Abstract
Defining how a glycan-binding protein (GBP) specifically selects its cognate glycan from among the ensemble of glycans within the cellular glycome is an area of intense study. Powerful insight into recognition mechanisms can be gained from 3D structures of GBPs complexed to glycans; however, such structures remain difficult to obtain experimentally. Here an automated 3D structure generation technique, called computational carbohydrate grafting, is combined with the wealth of specificity information available from glycan array screening. Integration of the array data with modeling and crystallography allows generation of putative co-complex structures that can be objectively assessed and iteratively altered until a high level of agreement with experiment is achieved. Given an accurate model of the co-complexes, grafting is also able to discern which binding determinants are active when multiple potential determinants are present within a glycan. In some cases, induced fit in the protein or glycan was necessary to explain the observed specificity, while in other examples a revised definition of the minimal binding determinants was required. When applied to a collection of 10 GBP–glycan complexes, for which crystallographic and array data have been reported, grafting provided a structural rationalization for the binding specificity of >90% of 1223 arrayed glycans. A webtool that enables researchers to perform computational carbohydrate grafting is available at www.glycam.org/gr (accessed 03 March 2016).
Keywords: binding determinant, glycan array screening, glycan-binding protein, lectin specificity, 3D structure
Introduction
The interactions between glycans and their cognate protein receptors are fundamental and profoundly important to biological function, with applications in both basic glycobiology research (Audfray et al. 2012; Varrot et al. 2013), and in the development of therapeutic and diagnostic reagents (Dube and Bertozzi 2005; Fuster and Esko 2005; Berthet et al. 2013; Ruhaak et al. 2013; Geissner et al. 2014). Discovering the targets of a glycan-binding protein (GBP), and understanding at a molecular level how these binding partners are specifically selected from among the vast array of heterogeneous glycan structures in a cellular glycome, are currently areas of intense study (Rillahanand Paulson 2011; Nycholat et al. 2012; Arnaud et al. 2013; Kuwabara et al. 2013; Promkuntod et al. 2013; Topin et al. 2013; Wang et al. 2013).
Glycan array screening has revolutionized the task of determining the specificity of a given GBP by allowing simultaneous screening of hundreds of glycans. Relative to other techniques, glycan microarrays allow an efficient and broader definition of the specificity of a GBP. Although these data have proved extremely useful it is often difficult to comprehensibly analyze why a particular glycan is bound, while other glycans that contain the same fragment or minimal binding determinant (MBD) are not.
Three-dimensional (3D) structures of protein-oligosaccharide co-complexes are of immense value in understanding the specificity of a GBP; however, large glycans are refractory to crystallization (Kosma and Müller-Loennies 2012; Owens and Nettleship 2011). Consequently, crystallographic data are generally limited to short oligosaccharide fragments that represent the biologically relevant MBD or a closely related structure. These co-complexes are frequently used to explain the affinity interaction between a GBP and the bound oligosaccharide, and it is also possible to rationalize how different substitutions found on intact glycans would be tolerated in the binding site (Kumar et al. 2013; Richichi et al. 2013; Tessier et al. 2013; Topin et al. 2013).
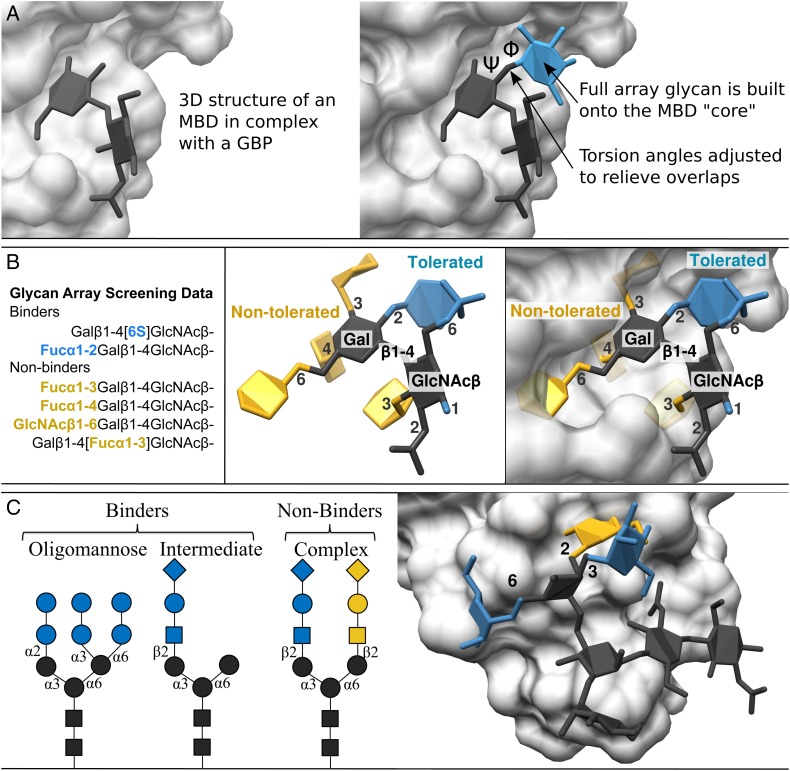
The current work builds on this concept; it is possible to compare glycan array screening data to a 3D structure and determine if the two data sets are consistent (Figure 1). A 3D structure of a co-complexed MBD should explain the observed specificity of binding on the array and, if not, can be iteratively adjusted until good agreement is achieved. If this is not possible, the set MBDs that were initially identified from the array data may then be re-evaluated until a consistent set of MBDs and their corresponding 3D structures are found.
Fig. 1.
An illustration of the grafting process. (A) A 3D structure of a bound MBD (dark gray) is used as a “core” upon which intact glycans are built. Each array glycan that contains this MBD is built into the binding site. Only vdW overlaps with the protein (light gray) are considered, and glycosidic torsion angles are adjusted within reasonable bounds to allow induced fit in the grafted branch (blue). If an intact glycan can physically fit in the binding site it is deemed to be a binder. The program compares these results to the glycan array data and reports on the level of agreement. (B) The structural origin of the specificity observed on a glycan array should be apparent from the orientation of the MBD in the 3D co-complex. Binders contain substitutions (blue) to the MBD that are physically tolerated in the binding site, whereas non-binders contain substitutions (yellow) which prevent recognition of the MBD. In this example, the 3D structure of ECL bound to Galβ1-4GlcNAcβ (MBD1) can be tested by grafting each observed binder and non-binder that contain MBD1. Grafting may be used to eliminate unlikely poses of the MBD in the binding site generated by modeling (Tessier et al. 2013). Good overall agreement between the 3D structure and the array data are only possible when the co-complex structure is correct and a set of MBDs that explain the observed binding pattern has been identified. (C) An illustration of the glycan structure classes that bind to UDA on the glycan array (oligomannose and intermediate) compared with those that do not (complex). The key structural difference is the branch shown in yellow. The experimental specificity data for UDA are consistent with the proposed 3D model; oligomannose and intermediate glycans can be bound (blue tolerated substitutions), whereas complex N-glycans collide with the surface (yellow non-tolerated substitution).
For the purposes of understanding the approach outlined here, specificity has been devolved into two components: the affinity for an MBD and the structural context within which this MBD can be recognized (Grant and Woods 2014). Due to the high degree of similarity between monosaccharides, the subset of molecular features that make up an MBD may be found within a variety of different glycans. For example, galactose and glucose differ only in the configuration of the 4-hydroxyl position, meaning they can only be differentiated if the 4-hydroxyl configuration is part of the MBD. The second component of specificity, structural context, is determined by a number of factors including differences in the substitution pattern from the MBD in different glycans. That is, although an MBD may be present in a glycan, extension at a particular position on the MBD may prevent recognition. In turn, the glycans may be conjugated to various biomolecules and biomolecular ensembles resulting in variations in glycan presentation (Gabius et al. 2011). Thus recognition is determined by the composition of the MBD, the position of the MBD within the overall 3D glycan structure, and the presentation of the glycan relative to its environment.
