.
We have also sequenced the 23S rRNA gene for a selected set of members of the B. cereus group. During 23S rRNA sequence alignment analysis of these and GenBank sequences, we did not find any conflicts between subgroupings based on 16S rRNA and subgroupings based on 23S rRNA (Tables 2 and 3 and Fig. 1 and 2). We also found that isolates from subgroup Cereus A, which have the same 16S rRNA sequence as B. anthracis Sterne (Fig. 1 and Table 2), have three subgroup-specific changes in the 23S rRNA sequence in comparison with B. anthracis Sterne (Fig. 2 and Table 3). Our study demonstrated that members of subgroups Anthracis and Cereus A may not be differentiated according to their 16S rRNA subgroup-specific signatures (Fig. 1; Table 2). However, we have demonstrated that B. cereus ATCC 10987, B. cereus NCTC 11143, B. thuringiensis B8, and B. cereus HER1414, which belong to the Cereus A subgroup, may be differentiated from members of subgroup Anthracis by using the 23S rRNA subgroup-specific changes Y/C(594), G/A(1559), and insertion G(1218-1219), in combination with 16S rRNA-targeted probes (Fig. 1 and 2; Tables 2 and 3). Strain-specific variations also may be used for these purposes (Tables 2 and 3). At the same time, because we do not have 23S rRNA sequences for most members of Cereus A subgroup we are not excluding the possibility that some nonpathogenic strains of B. anthracis, which have transcriptionally inactive toxin genes or which have lost their virulence plasmids (52), may be found besides the isolates that were placed in subgroup Cereus A according to their 16S rRNA sequences (Table 2).
Phylogenetic analysis of 16S rRNA sequences with two independent methods, minimum evolution and maximum parsimony, confirmed the presence of subgroups Mycoides A and B, Thuringiensis A and B, Cereus B, and joint subgroup Anthracis-Cereus A in the B. cereus group (Fig. 3A). Both methods generated subgroups with identical microbial content, which corresponded to the results of 16S and 23S rRNA alignment analyses (Tables 2 and 3 and Fig. 1 and 2). Low bootstrapping volumes for Anthracis-Cereus A (41%) and Cereus B (34%) subgroups obtained with the minimum evolution method reflect an absence of subgroup-specific differences between subgroups Anthracis and Cereus A and a strongly reproducible but small (one base only) difference between joint subgroup Anthracis-Cereus A and subgroup Cereus B in their 16S rRNA sequences (Table 2; Fig. 1). However, grouping of 16S rRNA sequences obtained with the maximum parsimony method strongly confirmed the data from the minimum evolution analysis (Fig. 3A).
In the rooted minimum evolution phylogenetic tree generated for 23S rRNA sequences of the B. cereus group, we found more significant differentiation of the Anthracis, Cereus A, Cereus B, Thuringiensis B, Mycoides A, and Mycoides B subgroups (Fig. 3B) according to their bootstrapping volumes because of the presence in these subgroups of a considerable amount of subgroup-specific changes in the 23S rRNA signatures (Table 3 and Fig. 2). The unrooted 23S rRNA phylogenetic tree obtained with the maximum parsimony method strongly confirmed this grouping (Fig. 3A and B). A rooted 23S rRNA tree also indicated that the B. cereus group divided into two clades, cluster I and cluster II, which included subgroups Anthracis, Cereus A, and Cereus B and Thuringiensis B, Mycoides A, and Mycoides B, respectively (Fig. 3B). The existence of microorganisms that sometimes shared their subgroup-specific alterations in 16S rRNA signatures between subgroups Thuringiensis B and Mycoides A (B. thuringiensis 82347, Bacillus sp. strain AH540, Bacillus sp. strain AH533, Bacillus sp. strain Termite isolate bac, bromate-reducing bacterium B6, glacial ice bacterium SB100-8-1, unidentified bacterium V, and B. cereus AH527) (Table 2), bacteria that shared some of their changes in 23S rRNA signatures between the Thuringiensis B and Mycoides A and B subgroups (B. cereus biovar toyoi and B. cereus 14893), and isolate B. cereus Tim-r01 (subgroup Cereus B), which shares a G(1218-1219) insertion in its 23S rRNA sequence with Cereus A subgroup (Table 3 and Fig. 2) confirm the division of the B. cereus group into these two clusters.
