Abstract
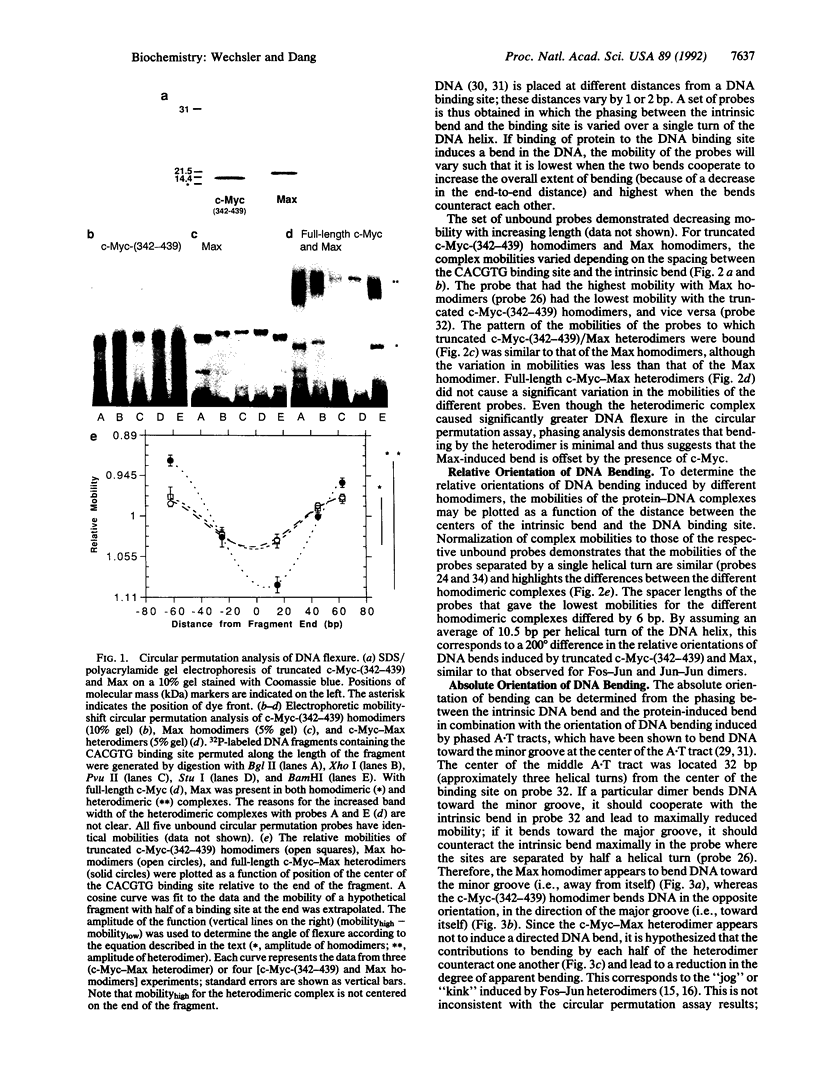
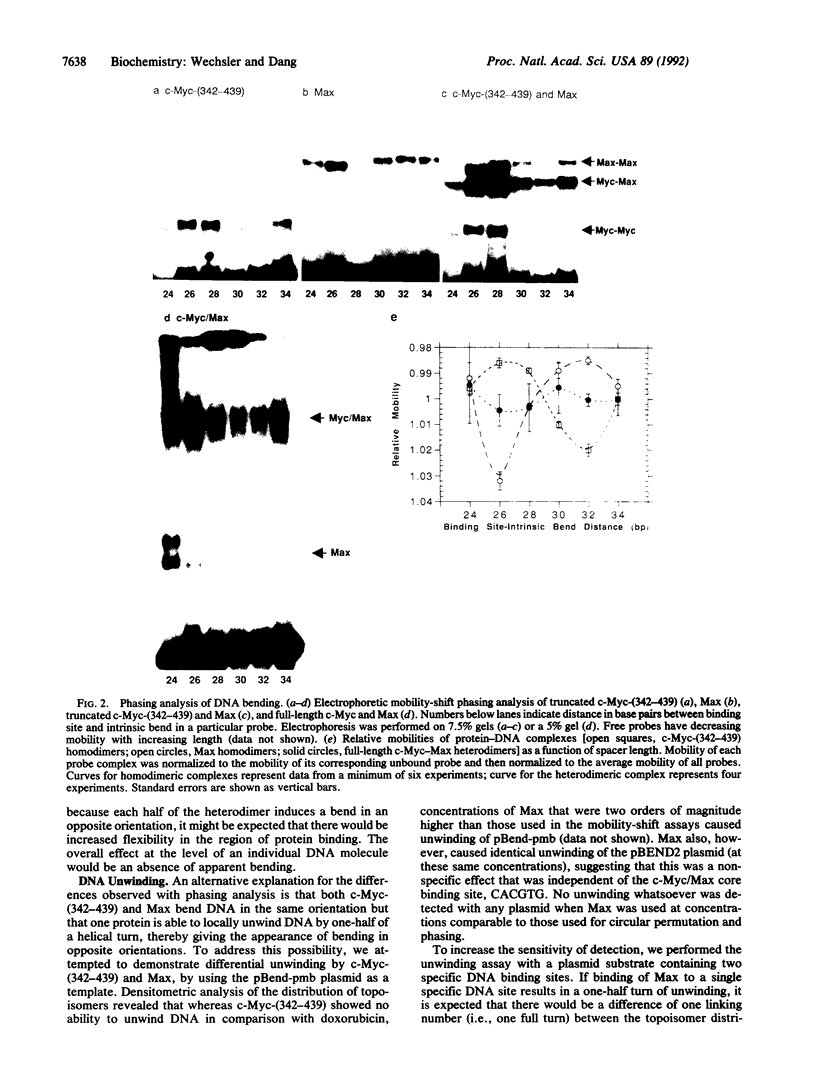
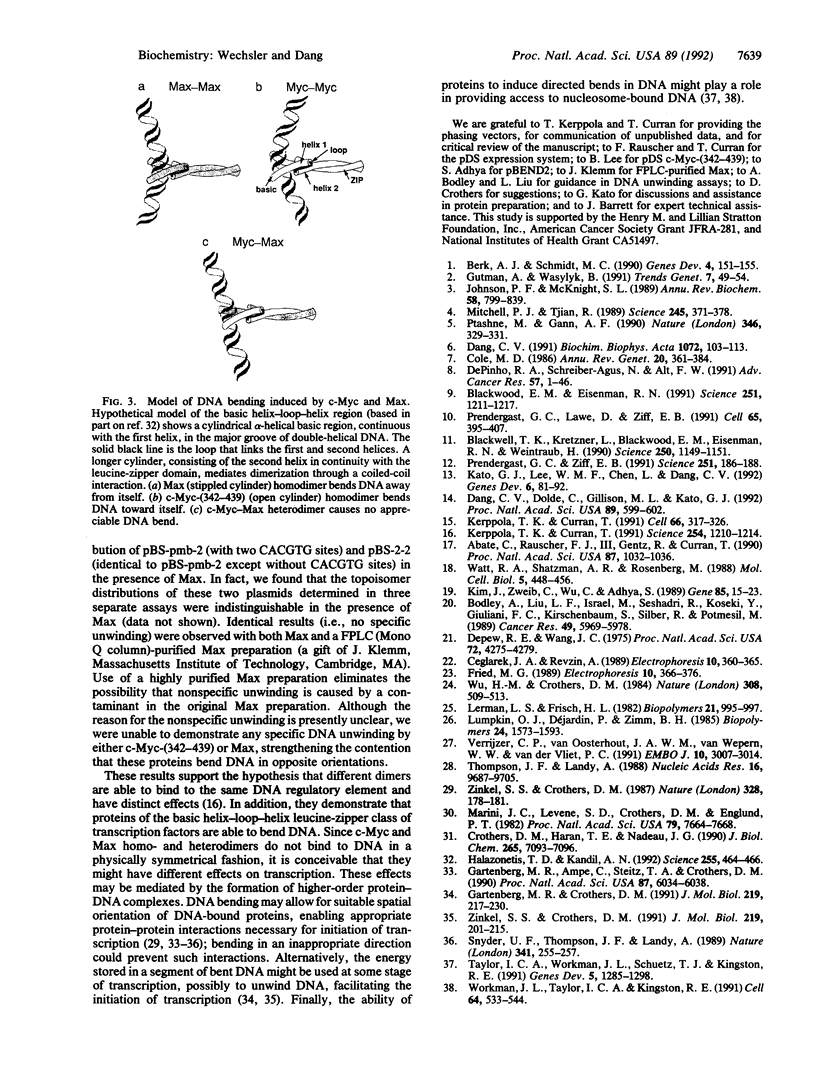
The control of gene transcription requires specific protein-protein and protein-DNA interactions. c-Myc, the protein product of the c-myc protooncogene, is a member of the basic helix-loop-helix leucine-zipper class of transcription factors. Although c-Myc is able to bind to a specific core hexanucleotide DNA sequence (CACGTG), its precise function in modulating transcription remains unclear. The recent discovery of Max, a basic helix-loop-helix leucine-zipper partner protein for c-Myc, suggests that the ability of c-Myc to regulate transcription is modulated by the presence of Max. By taking advantage of the altered mobility of protein-bound DNA in the mobility-shift assay, we demonstrate the homo- and heterodimeric complexes of c-Myc and Max are able to cause increased DNA flexure as measured by the circular permutation assay. Based on phasing analysis, c-Myc and Max homodimers bend DNA in opposite orientations, whereas c-Myc-Max heterodimers cause a smaller bend, in an orientation similar to that induced by Max homodimers. To address the possibility that the apparent opposite orientation of bending was the result of DNA unwinding by one of the proteins, we measured the ability of c-Myc and Max homodimers to affect DNA unwinding; we were unable to show any specific unwinding caused by c-Myc or Max. In addition to demonstrating that members of the basic helix-loop-helix leucine-zipper class of transcription factors are able to induce DNA bending, these results suggest that different transcription factor dimers are able to bind to identical DNA sequences and yet have distinct structural effects.