Computational carbohydrate grafting is a molecular modeling technique that builds 3D structures of full glycans from the MBD in a co-complex with a GBP, and assesses the ability of the full glycan to fit in the ligand-binding site (Tessier et al. 2013; Grant et al. 2014). In addition, non-bonded interaction energies, as well as solvation and entropic effects are purposefully excluded in grafting, as they are assumed to play a significant role only in the MBD region of the full glycan. This is a key concept of grafting; with array binders any extension from the MBD core is assumed to be passively tolerated in the binding site, with minimal induced fit, where it does not form affinity modulating interactions with the protein. With this assumption it is possible to generate putative co-complexes with each array glycan that contains an MBD by exploiting the observed conformational preferences of glycosidic linkages (Kirschner and Woods 2001; DeMarco and Woods 2008; Nivedha et al. 2013) (Figure 1A). If any steric overlaps occur with the protein surface that are not removed by allowing a reasonable level of induced fit in the glycan, the glycan is categorized as a non-binder. In order for an intact glycan to be bound by a GBP the full glycan structure must physically fit in the binding site, and when conjugated to an array surface, the glycan must be presented correctly for recognition by the GBP (Grant et al. 2014).
In contrast to automated ligand docking, grafting starts with a 3D structure of the MBD bound to the GBP. If the overall level of agreement with array data is poor it indicates that either the 3D structure of the glycan–GBP complex is incorrect, or the MBD(s) selected from the array data is misidentified. In these cases, iterative adjustment of both the co-complex 3D structure and the MBD definitions is performed until grafting reports that the two data sets are in optimal agreement (Figure 1B).
By exploiting the underlying connection between the structure and specificity data, it is possible to predict experimentally consistent 3D structures for protein–glycan complexes, and new specificities for GBPs. The present work highlights the insights that can be gained from the synergistic combination of modeling, glycan array data and 3D structure.
Results
The 10 GBPs selected for this study, along with their predicted MBDs, are reported in Table I. The MBDs are classified as being found in glycans that are either dominant or weak binders. For all 10 GBPs, a high overall level of agreement (92%) between the array data and 3D structure was achievable once the appropriate MBDs had been selected from the array data and their corresponding 3D structures generated. In the case of CTB, 100% agreement was achieved using the available co-complex structure and reported MBD (Merritt et al. 1994). In Polyporus squamosus lectin (PSL) and Burkholderia cenocepacia lectin (BC2L-A), the reported crystal structures and specificities (Merritt et al. 1994; Lameignere et al. 2008) were also sufficient to achieve good level of agreement (Table II). However, the remaining seven systems required some degree of modeling or redefinition of MBDs in order to generate 3D structures that were consistent with the experimental array data (Table II, Supplementary data, Table SI).
Table I.
The MBDs and PDB IDs for each of the 10 studied systems
| System name Abbreviation | PDB IDa |
MBDb,c | MBD ID |
|---|---|---|
|
Urtica dioica agglutinin UDA | 1EN2 |
Manβ1–4GlcNAcβ1–4GlcNAcβ | 1 |
| GlcNAcβ1–4GlcNAcβ1–4GlcNAcβ | 2 (X-ray) | |
|
Arachis hypogaea agglutinin PNA | 2DVA |
Galβ1–3GalNAcα | 1 (X-ray) |
| Galβ1–3GalNAcβ | 2 | |
| GlcNAcβ1–3Galβ | 3 | |
|
Erythrina crista-galli lectin ECL | 1GZC |
Galβ1–4Glcβ | –d (X-ray) |
| Galβ1–4GlcNAcβ | 1 | |
| GalNAcβ1–4GlcNAcβ | 2 | |
|
Dolichos biflorus agglutinin DBA | 1LU1 |
GalNAcα1–3GalNAcβ | 1 (X-ray) |
| GalNAcβ1–4[6S]GlcNAcβ | 2 | |
| GalNAcβ1–4[Neu5Acα2–3]Galβ | 3 | |
|
Clitocybene bularis lectin CNL | 3NBE |
GalNAcβ | 1 (X-ray) |
| GalNAcα | 2 | |
|
Marasmius oreades agglutinin MOA | 2IHO |
Galα1–3Galβ | 1 (X-ray) |
| Galα1–3GalNAcα | 2 | |
| Galα1–3GalNAcβ | 3 | |
|
Burkholderia cenocepacia lectin BC2L-A | 2VNV |
Manα | 1 (X-ray) |
|
Vibrio cholerae toxin subunit B CTB | 3CHB |
Galβ1–3GalNAcβ1–4[Neu5Acα2–3]Galβ1–4Glcβ | 1 (X-ray) |
|
Helix pomatia agglutinin HPA | 2CCV |
GalNAcα | 1 (X-ray) |
| GlcNAcα | 2 | |
|
Polyporus squamosus lectin PSL | 3PHZ |
Neu5Acα2–6Galβ | 1 (X-ray) |
| Neu5Gcα2–6Galβ | 2 |
aX-ray structures from the Protein Data Bank (Merritt et al. 1998; Hamelryck et al. 1999; Saul et al. 2000; Svensson et al. 2002; Natchiar et al. 2006; Sanchez et al. 2006; Grahn et al. 2007; Lameignere et al. 2008; Kadirvelraj et al. 2011; Pohleven et al. 2012).
bDominant binders are in boldface.
cCo-complexes for each MBD that did not have a crystal structure were modelled based on co-complexes of similar ligands using Maestro (Suite 2011) with glycan 3D structures built with GLYCAM-Web (Woods Group (2005–2015)). The 3D co-ordinates for each co-complexed MBD are available in Supplementary data, Files SF1–SF23.
dThe array data are not consistent with the co-crystallized structure being an MBD.
Table II.
The level of agreementa,b between the experimental array data and the 3D structures, based on grafting the array glycans
| System | Dominant | Weak | Non-binderb | Total |
|---|---|---|---|---|
| UDA | ||||
| Agreement | 100% | 86% | 98% | 97% |
| Exp. total | 9 | 7 | 125 | 141 |
| PNA | ||||
| Agreement | 100% | 100% | 87% | 90% |
| Exp. total | 20 | 17 | 141 | 178 |
| DBA | ||||
| Agreement | 100% | 100% | 85% | 90% |
| Exp. total | 4 | 3 | 13 | 20 |
| ECL | ||||
| Agreement | 100% | 100% | 85% | 87% |
| Exp. total | 35 | 13 | 272 | 320 |
| CNL | ||||
| Agreement | 100% | 97% | 90% | 91% |
| Exp. total | 7 | 33 | 143 | 183 |
| MOA | ||||
| Agreement | 100% | 100% | 73% | 93% |
| Exp. total | 24 | 9 | 11 | 44 |
| BC2L-A | ||||
| Agreement | 85% | 100% | 95% | 93% |
| Exp. total | 20 | 1 | 65 | 86 |
| CTB | ||||
| Agreement | 100% | 100% | 100% | 100% |
| Exp. total | 2 | 1 | 6 | 9 |
| HPA | ||||
| Agreement | 100% | 100% | 99% | 99% |
| Exp. total | 33 | 3 | 163 | 199 |
| PSL | ||||
| Agreement | 100% | 100% | 0% | 84% |
| Exp. total | 34 | 2 | 7 | 43 |
| Total | ||||
| Agreement | 98% | 98% | 90% | 92% |
| Exp. total | 188 | 89 | 946 | 1223c |
aAgreement reported for using a 10° range when rotating the glycosidic linkages.
bAgreement is based on combined data for each MBD. Glycans that contain two MBDs are counted only once.
cArray glycans which contain an MBD but do not bind to the GBP.
d3D structures are available for each grafted glycan upon request.
Of the 1223 glycans that contained an MBD, 272 were observed to bind. These binders are classified as either dominant or weak binders, while non-binders are glycans that contain an MBD, but were not observed to bind.
The level of agreement is reliant in part on the categorization criteria used for discerning binders (dominant or weak) from non-binders in the experimental array data. This dependence is highlighted in the specific cases of Erythrina crista-galli lectin (ECL), Helix pomatia agglutinin (HPA), PSL and Urtica dioica agglutinin (UDA) discussed below. A recent cross-platform analysis of glycan arrays also indicated that the binding properties of glycans containing weak MBDs are less consistent (Wang et al. 2014). Regardless of the categorization criteria, discrepancies between grafting predictions and glycan array data may reflect a false positive with regard to the modeling, or a false negative with regard to the array screening.