Comparative analysis of rRNA and gyrB sequences demonstrated excellent correlation for the grouping of bacteria from the B. cereus group. The single-copy gyrB gene is rather well conserved; however, for the B. cereus group, it displayed more nucleotide variations than the rRNA genes. The relatively high degree of variation in gyrB sequences probably explains the fact that analysis of gyrB sequences by four different methods (minimum evolution, neighbor-joining, UPGMA, and maximum parsimony [Fig. 4]) produced identical branching patterns (data not shown) and was supported with higher bootstrapping values than those obtained for rRNA analysis (Fig. 3). The rooted phylogenetic gyrB trees (Fig. 4) also showed the same organization of subgroups into two clusters that was found with the 23S rRNA phylogenetic analysis (Fig. 3B). At the same time, questions remain about the nature of isolates Bacillus sp. strains H-05, H-06, H-07, and H-17. Further investigation should be performed to clarify whether these microorganisms might represent strains of B. anthracis that have lost their virulence.
In summary, the existence of seven subgroups within the B. cereus group was confirmed by six independent methods: 16S rRNA, 23S rRNA, and gyrB sequence alignment analysis (Fig. 1, 2, and 5; Tables 2, 3, and 5) and phylogenetic analysis of all three sets of these sequences (Fig. 3 and 4).
In total we analyzed 183 16S and 74 23S rRNA sequences for 155 strains of B. cereus group bacteria, including 50 Bacillus isolates of unknown species and nine isolates of unknown genus (Tables 2 and 3). Identification of some unidentified microorganisms as members of the B. cereus group brought some interesting information. Particularly, we found that among B. cereus group bacteria there exist bromate-reducing (Bacillus sp. strain B6) and sulfur-degrading (Bacillus sp. strain KPU-0013) microorganisms. Some of the B. cereus group isolates were found in Cold Desert, India (Bacillus sp. strain Kaza-31 and Bacillus sp. strain Kaza-37), as intestinal symbionts (Bacillus sp. strain JJ-1, unidentified bacterium V, and unidentified bacterium sp6 [bovine rumen isolate]), on the surface of strawberry plants (Bacillus sp. strain Fa7), and even in the deepest sea mud of the Mariana Trench (unidentified bacterium HTA484). We also recognized that subgroup Cereus B was organized not later then 500,000 years ago, because glacial ice bacterium G500K-2 that belongs to this subgroup was isolated from more-than-500,000-year-old glacial ice from Gulia, China (Table 2; Fig. 3A).
Taxonomy of the B. cereus group.
Recent studies demonstrated that the species B. cereus, B. thuringiensis, and B. mycoides represent genetically heterogeneous groups of microorganisms (12-14, 21-23, 28, 40, 50, 55) which, according to their rRNA sequences, comprise a group of close relatives (5-7) (Tables 2 and 3; Fig. 1 and 2). These findings resulted in several suggestions to consider B. cereus and B. thuringiensis (8, 12, 58), or these two bacteria together with B. anthracis (14, 21), as one species. Results of our analysis of 16S and 23S rRNA sequences also show disagreement with the current taxonomic classification of species within the B. cereus group. Traditionally, classification of microorganisms in the B. cereus group has been based on morphological, physiological, and immunological data. However, recent data suggest that use these criteria for current B. cereus group species identification may have some obstacles. For example, B. thuringiensis has been traditionally distinguished from B. cereus by the production of a parasporal crystal of a protein that is toxic for Lepidoptera, Diptera, and Coleoptera larvae. The capacity to form crystals is plasmid encoded (3), however, and some plasmids may be lost by laboratory culturing (4, 45). Moreover, authentic cultures of B. cereus can acquire the ability to produce crystals as a result of growing in mixed culture with B. thuringiensis (19). Based on our findings, we suggested that plasmid exchange apparently exists inside and between subgroups Cereus B, Thuringiensis A, and Thuringiensis B only, because simultaneously B. cereus and B. thuringiensis strains were found only in these three subgroups (Tables 2 and 3 and Fig. 3). Thus, the discrimination of B. cereus from B. thuringiensis is a difficult task by any method, and the fact that they have grouped together in our analysis (Fig. 1 to 3; Tables 2 and 3) and in other recent studies (12-14, 21-23, 28, 40, 50, 55) is not surprising. At the same time, we do not exclude that after resequencing their 16S rRNAs some B. thuringiensis strains may be moved from subgroup Cereus B to subgroup Thuringiensis A, which differ from each other by only one subgroup-specific signature, C/T (192) (Table 2).