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Luk D., Gentz R., Rauscher F. J., 3rd, Curran T. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1032–1036. doi: 10.1073/pnas.87.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Bodley A., Liu L. F., Israel M., Seshadri R., Koseki Y., Giuliani F. C., Kirschenbaum S., Silber R., Potmesil M. DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res. 1989 Nov 1;49(21):5969–5978. [PubMed] [Google Scholar]
- Ceglarek J. A., Revzin A. Studies of DNA-protein interactions by gel electrophoresis. Electrophoresis. 1989 May-Jun;10(5-6):360–365. doi: 10.1002/elps.1150100514. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Dang C. V., Dolde C., Gillison M. L., Kato G. J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V. c-myc oncoprotein function. Biochim Biophys Acta. 1991 Dec 10;1072(2-3):103–113. doi: 10.1016/0304-419x(91)90009-a. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M. G. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis. 1989 May-Jun;10(5-6):366–376. doi: 10.1002/elps.1150100515. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli lac promoter. J Mol Biol. 1991 May 20;219(2):217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- Gutman A., Wasylyk B. Nuclear targets for transcription regulation by oncogenes. Trends Genet. 1991 Feb;7(2):49–54. doi: 10.1016/0168-9525(91)90231-E. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Predicted structural similarities of the DNA binding domains of c-Myc and endonuclease Eco RI. Science. 1992 Jan 24;255(5043):464–466. doi: 10.1126/science.1734524. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991 Jul 26;66(2):317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Frisch H. L. Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers. 1982 May;21(5):995–997. doi: 10.1002/bip.360210511. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J., Déjardin P., Zimm B. H. Theory of gel electrophoresis of DNA. Biopolymers. 1985 Aug;24(8):1573–1593. doi: 10.1002/bip.360240812. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A. A. Activators and targets. Nature. 1990 Jul 26;346(6282):329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- Snyder U. K., Thompson J. F., Landy A. Phasing of protein-induced DNA bends in a recombination complex. Nature. 1989 Sep 21;341(6239):255–257. doi: 10.1038/341255a0. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer C. P., van Oosterhout J. A., van Weperen W. W., van der Vliet P. C. POU proteins bend DNA via the POU-specific domain. EMBO J. 1991 Oct;10(10):3007–3014. doi: 10.1002/j.1460-2075.1991.tb07851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R. A., Shatzman A. R., Rosenberg M. Expression and characterization of the human c-myc DNA-binding protein. Mol Cell Biol. 1985 Mar;5(3):448–456. doi: 10.1128/mcb.5.3.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J. L., Taylor I. C., Kingston R. E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991 Feb 8;64(3):533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]