An analysis of the 3D structures associated with the grafted glycan–protein complexes led to the discovery of extended binding sites, new MBDs, the identification of key MBDs in glycans that contain multiple copies, and the prediction of induced fit upon complex formation. These observations are illustrated for specific systems in the following sections.
MBD discovery: Dolichos biflorus agglutinin
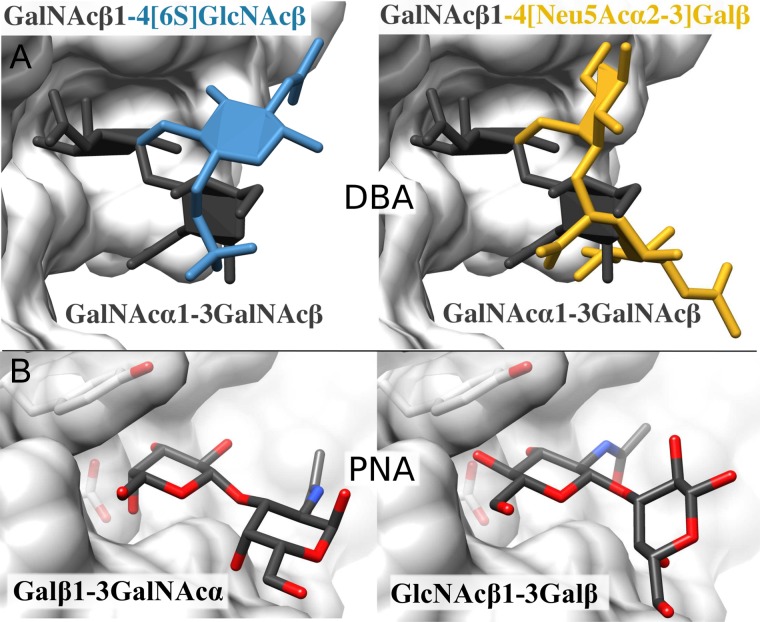
In some systems, it was not a straightforward task to achieve good agreement between the available 3D structures and the array data. In the case of DBA, the co-crystallized ligand was GalNAcα1–3GalNAcβ; however, it was not possible to completely explain the array data using this MBD alone. Two other structures were bound by DBA; GalNAcβ1–4[6S]GlcNAcβ and GalNAcβ1–4[Neu5Acα2–3]Galβ. It was initially difficult to reconcile these data, as the anomeric configuration of the glycosidic linkage gives rise to a different molecular shape, and other glycans containing GalNAcα or GalNAcβ were not observed to bind to DBA. A key observation was that glycans that lacked the sulfate moiety in the GalNAcβ1–4[6S]GlcNAcβ structure were not bound by DBA. After iteratively testing different possible binding modes, it was discovered that superimposing the non-reducing terminal GalNAc residues of each structure placed the 6S and Neu5Acα2–3 in the same coordinate space as the 3GalNAcβ of the co-crystallized MBD (Figure 2A). Grafting to these three binding modes results in 90% agreement with the array data, providing support for both the MBD selection and the proposed 3D structures.
Fig. 2.
The structural similarities between bound MBDs. (A) In the binding site of DBA (surface), the orientation the 6-sulfate in MBD2 (left) and the carboxylate group in MBD3 (right) occupy the same spatial region as the 3GalNAc in MBD1 (both panels). (B) The proposed orientation of MBD3 in the PNA-binding site (surface) was modelled on the crystal structure of MBD1 (left). Confidence in the prediction of this new MBD for PNA is driven by the high level of agreement (90%) between grafting and the array data, when MBD3 is included. This figure is available in black and white in print and in color at Glycobiology online.
Extended binding site discovery: Urtica dioica (Stinging nettle) agglutinin
UDA is known as a superantigen as it stimulates T-cells by mediating the interaction between MHC I/II and the β chain of the Vβ8.3+ lymphocytes (Rovira et al. 1999). UDA can bind to N-glycans present on the MHC I/II glycoprotein (Ryan and Cobb 2012), and simultaneously to those present on the T-cell surface (Rovira et al. 1999; Saul et al. 2000). Glycan array screening data show that UDA only interacts with oligomannose or certain under-processed N-glycans, and not with complex N-glycans (Figure 1C).
The endogenous function of UDA is to protect the plant against fungal infection by binding to fungal cell wall chitin (polymers of 4GlcNAcβ1–4GlcNAcβ) (Broekaert et al. 1989) and possibly also to the chitobiose core of N-linked glycans, as these are also present on fungal cell surfaces (Deshpande et al. 2008). Previous structural studies have shown how binding sites similar to that found in UDA can bind both chitin and N-glycan cores (Hernandez-Gay et al. 2010). In this case, the challenge was to understand how UDA can bind to the chitobiose core common to all N-linked glycans, but not bind to complex-type N-linked glycans.
Grafting of oligomannose glycans onto a chitobiose fragment in the UDA crystal structure (Hernandez-Gay et al. 2010) led to the conclusion that a single conformation (the gauche-gauche (gg) rotamer) of the flexible 1–6 branch was being specifically recognized by UDA. Grafting with this conformation successfully reproduced 98% of the relevant glycan array data. An examination of the proposed 3D structures for the oligomannose glycans in the binding site of UDA provided a clear structural rationalization for the observed inability of UDA to bind to complex-type glycans. Complex-type glycans are modified with extensions at the 2 position of the 1–6 branch, which according to grafting would not be tolerated in the binding site of UDA (Figure 1C).
By combining grafting with the glycan array data, it is possible to infer that there must be an extended binding site which binds the gg rotamer of the Manα1–6Manβ linkage. An extended binding site would explain the general increase in signal observed for high mannose glycans over chitin containing glycans in the array data. The proposed site is composed of Arg16, Asp26, Ser27 and Tyr30. This 3D structure matches the observed specificity, in that substitution at the 3- and 6-position of the Manα is physically tolerated, whereas substitution at the 2 position it is prevented by the backbone and side chain of Asp26 (Figure 3A).
Fig. 3.
(A) The proposed extended binding site of UDA is made up of Arg16, Asp26, Ser27 and Tyr30. (B) The co-crystal of GlcNAcβ1–4GlcNAcβ1–4GlcNAcβ (MBD2) and (C) proposed alignment of Galβ1–4GlcNAcβ1–3Galβ in UDA. Some of the molecular features required for recognition in MBD2 are also found in Galβ1–4GlcNAcβ1–3Galβ, which may explain the low-level binding by UDA to array glycans containing Galβ1–4GlcNAcβ1–3Galβ. This figure is available in black and white in print and in color at Glycobiology online.
The level of agreement with the array data for this 3D structure of MBD1 is excellent (98%). However, the binder categorization method used here also predicts a third MBD (Galβ1–4GlcNAcβ1–3Galβ) that is weakly bound by UDA, which, when included in the grafting analysis, does not produce results consistent with the array data. This is discussed in the Issues associated with binding categorization section.
MBD discovery: Arachis hypogaea (Peanut) agglutinin
The hemagglutinating activity of Arachis hypogaea (peanut) agglutinin (PNA) has been reported to be most potently inhibited by glycans containing Galβ1-3GalNAc (Lotan et al. 1975; Swamy et al. 1991). Inhibition by α1-acid glycoprotein, which lacks such Gal-GalNAc structures, but does contain Gal-GlcNAc structures (Jeanloz 1972), was also noted, but shown to be ∼50-fold less potent (Lotan et al. 1975). The disaccharides lactose (Galβ1–4Glc) and lactosamine (Galβ1–4GlcNAc) were shown to inhibit PNA hemagglutinating activity, again with ∼50-fold less potency than the Galβ1–3GalNAc disaccharide.
The experimental glycan array screening data show that, as expected, Galβ1–3GalNAc containing structures were present in dominant binders. These glycans contain either Galβ1–3GalNAcα (MBD1, also known as the cancer associated TF-antigen) or Galβ1–3GalNAcβ (MBD2). Contrary to expectation, grafting did not support lactose or lactosamine being an MBD. Unsubstituted lactose or lactosamine is present at the non-reducing termini of 74 glycans on the array, 37 of which are N-glycans. However, with the exception of three glycans that also contain MBD1, none of these glycans displayed significant binding to PNA on the array. This suggests that the affinity of lactose/lactosamine for PNA was insufficient for detection. However, a previously unreported MBD comprised of GlcNAcβ1–3Galβ (MBD3) lead to a high level (90%) of consistency between the grafting and array data. This MBD is present at the reducing termini of 13-weak binders, and a further 4-weak binders that also contain MBD1. It appears that avidity may be required for MBD3-containing glycans to generate a significant glycan array signal, as all but one of the experimentally observed MBD3 binders contains more than one instance of the determinant. This may explain why this glycan was not previously identified as a ligand for PNA using traditional means of determining affinity that neglect avidity effects, although to our knowledge its relative affinity has never been tested (Swamy et al. 1991).