For B. mycoides, sporadic loss of the ability to form rhizoid colonies has been observed in several strains (45), indicating an instability of morphology in this species. In this case, B. mycoides becomes morphologically similar to B. cereus. Bacterial isolates can also undergo physiological changes after the loss or acquisition of plasmids coding for toxins, sporulation, or antibiotic resistance (48). Therefore, flexible colony morphology may be a reason why strains of B. mycoides and B. cereus are present together in subgroups Mycoides A and Mycoides B (Tables 2 and 3 and Fig. 2 and 3).
Therefore, the current division of the B. cereus group into seven species (B. anthracis, B. cereus, B. thuringiensis, B. mycoides, B. pseudomycoides, B. weihenstephanensis, and B. medusa) seems to be insolvent from genomic as well as phenotypic points of view.
Demonstrating that 16S rRNA and 23S rRNA sequences independently confirm separation of the B. cereus group into seven distinct subgroups, we tried to find additional confirmation for this division among other genomic sequences and morpho-physiological evidence. The Anthracis subgroup can be taken as an example. This subgroup is represented by one species only, B. anthracis, which is distinguishable from members of other subgroups based on its plasmid content and ability to induce anthrax (36, 42, 49, 51, 52).
We found some unique properties of subgroup Thuringiensis B in the literature. Two recent studies (13, 55) exploited differences in the gyrB gene as well as a 16S rRNA signature of A/G (77), T/C (90), and T/A (92) (unique for the Thuringiensis B subgroup in our classification) for discrimination of B. cereus and B. thuringiensis. Both studies demonstrated a good correlation between these two parameters but not with the current nomenclature, which is based on phenotypic characterization. We also analyzed gyrB sequences for a set of 30 B. cereus group isolates for which both 16S rRNA and gyrB gene sequences were deposited recently in GenBank (Table 4). This provided the unique possibility to identify a number of strong phylogenetic markers represented by gyrB subgroup-specific signatures (Table 5) for most of the described subgroups (Tables 2 and 3; Fig. 3 and 4). Each signature contained between 12 and 32 unique subgroup-specific differences and strongly differentiated its own subgroup from all others, with the exception of subgroups Cereus B and Thuringiensis A (Table 5). Because gyrB sequences were available for only two members of subgroup Thuringiensis A (Table 4; Fig. 4) and because the published gyrB sequences represent only about 65% of the whole length of the gyrB gene, we are unable to draw a final conclusion about the relationship between subgroups Cereus B and Thuringiensis A. However, if more-thorough sequencing of 23S rRNA, gyrB, and other genes does not reveal more differences between subgroups Cereus B and Thuringiensis A, this would suggest that subgroup Thuringiensis A should not be considered as a separate subgroup but should instead be characterized as a variation of subgroup Cereus B. At the same time, the 15 subgroup-specific differences found in the gyrB gene for subgroup Anthracis and the 12 differences observed for subgroup Cereus A provide us numerous possibilities for differentiation of members of these two subgroups (Table 5). Subgroups Thuringiensis B and Mycoides A and the joint Cereus B-Thuringiensis A subgroup also included a number of differences (13, 32, and 18, respectively) which could be useful for discrimination of these subgroups (Table 5).
Another distinguishing characteristic that relates to B. mycoides is psychrotolerance. Recent DNA relatedness studies have indicated that the B. mycoides species actually consists of two genetically distinct groups (37, 38). The fact that our study placed B. mycoides strains into two subgroups, Mycoides A and Mycoides B, supports this finding. The type strain of B. mycoides group 1 (B. mycoides sensu stricto), B. mycoides ATCC 6462 (37, 38), was included in our Mycoides A subgroup (Table 2; Fig. 3A), and the type strain of B. mycoides group 2 (B. pseudomycoides), B. mycoides NRRL B-617 T (B. pseudomycoides sp. nov.), is a member of our Mycoides B subgroup (Table 2; Fig. 3A). According to our classification scheme (Table 2), four representatives of psychrotolerant strains of B. cereus (WSBC10201, WSBC10204, WSBC10206, and WSBC10210), which were recently named as the new species B. weihenstephanensis (33), fall under subgroup Mycoides A. This finding suggests that species B. weihenstephanensis may be one of the B. mycoides strains that belongs to the subgroup Mycoides A. This suggestion was confirmed by the high degree of similarity of genomic DNA sequences (85 to 88%) between B. cereus strains WSBC10201, WSBC10204, and WSBC10206 and B. mycoides DSM2048 (33), which is also located in subgroup Mycoides A. In addition, based on the ability to grow at low temperature, B. mycoides is the most closely related species to B. weihenstephanensis in the B. cereus group (33). At the same time, the mesophilic strains B. cereus WSBC10030, B. cereus WSBC10037, and B. cereus HER1414 (33) were included in subgroup Cereus A, and mesophilic B. cereus ATCC 27877 (33) and B. cereus ATCC14579 (17) were found in subgroup Cereus B (Table 2; Fig. 3A). However, mesophilic strains (17, 41) are also widely represented in other subgroups. Thus, B. thuringiensis WS2614, B. thuringiensis WS2617, B. thuringiensis WS2618, and B. thuringiensis WS2626 (41) were included in subgroup Cereus B, B. thuringiensis WS2625 (41) is a member of subgroup Thuringiensis A, and B. thuringiensis ATCC 10792 (17) is situated in subgroup Thuringiensis B. Interestingly, representative of Mycoides B subgroup, B. pseudomycoides sp. nov. has a minimum growth temperature of 15°C (38) and is therefore a mesophilic strain. Summarizing the above results, we can stress that currently we did not find any mesophilic representatives in the Mycoides A subgroup, nor did we find any psychrotolerant strains outside of this subgroup.