As an experimental 3D structure for MBD3 bound to PNA was unavailable, a model based on MBD1 was built. In the MBD1 and MBD2 complexes, a hydrogen bond exists between the axial-O4 group in the non-reducing terminal Gal residue, which is distorted if not completely abrogated when the Gal residue is replaced by a GlcNAc residue in the MBD3 complex (Figure 2B). Interpretation of the glycan array data in light of the grafting results suggests that the interaction with the O4 group modulates affinity, with an axial configuration being strongly preferred but not essential. However, it should be noted that this conclusion assumes that the PNA lectin sample was pure. PNA is isolated from a complex mixture of isolectins using lactose. The affinity observed for MBD3 could be a result of another isolectin co-eluting with PNA.
The present grafting analysis suggests that PNA is less specific than previously thought, and provides a structural rationale for a new binding determinant (GlcNAcβ1–3Galβ, MBD3). This is particularly significant in light of the fact that PNA is used as a reagent thought to be able to selectively detect glycans which contain the cancer associated Galβ1–3GalNAcα (MBD1, TF-antigen) and Galβ1–3GalNAcβ (MBD2) (Cooper 1984; Gabius et al. 1990; Wu et al. 1997) motifs. Thus, the use of PNA as a reagent to exclusively detect Galβ1–3GalNAc-containing structures should be reconsidered, especially when studying biological samples, as avidity effects may be anticipated that would enhance binding to MBD3-containing glycans.
Detecting induced Fit in the glycan: Clitocybe nebularis lectin
CNL has been reported as having specificity for GalNAcβ1–4GlcNAc and GalNAcα1–3(Fucα1–2)Galβ (human blood group A determinant) and has been shown to inhibit leukemic T-cells (Pohleven et al. 2009) and stimulate dendritic cells (Svajger et al. 2011).
Analysis revealed two MBDs in the experimental glycan array data for CNL: GalNAcβ (MBD1) appeared in glycans in the dominant binder category, while GalNAcα (MBD2) only occurred in weak binders. The overall level of agreement when grafting to these MBDs was 91%, when using the default 10° range of rotational motion allowed in the grafted glycosidic linkages. However, it was noted that in the case of CNL the level of agreement was dependent on the maximal range of motion allowed. As might be expected, allowing greater flexibility in the glycosidic linkages of the grafted glycans generally enables more glycans to fit into the binding site. This enhances the number of correctly predicted binders, but conversely increases the number of non-binders that are incorrectly predicted to be binders. When this trend is observed in the grafting data, it indicates that the grafted branches are forming interactions with the protein that influence binding, rather than the branches being merely passively tolerated by the binding site topology. With this trend, and using the binders as a guide, grafting can predict when there must be an induced fit in either the glycan or the protein, or both, upon binding. For example, 20 of the 22 CNL binders contain the structure GalNAcα1–3(Fucα1–2)Galβ, and only when linkage flexibility was enabled were these correctly predicted to bind (see Table III). Closer examination indicated that flexibility removed initial overlaps between the Fucα branch and the protein surface. Thus the ability to score these branches as they are grafted, and allow flexibility in the protein sidechains, could increase the overall accuracy of grafting.
Table III.
Influence of maximum glycosidic rotation range on the level of agreement between grafting and the array data for CNL
| GBP name | Binding category | Rotation range |
Exp. total | ||
|---|---|---|---|---|---|
| 0° | 10° | 20° | |||
| Percentage agreement | |||||
| CNL | Dominant | 100 | 100 | 100 | 7 |
| Weak | 33 | 97 | 100 | 33 | |
| Non-binder | 92 | 90 | 76 | 143 | |
There is a general trend in the data showing that wiggling allows more binders and non-binders to fit. In such cases, this indicates that induced fit in the glycan or protein is required to optimize affinity. By generating 3D models for each glycan, grafting allows the user to identify the region of the protein responsible for the induced fit. With this insight it is possible, for example, to predict that a co-crystal of CNL with a glycan containing a Fucα1–2 branch would show induced fit in the Fuc and/or the His35, His36 and Asn122 region of the protein (Figure 4A).
Fig. 4.
(A) The Fucα1–2 substitution is tolerated experimentally and requires induced fit in the glycosidic linkage in order to physically fit in the 3D structure. Induced fit in the area of CNL made up of His35, His36, Asn122 and Ser123 could also explain the observed specificity. (B) Grafting to the ligand present in MOA structure 3EF2 indicated that a reorientation of the side chain of Asp23 would be required in order to tolerate a 1–4GlcNAcβ-substitution. A co-complex of MOA (PDBID 2IHO) with a 1–4GlcNAcβ-substituted oligosaccharide confirmed this prediction. This figure is available in black and white in print and in color at Glycobiology online.
Detecting induced fit in the protein: Marasmius oreades agglutinin
MOA is known as a blood group B-specific lectin that binds to glycans containing Galα1–3Gal(NAc) (Loganathan et al. 2003). In agreement with this, the experimental array data show that MOA has two dominant motifs: Galα1–3Galβ (MBD1) and Galα1–3GalNAcα (MBD2), and an additional weak motif: Galα1–3GalNAcβ (MBD3). The level of agreement between the array data and grafting for the binders was 100%, but it was noted that glycans containing a 4GlcNAcβ on the reducing terminal required wiggling to remove a collision of the carbonyl of the NAc group of the adjacent 4GlcNAcβ with Asp23.
The grafting analysis of the α-site in MOA is extremely sensitive to the position of Asp23. When co-crystallized with Galα1–3Galβ1–4GlcNAcβ in (PDBID: 2IHO) the position of Asp23 shifts, relative to its position in the co-complex with Galα1–3Galβ (PDBID: 3EF2; Grahn et al. 2009), so as to accommodate the 4GlcNAcβ in the former glycan (Figure 4B). Additionally, comparing with the optimal theoretical conformation of the Galα1–3Galβ1–4GlcNAcβ glycan, the crystallographic data reveal distortions of +29° and −13° in the glycosidic φ and ψ torsions of the Galβ1–4GlcNAcβ linkage, suggesting an induced fit in this linkage is required for binding by MOA. Thus although grafting can be sensitive to the initial 3D structure of the MBD co-complex, the grafting analysis can also identify the structural source of subsequent overlap-related discrepancies (such as the orientation of Asp23 in MOA). Thus the array data can be combined with grafting to highlight which residues in the glycan or protein require induced fit in order to agree with the array data.
Determining functional motifs: Burkholderia cenocepacia lectin
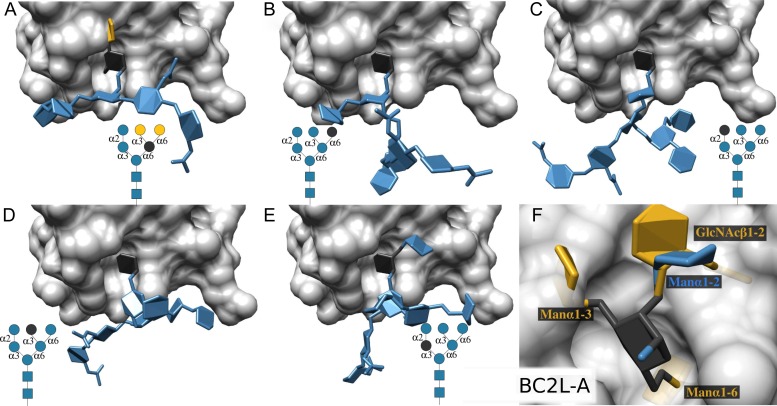
BC2L-A binds to Manα (Lameignere et al. 2008) which was the single MBD observed in the glycan array data. Each of the 20 dominant binders has multiple copies of the Manα-binding motif within its structure, suggesting an avidity component to the binding responses.