Experience with thousands of strains from several hundred species led taxonomists to conclude that 70% or higher DNA-DNA relatedness with 5% or less divergence within related sequences, together with 97% or higher 16S rRNA sequence similarity, is the best means of defining a species (10). Our findings indicated that 16S rRNA of B. cereus from subgroup Cereus B may differ from that of B. thuringiensis from subgroup Thuringiensis A in one base only (Table 2). Even B. cereus from the Mycoides A subgroup may have only nine substitutions (equivalent to 0.6% of 16S rRNA) in comparison with B. thuringiensis from Thuringiensis A subgroup (Table 2), in spite of a great difference between representatives of these two subgroups in phenetic (46) relationship that includes differences both in genotype and in phenotype. However, we found B. cereus strains in six of the seven subgroups and B. thuringiensis in three of the subgroups, whereas members of the different subgroups are considerably different both in phenotype and in rRNA sequences. For example, B. cereus strains are present in Thuringiensis A and B as well as in Mycoides A and B subgroups. At the same time, DNA-DNA relatedness between B. thuringiensis and B. cereus may range from 54.3 to 96.4% (33), with divergence inside of these two species of up to 45% (33, 37). In these terms, unification of all strains of B. cereus with all strains of B. thuringiensis, even without B. anthracis, as one species is rather questionable. Apparently, this is a case when only polyphasic (consensus) taxonomy (10), which integrates all available data, may provide the best opportunity to find the right solution.
The results of this work demonstrated the potential for the use of 16S rRNA, 23S rRNA, and gyrB gene sequences for identification of the members of the B. cereus group and, especially, differentiation of B. anthracis from other relatives, something which has for a long time been considered impossible by using rRNA sequences (5-7). Further work is needed to determine how the differences in rRNA genes, which have been revealed in our study, relate to differences in phenotypic traits and to determine what kind of revisions are necessary in the taxonomy of the B. cereus group.
Acknowledgments
We express our gratitude to Wentso Liu for his help in the early stages of this project and Felicia King for excellent assistance in manuscript preparation.
This research was supported by the Defense Advanced Research Project Agency under interagency agreement AO-E428 and by the Russian Human Genome Program, grant 5/2000.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson, A. I. 1993. Insecticidal toxins, p. 953-963. In A. B. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 4.Aronson, A. I., W. Beckman, and P. Dunn. 1986. Bacillus thuringiensis and related insect pathogens. Microbiol. Rev. 50:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ash, C., and M. D. Collins. 1992. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol. Lett. 94:75-80. [DOI] [PubMed] [Google Scholar]
- 6.Ash, C., J. A. E. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 7.Ash, C., J. A. E. Farrow, W. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 8.Baumann, L., K. Okamoto, B. M. Unterman, M. J. Lynch, and P. Baumann. 1984. Phenotypic characterization of B. thuringiensis and B. cereus. J. Invertebr. Pathol. 44:329-341. [Google Scholar]
- 9.Beyer, W., P. Glockner, J. Otto, and R. Bohm. 1996. A nested PCR and DNA-amplification-fingerprinting method for detection and identification of Bacillus anthracis in soil samples from former tanneries. Salisbury Med. Bull. Sp. Suppl. 87:47-49. [Google Scholar]
- 10.Brenner, D. J., J. T. Staley and N. R. Krieg. 2001. Classification of procaryotic organisms and the concept of bacterial speciation, p. 27-31. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, N.Y.