BC2L-A was predicted to bind not only to terminal Manα but also to a penultimate Manα depending on the substitution at the 2-position in the inner Manα. Specifically, BC2L-A may bind to Manα1–2Manα via either Manα, but cannot bind to the Manα in GlcNAcβ1–2Manα. Grafting also predicts that substitution at the 3- or 6-position of the bound Manα would not be tolerated. Taking one of the highest ranked N-glycan binders (213: manα1–6[Manα1–3]Manα1–6[Manα1–2Manα1–3]Manβ1–4GlcNAcβ1–4GlcNAcβ-Sp12) as an example, grafting is able to predict that BC2L-A can interact with four of the five Manα residues (Figure 5), although presumably each mode would have a unique affinity.
Fig. 5.
(A)–(E) Grafting of each Manα (indicted in dark grey) contained within glycan 213 to BC2L-A (transparent light gray surface). (A) The Manα which is branched at the 3- and 6-position is not tolerated in the BC2L-A binding site (non-tolerated residues in yellow). (B, C) The Manα1–6 on the non-reducing terminus of the 6-arm, and the Manα1–2 on the non-reducing terminus of the 3-arm, are predicted to be bound. (D, E) The Manα1–3 non-reducing terminus of the 6-arm, and the inner branched Manα residue on the 3-arm, are predicted to fit if induced fit is allowed in the glycosidic linkages. (F) A graphical summary of the specificity of BC2L-A; tolerated substitutions to a bound Manα are shown in blue, and non-tolerated substitutions are shown in yellow.
Examples illustrating issues associated with binding categorization
The definition of the boundary between dominant, weak and non-binder can significantly impact the accuracy of the predictions, as seen in the cases of HPA, PSL, ECL and UDA. These examples illustrate the complex case of assessing whether inconsistencies between the predicted and observed specificities represent a false positive on the behalf of grafting, or a false negative in the glycan array screening, or a mixture of both. Such discrepancies can arise from factors that are not currently modeled by grafting, such as induced fit in the protein surface, or electrostatic interactions. In addition, multimeric (avidity) effects are not accounted for in the modeling and may be required in order to amplify glycan array responses above the detection limit (Rillahan and Paulson 2011; Drickamer and Taylor 2014).
HPA binds to glycans containing GalNAc and GlcNAc on the array, which is consistent with reported specificity (Sanchez et al. 2006; Rambaruth et al. 2012). Considering only the two dominant MBDs, GalNAcα (MBD1) and GlcNAcα (MBD2), grafting resulted in up to 99% agreement with the array data. Weak binding responses were also observed for two glycans containing GalNAcβ and GlcNAcβ, but glycans containing these MBDs were not predicted by grafting to be binders. In this case, the MBDs are rejected by grafting as the level of agreement for the binders is <75%. The structures are not reported in Table I or included in the results for Table II.
PSL is observed to be specific for Neu5Acα2–6Galβ1–4Glc/GlcNAcβ structures (Mo et al. 2000) and also bound to Neu5Gcα2–6Galβ structures on the CFG array. Grafting identified all of the 36 binders, but also appeared to incorrectly predict that seven others should bind. Notably, four glycans (319, 488, 457 and 521, see Supplementary data, collated microarray data) had relative fluorescence unit (RFU) responses >10%, at the highest protein concentration, and two glycans (273 and 319) have been shown by FAC/MS to bind to PSL (Zhang et al. 2001). This suggests that at least five of the seven predicted ligands may indeed be capable of binding to PSL, but have RFU signals that are below the cut-off for definition as a binder.
ECL has been shown to bind Gal/GalNAcβ1–4GlcNAcβ (Wu et al. 2007), which is consistent with the observed binding pattern on the array. Grafting predicted all of the 48 binders and 85% of the 272 non-binders. The glycans with the highest RFU signals all contain multiple branches terminating in Galβ1–4GlcNAcβ, suggesting that avidity effects may be enhancing the array signal.
Of the 42 glycans predicted to bind, but not observed to, 14 showed binding responses >10% of the maximum at the highest protein concentration, but were not categorized as binders. A further four non-binders are sulfated at the 6 position of the non-reducing terminal residue of the MBD. Grafting placed the sulfate in close contact with the protein surface where it may form unfavorable electrostatic interactions with Asp89. The remaining 24 outliers are generally short glycans that contain only a single MBD in their structure, and may therefore lack the ability to form avidity-enhancing interactions with ECL.
UDA was discussed above in the Extended binding site discovery. Grafting to the proposed conformation of MBD1 (Manβ1–4GlcNAcβ1–4GlcNAcβ) and the crystal structure of MBD2 (GlcNAcβ1–4GlcNAcβ1–4GlcNAcβ) resulted in 97% agreement with the relevant glycan array data. However, weak binding was also observed for structures containing Galβ1–4GlcNAcβ1–3Galβ using the current categorization criteria. The proposed alignment of this oligosaccharide based on the central GlcNAcβ in each MBD reproduces the overall shape and some of the contacts formed by the dominant MBDs (Figure 3).
However, the majority of glycans that contain MBD3 did not display binding on the array, although grafting predicted that they would fit in the binding site. This led to a poor level of agreement (25%) when grafting to the 3D structure of Galβ1–4GlcNAcβ1–3Galβ suggesting that the interaction with UDA is too weak to produce a consistent binding pattern on the glycan array. While it is possible that the observed binding has a structural rationale (Figure 3), the lack of consistent binding on the array could indicate that the structure is not biologically relevant.
In this case, where it was not possible to achieve good agreement (>75%) between the 3D structure of a predicted MBD and the array data, the structure is rejected as an MBD. Thus, the structure is not reported in Table I or included in the results for Table II.
Discussion
We have outlined an approach to integrating 3D structural modeling with publically available specificity data from glycan array screening. The technique allows a unique exploitation of the array data to generate 3D structures, while simultaneously providing in-depth analysis of the array data from a 3D perspective. Putative 3D models are built based on the MBDs observed in the array data. Each of the binders and non-binders are grafted onto this model to check if they are physically tolerated in the binding site. Only binders should fit, and therefore it is possible to iteratively refine the 3D model of the bound MBD until a high level of agreement with the experimental data is found.
The grafting technique works best when using data from glycans containing a dominant MBD, but overall the modeling agreed with 92% of the 1223 data points from glycan array screening data. Selecting cut-offs or a set of criteria for discerning binders from non-binders in glycan array data sets is difficult. A simple categorization method was adopted here, as the qualitative or semi-quantitative nature of the array data precludes precise quantitative analysis. There are multiple potential sources of disagreement between the modeling and the experimental data. The cut-offs employed in discerning binders from non-binders may reduce accuracy when the experimental responses are close to the cut-off values. Both false positives and false negatives may arise from linker effects (Grant et al. 2014), printing efficiency, density effects, etc. (Heimburg-Molinaro et al. 2011). Disagreements may also arise from phenomena not included in the modeling, such as the difficulty of detecting low-affinity (monomeric) interactions in array screening (Rillahan and Paulson 2011; Taylor and Drickamer 2009). In our previous work, we provided a rationale for linker effects observed for an mAb binding to a disaccharide conjugated directly to the slide surface (Grant et al. 2014). However, the grafting program does not account for avidity effects (from multimeric interactions) that may be required for detectable binding on the microarray.
The high-level of agreement between the predicted and observed binders attests to the accuracy of the modeling, and indicates a need for revision of some MBD definitions, as in the case of PNA. However, a real source of concern when drawing conclusions regarding GBPs extracted from natural sources is whether or not they have been completely purified. For example in the case of PNA, the signals observed for MBD3 may result from a different lectin which co-elutes with PNA. Regardless, the insight from grafting provides an impetus for additional experimental analysis to determine if MBD3 is a biologically relevant ligand for PNA.
Text-based algorithms for the detection of MBDs (Porter et al. 2010; Cholleti et al. 2012; Kletter et al. 2013; Agravat et al. 2014) are extremely useful as a first pass at identifying binding motifs. However, these methods rely on carbohydrate nomenclature, which does not reflect the underlying 3D structural similarity possible between different sequences. As such, they cannot recognize structurally related 3D motifs that are not also composed of the same monosaccharides. As an example, wheat germ agglutinin binds both GlcNAc and Neu5Ac in identical manners, in which the binding motif is a subset of the molecular structures of these two monosaccharides. Such a relationship cannot be detected by algorithms that attempt to identify motifs based on nomenclature.
A further point to note is that when representing MBDs using just residue nomenclature, less than ∼20% of the glycans containing an MBD will be bound by a given GBP (Table II). The apparent high percentage of non-binders is also due to deficiencies in the nomenclature used to define MBDs, which ignore ∼80% of the rules for determining binding. Thus, a true definition of what constitutes an MBD should include both the underlying 3D structural properties that determine the affinity and also a definition of the tolerated and non-tolerated substitution patterns.
Grafting presents an opportunity for researchers to perform an in-depth specificity analysis of glycan array data and concurrently build experimentally consistent 3D structures of GBP–glycan co-complexes. A key feature of the approach presented here is that it enables the rationalization of glycan specificity in terms of the 3D structure of the glycan–protein complex. Such a putative 3D structure can potentially be validated subsequently using traditional structural biology approaches, but can also be employed for protein engineering and inhibitor design.
Materials and methods
Glycan microarray analysis
Biotinylated UDA, PSL and MOA were purchased from E.Y. Laboratories, PNA, and DBA from Vector Laboratories, and CTB and HPA from Sigma (see screen details in Supplementary data, collated microarray data for details). Lectins were analyzed by the Consortium for Functional Glycomics (CFG) Core D on the CFG Mammalian Glycan Array v5.0 using standard protocols (Blixt et al. 2004). The data for ECL and CNL were reported previously (Pohleven et al. 2012; Wang et al. 2014). To our knowledge, the data set for BC2L-A used in this study remains unreported; however, the original data from each glycan array screen may be reviewed in Supplementary data, collated microarray data or online at the CFG website (www.functionalglycomics.org, accessed 03 March 2016).
Criteria for system selection
As a quality control for the experimental specificity data, each of the GBPs had to have been screened at more than one concentration against the CFG glycan arrays (Blixt et al. 2004; Blixt and Razi 2006; Paulson et al. 2006) v4.0 or above (see Table I). Only systems for which a crystal structure was available for the GBP in complex with a fragment of a glycan, preferably the MBD(s) observed in the array data, were selected for analysis.
Classification of glycan array data into binding categories
Glycan array screening is frequently carried out at a series of GBP concentrations, with specificities being inferred from the lowest concentration that results in a significant signal (Smith et al. 2010; Rillahan and Paulson 2011). GBPs frequently have a range of affinities for different binding motifs with small structural alterations (Costello and Bundle 2012; Wang et al. 2014). At higher concentrations, it is common to observe an increase in both the number and intensity of signals, and in the range of apparent binding motifs (Rillahan and Paulson 2011). Unexpected, or anomalous motifs may be attributed to non-specific interactions, but may also represent low-affinity ligands that appear to bind only at high protein concentrations (Wang et al. 2014). In order to interpret the glycan array data, it is useful to divide the experimental data into categories that likely represent high-affinity motifs, or “dominant” binders, and any other motifs that are found in weaker binders, as well as non-binders. The data set for a particular protein concentration was considered unsuitable for analysis if the maximum signal was <2000 RFU. Also disregarded were single data points from a particular concentration for any binder whose signal variance was too high, as defined by having a percentage coefficient of variation >50%.
Dominant binders were defined as those that gave rise to signals >10% of the maximum signal at each analyzed GBP concentration. Weak binders were glycans whose signal was not >10% of maximum for the lower protein concentrations, but did show signals >10% at the top two GBP concentrations, or showed >50% of maximal binding at only the top concentration. When calculating the overall agreement between theoretical and experimental specificities, experimental non-binders are defined as glycans which contain at least one MBD, but do not display detectable binding to the GBP on the array. While these criteria are inherently arbitrary, they attempt to define reasonable boundaries.
MBD classification and modeling
Once the data were sorted into dominant, weak and non-binders, putative MBDs were manually identified as the smallest common motifs present in each of the binding glycans. The details of each system and any modeling required to convert the bound glycan fragment into the appropriate MBD are included in Supplementary data, Table SI.
Grafting
The concept of grafting and its methodology has been introduced previously (Tessier et al. 2013) and was recently extended so that the exact linker and array surface can be modelled (Grant et al. 2014). Briefly, virtual 3D structural libraries were constructed that contain glycans present on the CFG arrays. The list of array glycans was parsed into IUPAC format so that 3D structures could be generated and energy minimized using an automated version of GLYCAM-Web. Glycans were excluded from analysis if their structure was ambiguous due to annotation errors on the array or if it contained a residue not parameterized in GLYCAM06 (see Supplementary Information). This 3D library was searched via the GLYCAM residue nomenclature LINK cards contained within each generated file. Using the 3D structure of a GBP in complex with a glycan fragment, the grafting program first searches the 3D library for glycans that contain this fragment and then builds these larger glycans into the protein binding site. The program achieves this by superimposing the appropriate “branches” of the larger glycan onto the bound fragment. If the aligned glycan can be accommodated without significant steric overlap with the protein surface, then it is predicted to be a binder. In contrast to similar methods such as the “anchor and grow” approach available in certain computational docking software (Turton et al. 2004), grafting does not attempt to calculate affinity. Contributions from electrostatics, desolvation and entropy are purposefully excluded. Rather the program incorporates knowledge of observed binders and non-binders from glycan array screening data allowing it to report on the relative level of agreement between different 3D models and the array data.
Previously, the glycan and the GBP were maintained in a rigid conformation during the grafting analysis. The grafting program has been further modified to incorporate the natural flexibility of glycosidic linkages (DeMarco and Woods 2008; Nivedha et al. 2013) allowing them to adapt to the binding site topology. If the atoms of a grafted branch overlap with the binding site, grafting attempts to improve the fit of the glycan by “wiggling” each glycosidic torsion angle in 5° increments up to a maximum of 20°. The algorithm starts with the linkage closest to the core and proceeds along the branch towards the residue that is causing the overlap. Failure to reduce steric overlap to below the equivalent surface area of a buried carbon atom results in the glycan being classified as a non-binder. The grafting program then reports on the level of agreement between the predicted specificity the 3D structure and the results from glycan array screening.
Workflow - Integrating glycan array data to guide generation of GBP-glycan 3D structures
Experimental glycan array data are categorized into binders and non-binders.
A set of MBDs that collectively account for all the array binders are proposed.
3D structures for each MBD are taken from available crystal structures or modelled
A prebuilt 3D library of the glycan array is searched for the MBDs
- The 3D library glycans are grafted onto the co-complex from Step 3
- A 3D library glycan which fits in the binding site is deemed a binder
- Library glycans that contain an MBD but do not fit are deemed non-binders.
- Grafting reports on the level of agreement with experiment
- If agreement is poor: iteratively refine models generated in Step 3
- If iterative modeling fails to reproduce array data: re-evaluate MBDs defined in Step 2.
An MBD is disregarded in cases where iterative modeling fails to generate a 3D structure that is in good agreement (>75%) with the observed binding for the array binders and non-binders.
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
The authors are grateful to the National Institutes of Health (R01 GM100058 , GM094919 (EUREKA) and P41 GM103390) for financial support.
Conflict of interest statement
None declared.
Abbreviations
BC2L-A, Burkholderia cenocepacia lectin; CFG, consortium for functional glycomics; CNL, Clitocybene bularis lectin; CTB, Vibrio cholerae toxin subunit B; DBA, Dolichos biflorus agglutinin; ECL, Erythrina crista-galli lectin; GBP, glycan-binding protein; HPA, Helix pomatia agglutinin; MBD, minimal binding determinant; MOA, Marasmius oreades agglutinin; PNA, Arachis hypogaea agglutinin; PSL, Polyporus squamosus lectin; RFU, relative fluorescence units; UDA, Urtica dioica agglutinin.
Supplementary Material
Acknowledgements
The authors recognize the use of both Glyco3D (Pérez et al. 2015) and sugarbindDB (Shakhsheer et al. 2013). Molecular graphic images were produced using the UCSF Chimera (Pettersen et al. 2004) package from the Computer Graphics Laboratory, University of California, San Francisco (supported by NIH P41 RR01081). The computational carbohydrate grafting program is available online through the GLYCAM-Web suite of modelling tools at www.glycam.org/gr (accessed 03 March 2016).