- 11.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson, C. R., D. A. Caugant, and A.-B. Kolstǿ. 1994. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, M. L., and H. Y. Tsen. 2002. Discrimination of Bacillus cereus and Bacillus thuringiensis with 16S rRNA and gyrB gene based PCR primers and sequencing of their annealing sites. J. Appl. Microbiol. 92:912-919. [DOI] [PubMed] [Google Scholar]
- 14.Daffonchio, D., A. Cherif, and S. Borin. 2000. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group.” Appl. Environ. Microbiol. 66:5460-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaporte, M. 1969. Description de Bacillus medusa nov. sp. C. R. Acad. Sci. Ser. D 269:1129-1131. [PubMed] [Google Scholar]
- 16.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis, K. P., R. Mayr, F. von Stetten, G. S. A. B. Stewart, and S. Scherer. 1988. Discrimination of psychrotrophic and mesophilic strains of the Bacillus cereus group by PCR targeting of major cold shock protein genes. Appl. Environ. Microbiol. 64:3525-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giffel, M. C., R. R. Beumer, N. Klijn, A. Wagendorp, and F. M. Rombouts. 1997. Discrimination between Bacillus cereus and Bacillus thuringiensis using specific DNA probes based in variable regions of 16S rRNA. FEMS Microbiol. Lett. 146:47-51. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, J. M., Jr., B. J. Brown, and B. C. Carlton. 1982. Transfer of Bacillus thuringiensis plasmids coding for δ-endotoxin among strains of B. thuringiensis and B. cereus. Proc. Natl. Acad. Sci. USA 79:6951-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrell, L. J., G. L. Andersen, and K. H. Wilson. 1995. Genetic variability of Bacillus anthracis and related species. J. Clin. Microbiol. 33:1847-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helgason, E., D. A. Caugant, M.-M. Lecadet, Y. Chen, J. Mahillon, A. Lövgren, I. Hegna, K. Kvalǿy, and A.-B. Kolstǿ. 1998. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 37:80-87. [DOI] [PubMed] [Google Scholar]
- 23.Helgason, E., D. A. Caugant, I. Olsen, and A.-B. Kolstǿ. 2000. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 38:1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson, I. 1996. Fingerprinting Bacillus anthracis strains. Salisbury Med. Bull. Sp. Suppl. 87:55-58. [Google Scholar]
- 25.Henderson, I., C. J. Duggleby, and P. C. B. Turnbull. 1994. Differentiation of Bacillus anthracis from other Bacillus cereus group bacteria with the PCR. Int. J. Syst. Bacteriol. 44:99-105. [DOI] [PubMed] [Google Scholar]
- 26.Henderson, I., Y. Dongzheng, and P. C. B. Turnbull. 1995. Differentiation of Bacillus anthracis and other Bacillus cereus group bacteria using IS231-derived sequences. FEMS Microbiol. Lett. 128:113-118. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, P. J., K. K. Hill, M. T. Laker, L. O. Ticknor, and P. Keim. 1999. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J. Appl. Microbiol. 87:263-269. [DOI] [PubMed] [Google Scholar]
- 28.Joung, K.-B., and J.-C. Côte. 2001. Phylogenetic analysis of Bacillus thuringiensis serovars based on 16S rRNA gene restriction fragment length polymorphism. J. Appl. Microbiol. 90:115-122. [DOI] [PubMed] [Google Scholar]
- 29.Kersters, K., W. Ludwig, M. Vancannyet, P. de Vos, M. Gillis, and K. H. Schleifer. 1996. Recent changes in the classification of the pseudomonas: an overview. Syst. Appl. Microbiol. 19:465-476. [Google Scholar]
- 30.Kiem, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, I. B. Jakobsen, and Masatoshi Nei. 2001. MEGA2: molecular evolutionary genetic analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-176. In E. Stackenbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, England.
- 33.Lechner, S., R. Mayr, K. P. Fransis, B. M. Prub, T. Kaplan, E. Wiebner-Gunkel, G. S. Stewart, and A. B. Scherer. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48:1373-1382. [DOI] [PubMed] [Google Scholar]
- 34.Liang, X., and D. Yu. 1999. Identification of Bacillus anthracis strains in China. J. Appl. Microbiol. 87:200-203. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig, W., and H.-P. Klenk. 2001. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics, p. 49-65. In G. M. Garrity (ed.) Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, N.Y.
- 36.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-677. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, L. K., and M. A. Jackson. 1995. Clarification of the taxonomy of Bacillus mycoides. Int. J. Syst. Bacteriol. 45:46-49. [Google Scholar]
- 38.Nakamura, L. K. 1998. Bacillus pseudomycoides sp. nov. Int. J. Syst. Bacteriol. 48:1031-1034. [DOI] [PubMed] [Google Scholar]
- 39.Patra, G., P. Sylvestre, V. Ramisse, J. Therasse, and J. L. Guesdon. 1996. DNA fingerprinting of Bacillus anthracis strains. Salisbury Med. Bull. Sp. Suppl. 87:59. [Google Scholar]
- 40.Priest, F. G., D. A. Kaji, Y. B. Rosato, and V. P. Canhos. 1994. Characterization of Bacillus thuringiensis and related bacteria by ribosomal RNA gene restriction fragment length polymorphisms. Microbiology 140:1015-1022. [DOI] [PubMed] [Google Scholar]
- 41.Prub, B. M., K. P. Francis, F. von Stetten, and S. Scherer. 1999. Correlation of 16S ribosomal DNA signature sequences with temperature-dependent growth rates of mesophilic and psychrotolerant strains of the Bacillus cereus group. J. Bacteriol. 181:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinn, P. C., and P. C. B. Turnbull. 1998. Anthrax, p. 799-818. In W. J. Hausler, Jr., and M. Sussman (ed.), Topley and Wilson's microbiology and microbial infections, vol. 3. Arnold, London, England.
- 43.Ramisse, V., G. Patra, H. Garrigue, J.-L. Guesdon, and M. Mock. 1996. Identification and characterization of Bacillus anthracis by multiplex PCR analysis of sequences on plasmids pXO1 and pXO2 and chromosomal DNA. FEMS Microbiol. Lett. 145:9-16. [DOI] [PubMed] [Google Scholar]
- 44.Shangkuan, Y.-H., J.-F. Yang, H.-C. Lin, and M.-F. Shaio. 2000. Comparison of PCR-RFLP, ribotyping and ERIC-PCR for typing Bacillus anthracis and Bacillus cereus strains. J. Appl. Microbiol. 89:452-462. [DOI] [PubMed] [Google Scholar]
- 45.Sneath, P. H. A. 1986. Endospore-forming gram-positive rods and cocci, p. 1104-1139. In J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins Co., Baltimore, Md.
- 46.Sneath, P. H. A. 2001. Numerical taxonomy, p. 39-42. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, Springer, New York, N.Y.
- 47.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley and Sons, Chichester, England.
- 48.Thorne, C. B. 1985. Genetics of Bacillus anthracis, p. 56-62. In L. Leive (ed.), Microbiology—1985. American Society for Microbiology, Washington, D.C.
- 49.Thorne, C. B. 1993. Bacillus anthracis, p. 113-124. In A. L. Soneshein (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 50.Ticknor, L. O., A.-B. Kolsto, K. K. Hill, P. Keim, M T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbull, P. C. B. 1999. Definitive identification of Bacillus anthracis—a review. J. Appl. Microbiol. 87:237-240. [DOI] [PubMed] [Google Scholar]
- 52.Turnbull, P. C. B., R. A. Hutson, M. J. Ward, M. N. Jones, C. P. Quinn, N. J. Finnie, C. J. Duggleby, J. M. Kramer, and J. Melling. 1992. Bacillus anthracis but not always anthrax. J. Appl. Bacteriol. 72:21-28. [DOI] [PubMed] [Google Scholar]
- 53.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wunschel, D., K. F. Fox, G. E. Black, and A. Fox. 1994. Discrimination among the Bacillus cereus group, in comparison to B. subtilis, by structural carbohydrate profiles and ribosomal RNA spacer region PCR. Syst. Appl. Microbiol. 17:625-635. [Google Scholar]
- 55.Yamada, S., E. Ohashi, N. Agata, and K. Venkateswaran. 1999. Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the deletion of B. cereus in rice. Appl. Environ. Microbiol. 65:1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamakama, I., D. Nakajama, and O. Ohara. 1996. Identification of sequence motifs causing band compressions on human cDNA sequencing. DNA Res. 3:81-86. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto, S., and S. Harayama. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zahner, V., H. Momen, C. A. Salles, and L. Rabinovitch. 1989. A comparative study of enzyme variation in Bacillus cereus and Bacillus thuringiensis. J. Appl. Bacteriol. 67:275-282. [DOI] [PubMed] [Google Scholar]