References
- Agravat SB, Saltz JH, Cummings RD, Smith DF. 2014. GlycoPattern: A web platform for glycan array mining. Bioinformatics. 30:3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud J, Audfray A, Imberty A. 2013. Binding sugars: From natural lectins to synthetic receptors and engineered neolectins. Chem Soc Rev. 42:4798–4813. [DOI] [PubMed] [Google Scholar]
- Audfray A, Claudinon J, Abounit S, Ruvoen-Clouet N, Larson G, Smith DF, Wimmerova M, Le Pendu J, Romer W, Varrot A et al. 2012. Fucose-binding lectin from opportunistic pathogen Burkholderia ambifaria binds to both plant and human oligosaccharidic epitopes. J Biol Chem. 287:4335–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet N, Thomas B, Bossu I, Dufour E, Gillon E, Garcia J, Spinelli N, Imberty A, Dumy P, Renaudet O. 2013. High affinity glycodendrimers for the lectin LecB from Pseudomonas aeruginosa. Bioconjug Chem. 24:1598–1611. [DOI] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J et al. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 101:17033–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Razi N. 2006. Chemoenzymatic synthesis of glycan libraries. Methods Enzymol. 415:137–153. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Van Parijs J, Leyns F, Joos H, Peumans WJ. 1989. A chitin-binding lectin from stinging nettle rhizomes with antifungal properties. Science. 245:1100–1102. [DOI] [PubMed] [Google Scholar]
- Cholleti SR, Agravat S, Morris T, Saltz JH, Song X, Cummings RD, Smith DF. 2012. Automated motif discovery from glycan array data. OMICS. 16:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper HS. 1984. Lectins as probes in histochemistry and immunohistochemistry: The peanut (Arachis hypogaea) lectin. Hum Pathol. 15:904–906. [DOI] [PubMed] [Google Scholar]
- Costello C, Bundle DR. 2012. Synthesis of three trisaccharide congeners to investigate frame shifting of beta1,2-mannan homo-oligomers in an antibody binding site. Carbohydr Res. 357:7–15. [DOI] [PubMed] [Google Scholar]
- DeMarco ML, Woods RJ. 2008. Structural glycobiology: A game of snakes and ladders. Glycobiology. 18:426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande N, Wilkins MR, Packer N, Nevalainen H. 2008. Protein glycosylation pathways in filamentous fungi. Glycobiology. 18:626–637. [DOI] [PubMed] [Google Scholar]
- Drickamer K, Taylor ME. 2014. Convergent and divergent mechanisms of sugar recognition across kingdoms. Curr Opin Struct Biol. 28:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. 2005. Glycans in cancer and inflammation – Potential for therapeutics and diagnostics. Nat Rev Drug Discov. 4:477–488. [DOI] [PubMed] [Google Scholar]
- Fuster MM, Esko JD. 2005. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer. 5:526–542. [DOI] [PubMed] [Google Scholar]
- Gabius HJ, Andre S, Jimenez-Barbero J, Romero A, Solis D. 2011. From lectin structure to functional glycomics: Principles of the sugar code. Trends Biochem Sci. 36:298–313. [DOI] [PubMed] [Google Scholar]
- Gabius HJ, Schroter C, Gabius S, Brinck U, Tietze LF. 1990. Binding of T-antigen-bearing neoglycoprotein and peanut agglutinin to cultured tumor cells and breast carcinomas. J Histochem Cytochem. 38:1625–1631. [DOI] [PubMed] [Google Scholar]
- Geissner A, Anish C, Seeberger PH. 2014. Glycan arrays as tools for infectious disease research. Curr Opin Chem Biol. 18:38–45. [DOI] [PubMed] [Google Scholar]
- Grahn E, Askarieh G, Holmner A, Tateno H, Winter HC, Goldstein IJ, Krengel U. 2007. Crystal structure of the Marasmius oreades mushroom lectin in complex with a xenotransplantation epitope. J Mol Biol. 369:710–721. [DOI] [PubMed] [Google Scholar]
- Grahn EM, Winter HC, Tateno H, Goldstein IJ, Krengel U. 2009. Structural characterization of a lectin from the mushroom Marasmius oreades in complex with the blood group B trisaccharide and calcium. J Mol Biol. 390:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant OC, Smith HM, Firsova D, Fadda E, Woods RJ. 2014. Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data. Glycobiology. 24:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant OC, Woods RJ. 2014. Recent advances in employing molecular modelling to determine the specificity of glycan-binding proteins. Curr Opin Struct Biol. 28:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelryck TW, Loris R, Bouckaert J, Dao-Thi MH, Strecker G, Imberty A, Fernandez E, Wyns L, Etzler ME. 1999. Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus. J Mol Biol. 286:1161–1177. [DOI] [PubMed] [Google Scholar]
- Heimburg-Molinaro J, Song X, Smith DF, Cummings RD. 2011. Preparation and analysis of glycan microarrays. Curr Protoc Protein Sci. Chapter 12:Unit12 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gay JJ, Arda A, Eller S, Mezzato S, Leeflang BR, Unverzagt C, Canada FJ, Jimenez-Barbero J. 2010. Insights into the dynamics and molecular recognition features of glycopeptides by protein receptors: The 3D solution structure of hevein bound to the trisaccharide core of N-glycoproteins. Chemistry. 16:10715–10726. [DOI] [PubMed] [Google Scholar]
- Jeanloz R. 1972. Α 1-acid Glycoprotein. Amsterdam: Elsevier. [Google Scholar]
- Kadirvelraj R, Grant OC, Goldstein IJ, Winter HC, Tateno H, Fadda E, Woods RJ. 2011. Structure and binding analysis of Polyporus squamosus lectin in complex with the Neu5Ac{alpha}2-6Gal{beta}1-4GlcNAc human-type influenza receptor. Glycobiology. 21:973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KN, Woods RJ. 2001. Solvent interactions determine carbohydrate conformation. Proc Natl Acad Sci USA. 98:10541–10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletter D, Cao Z, Bern M, Haab B. 2013. Determining lectin specificity from glycan array data using motif segregation and GlycoSearch software. Curr Protoc Chem Biol. 5:157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma P, Müller-Loennies S. 2012. Anticarbohydrate antibodies: from molecular basis to clinical application. Wien: Springer-Verlag. [Google Scholar]
- Kumar S, Frank M, Schwartz-Albiez R. 2013. Understanding the specificity of human Galectin-8C domain interactions with its glycan ligands based on molecular dynamics simulations. PLoS ONE. 8:e59761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N, Hu D, Tateno H, Makyio H, Hirabayashi J, Kato R. 2013. Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity. FEBS Lett. 587:3620–3625. [DOI] [PubMed] [Google Scholar]
- Lameignere E, Malinovska L, Slavikova M, Duchaud E, Mitchell EP, Varrot A, Sedo O, Imberty A, Wimmerova M. 2008. Structural basis for mannose recognition by a lectin from opportunistic bacteria Burkholderia cenocepacia. Biochem J. 411:307–318. [DOI] [PubMed] [Google Scholar]
- Loganathan D, Winter HC, Judd WJ, Petryniak J, Goldstein IJ. 2003. Immobilized Marasmius oreades agglutinin: use for binding and isolation of glycoproteins containing the xenotransplantation or human type B epitopes. Glycobiology. 13:955–960. [DOI] [PubMed] [Google Scholar]
- Lotan R, Skutelsky E, Danon D, Sharon N. 1975. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J Biol Chem. 250:8518–8523. [PubMed] [Google Scholar]
- Merritt EA, Kuhn P, Sarfaty S, Erbe JL, Holmes RK, Hol WG. 1998. The 1.25 A resolution refinement of the cholera toxin B-pentamer: Evidence of peptide backbone strain at the receptor-binding site. J Mol Biol. 282:1043–1059. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Sarfaty S, van den Akker F, L'Hoir C, Martial JA, Hol WG. 1994. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 3:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H, Winter HC, Goldstein IJ. 2000. Purification and characterization of a Neu5Acalpha2-6Galbeta1-4Glc/GlcNAc-specific lectin from the fruiting body of the polypore mushroom Polyporus squamosus. J Biol Chem. 275:10623–10629. [DOI] [PubMed] [Google Scholar]
- Natchiar SK, Srinivas O, Mitra N, Surolia A, Jayaraman N, Vijayan M. 2006. Structural studies on peanut lectin complexed with disaccharides involving different linkages: Further insights into the structure and interactions of the lectin. Acta Crystallogr D Biol Crystallogr. 62:1413–1421. [DOI] [PubMed] [Google Scholar]
- Nivedha AK, Makeneni S, Foley BL, Tessier MB, Woods RJ. 2013. The importance of ligand conformational energies in carbohydrate docking: Sorting the wheat from the chaff. J Comput Chem. 35:526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA et al. 2012. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem Int Ed Engl. 51:4860–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R, Nettleship J. 2011. Functional and Structural Proteomics of Glycoproteins. The Netherlands: Springer. [Google Scholar]
- Paulson JC, Blixt O, Collins BE. 2006. Sweet spots in functional glycomics. Nat Chem Biol. 2:238–248. [DOI] [PubMed] [Google Scholar]
- Pérez S, Sarkar A, Rivet A, Breton C, Imberty A. 2015. Glyco3D: A portal for structural glycosciences. In: Lütteke T, Frank M, editors. Glycoinformatics. New York: Springer; p. 241–258. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera – A visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Pohleven J, Obermajer N, Sabotic J, Anzlovar S, Sepcic K, Kos J, Kralj B, Strukelj B, Brzin J. 2009. Purification, characterization and cloning of a ricin B-like lectin from mushroom Clitocybe nebularis with antiproliferative activity against human leukemic T cells. Biochim Biophys Acta. 1790:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohleven J, Renko M, Magister S, Smith DF, Kunzler M, Strukelj B, Turk D, Kos J, Sabotic J. 2012. Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N’-diacetyllactosediamine (GalNAcbeta1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis. J Biol Chem. 287:10602–10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A, Yue T, Heeringa L, Day S, Suh E, Haab BB. 2010. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 20:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promkuntod N, Wickramasinghe IN, de Vrieze G, Grone A, Verheije MH. 2013. Contributions of the S2 spike ectodomain to attachment and host range of infectious bronchitis virus. Virus Res. 177:127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaruth ND, Greenwell P, Dwek MV. 2012. The lectin Helix pomatia agglutinin recognizes O-GlcNAc containing glycoproteins in human breast cancer. Glycobiology. 22:839–848. [DOI] [PubMed] [Google Scholar]
- Richichi B, Imberty A, Gillon E, Bosco R, Sutkeviciute I, Fieschi F, Nativi C. 2013. Synthesis of a selective inhibitor of a fucose binding bacterial lectin from Burkholderia ambifaria. Org Biomol Chem. 11:4086–4094. [DOI] [PubMed] [Google Scholar]
- Rillahan CD, Paulson JC. 2011. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 80:797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira P, Buckle M, Abastado JP, Peumans WJ, Truffa-Bachi P. 1999. Major histocompatibility class I molecules present Urtica dioica agglutinin, a superantigen of vegetal origin, to T lymphocytes. Eur J Immunol. 29:1571–1580. [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Koeleman CA, Uh HW, Stam JC, van Heemst D, Maier AB, Houwing-Duistermaat JJ, Hensbergen PJ, Slagboom PE, Deelder AM et al. 2013. Targeted biomarker discovery by high throughput glycosylation profiling of human plasma alpha1-antitrypsin and immunoglobulin A. PLoS ONE. 8:e73082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SO, Cobb BA. 2012. Roles for major histocompatibility complex glycosylation in immune function. Semin Immunol. 34:425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Lescar J, Chazalet V, Audfray A, Gagnon J, Alvarez R, Breton C, Imberty A, Mitchell EP. 2006. Biochemical and structural analysis of Helix pomatia agglutinin. A hexameric lectin with a novel fold. J Biol Chem. 281:20171–20180. [DOI] [PubMed] [Google Scholar]
- Saul FA, Rovira P, Boulot G, Damme EJ, Peumans WJ, Truffa-Bachi P, Bentley GA. 2000. Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II. Structure. 8:593–603. [DOI] [PubMed] [Google Scholar]
- Shakhsheer B, Anderson M, Khatib K, Tadoori L, Joshi L, Lisacek F, Hirschman L, Mullen E. 2013. SugarBind database (SugarBindDB): A resource of pathogen lectins and corresponding glycan targets. J Mol Recognit. 26:426–431. [DOI] [PubMed] [Google Scholar]
- Smith DF, Song X, Cummings RD. 2010. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 480:417–444. [DOI] [PubMed] [Google Scholar]
- Suite 2011. Maestro, Version 9.2. New York, NY: Schrödinger, LLC. [Google Scholar]
- Svajger U, Pohleven J, Kos J, Strukelj B, Jeras M. 2011. CNL, a ricin B-like lectin from mushroom Clitocybe nebularis, induces maturation and activation of dendritic cells via the toll-like receptor 4 pathway. Immunology. 134:409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C, Teneberg S, Nilsson CL, Kjellberg A, Schwarz FP, Sharon N, Krengel U. 2002. High-resolution crystal structures of Erythrina cristagalli lectin in complex with lactose and 2’-alpha-L-fucosyllactose and correlation with thermodynamic binding data. J Mol Biol. 321:69–83. [DOI] [PubMed] [Google Scholar]
- Swamy MJ, Gupta D, Mahanta SK, Surolia A. 1991. Further characterization of the saccharide specificity of peanut (Arachis hypogaea) agglutinin. Carbohydr Res. 213:59–67. [DOI] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. 2009. Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology. 19:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier MB, Grant OC, Heimburg-Molinaro J, Smith D, Jadey S, Gulick AM, Glushka J, Deutscher SL, Rittenhouse-Olson K, Woods RJ. 2013. Computational screening of the human TF-glycome provides a structural definition for the specificity of anti-tumor antibody JAA-F11. PLoS ONE. 8:e54874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topin J, Arnaud J, Sarkar A, Audfray A, Gillon E, Perez S, Jamet H, Varrot A, Imberty A, Thomas A. 2013. Deciphering the glycan preference of bacterial lectins by glycan array and molecular docking with validation by microcalorimetry and crystallography. PLoS ONE. 8:e71149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton K, Natesh R, Thiyagarajan N, Chaddock JA, Acharya KR. 2004. Crystal structures of Erythrina cristagalli lectin with bound N-linked oligosaccharide and lactose. Glycobiology. 14:923–929. [DOI] [PubMed] [Google Scholar]
- Varrot A, Basheer SM, Imberty A. 2013. Fungal lectins: Structure, function and potential applications. Curr Opin Struct Biol. 23:678–685. [DOI] [PubMed] [Google Scholar]
- Wang L, Cummings RD, Smith DF, Huflejt M, Campbell CT, Gildersleeve JC, Gerlach JQ, Kilcoyne M, Joshi L, Serna S et al. 2014. Cross-platform comparison of glycan microarray formats. Glycobiology. 24:507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chinoy ZS, Ambre SG, Peng W, McBride R, de Vries RP, Glushka J, Paulson JC, Boons GJ. 2013. A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans. Science. 341:379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods Group. 2005. –2015. GLYCAM Web. Athens, GA: Complex Carbohydrate Research Center, University of Georgia; (http://www.glycam.org). [Google Scholar]
- Wu AM, Wu JH, Tsai MS, Yang Z, Sharon N, Herp A. 2007. Differential affinities of Erythrina cristagalli lectin (ECL) toward monosaccharides and polyvalent mammalian structural units. Glycoconj J. 24:591–604. [DOI] [PubMed] [Google Scholar]
- Wu W, Punt JA, Granger L, Sharrow SO, Kearse KP. 1997. Developmentally regulated expression of peanut agglutinin (PNA)-specific glycans on murine thymocytes. Glycobiology. 7:349–356. [DOI] [PubMed] [Google Scholar]
- Zhang B, Palcic MM, Mo H, Goldstein IJ, Hindsgaul O. 2001. Rapid determination of the binding affinity and specificity of the mushroom Polyporus squamosus lectin using frontal affinity chromatography coupled to electrospray mass spectrometry. Glycobiology. 11:141–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.