Abstract
Background:
One of the most common causes of hospital-acquired secondary infections in hospitalized patients is Pseudomonas aeruginosa. The aim of this study is to evaluate the expression of IMP and VIM in Pseudomonas aeruginosa strains (carbapenem resistant and producer MBL enzyme) in patients with secondary immunodeficiency.
Materials and Methods:
In a cross sectional study, 96 patients with secondary immunodeficiency hospitalized in the Al-Zahra hospital were selected. Carbapenem resistant strains isolated and modified Hodge test was performed in order to confirm the presence of the metallo carbapenemase enzyme. Under the standard conditions they were sent to the central laboratory for investigating nosocomial infection Multiplex PCR.
Results:
Of 96 samples 28.1% were IMP positive, 5.2% VIM positive and 3.1% both VIM and IMP positive. The prevalence of multidrug resistance in the IMP and/or VIM negative samples was 29%, while all 5 VIM positive samples have had multidrug resistance. Also the prevalence of multi-drug resistance in IMP positive samples were 96.3% and in IMP and VIM positive samples were 100%. According to Fisher’s test, the prevalence of multi-drug resistance based on gene expression has significant difference (P < 0.001).
Conclusion:
Based on the results of this study it can be concluded that, a significant percentage of patients with secondary immunodeficiency that suffer nosocomial infections with multidrug resistance, especially Pseudomonas aeruginosa, are probably MBL-producing gene positive. Therefore the cause of infection should be considered in the hospital care system to identify their features, the presence of genes involved in the development of multi-drug resistance and antibiotic therapy.
Keywords: Metallo-beta-lactamase, Pseudomonas aeruginosa, VIM, IMP
INTRODUCTION
One of the most common causes of hospital-acquired secondary infections in inpatients is Pseudomonas aeruginosa. Pseudomonas aeruginosa is a gram-negative, motile, aerobic and opportunistic hospital pathogen, which is sometimes resulting in deadly infections in their own host.[1] In addition, it is one of the infectious factors in patients with cystic fibrosis and it can also cause cancer and other diseases.[2,3,4]
Immunocompromised patients, including cancer patients, patients undergoing kidney transplant and immunosuppressive drug users due to general weakness and continuous use of immune-suppressing drugs are prone to different opportunistic infections, particularly Pseudomonas. Treatment of these infections is very difficult because of the resistance of these strains to antibiotics. However, several studies have shown that these strains have genes that produce metallo-beta-lactamase (MBL) and provide resistance to various antibiotics, especially carbapenems.
Antibiotic resistance in Pseudomonas aeruginosa can be originated or acquired chromosomal or with plasmid origin which in this case it can easily transferred between different strains and species of bacteria even between related genera. Recently, a strain of multidrug resistance (MDR) has been seen. Carbapenems, such as imipenem and meropenem are the most important antibiotics used to treat resistant strains of Pseudomonas aeruginosa.[5] Metallo-beta-lactamase hydrolyses wide range of beta-lactamase (antibiotics, anti-cell wall) including cephalosporins, carbapenems and penicillins.[1,6] It also causes resistance to aminoglycosides. However, it is unable to hydrolyse monobactem like Aztreonam.[1] Metallo-beta-lactamase requires the Zn metal cofactor for its catalytic activity that is inhibited by EDTA and thiol compounds. Carbapenamase class B are designed of metallo-beta-lactamase and are divided in 6 groups based on the molecular structure including AIM, APM, GIM, SIM, IMP and VIM; which the most common ones are IMP and VIM.
Due to the mobile genetic nature, Pseudomonas aeruginosa can transmit these enzymes and transfer a variety of genes and transposons into their own genome. It results increasing in resistant strains.[6] Genes encoding MBL are located on integron class 1, on plasmids which are able to transfer them.[7,8] There are several available phenotypic methods which can identify and isolate MBLs. However, these days PCR is used as a sensitive and reliable method.[9,10]
The organisms which produce Metallo carbapenemase are often Multidrug Resistant. Therefore, there are many limitation in choosing appropriate antibiotic to care it, even though the mortality rate of carbapenemase producing organisms infection is high.[11,12] In fact metallo carbapenemase producing organisms can cause resistance to a wide range of antibiotics. This occurs because of hydrolysing carbapenem as well as broad-spectrum penicillin such as oxymino - cephalosporins and cephamycins.[13]
According to microbiological methods and antibiotic susceptibility tests, Pseudomonas aeruginosa resistant to carbapenems were reported in patients suffered immune deficiency. Identification of these enzymes in microbiology laboratories either by conventional susceptibility testing or automatic methods is complicated. Awareness of the genetic mechanisms responsible for resistance to antibiotics can effectively facilitate prevention and control of infectious diseases in immunocompromised patients who are highly susceptible to infection. This study is performed using molecular PCR methods and standard identification method based on molecular techniques including PCR based Metallo carbapenemase.[14,15] The aim of this study is to evaluate the expression of IMP and VIM in Pseudomonas aeruginosa strains (carbapenem resistant and producer MBL enzyme) in patients with secondary immunodeficiency in the Al-Zahra hospital, Isfahan, Iran.
MATERIALS AND METHODS
Study design
This is a cross-sectional study which was conducted in 2014 - 2015 in the Al-Zahra hospital, Isfahan, Iran. The study population is Pseudomonas aeruginosa isolated from patients with secondary immune deficiency. Inclusion criteria were Pseudomonas aeruginosa strains (carbapenem resistantant and producer MBL enzyme) and good quality samples for PCR. Lack of good quality samples for PCR and samples contaminated with psudomonance were considered as exclusion criteria. The required sample size was estimated 96 patients based on 95% confidence level, the prevalence of the most common gene (VIM) in the same study as 52%[16] and considering acceptance error rate of 0.1. This study was approved by Ethical Committee of Isfahan University of Medical Sciences.
Bacterial isolates
Pseudomonas aeruginosa isolates were obtained from patients with organ transplantation, receiving immunosuppressive drugs, chemotherapy and corticosteroids (with immunosuppressive dose 15 mg of prednisolone for at least one month). To identify the isolates of Gram-negative bacteria and ensure the genera and species of bacteria, standard biochemical tests were performed and antibiotic resistance of clinical isolates were determined using Kirby Bauer disk diffusion test.
Antibiotic susceptibility tests
Carbapenem resistant strains were isolated and modified Hodge test was performed on them to confirm the presence of the metallo carbapenem enzyme. Under the standard conditions they were sent to the central laboratory in order to evaluate nosocomial infection Multiplex PCR.
DNA extraction
To extract bacterial DNA (plasmid and chromosomal) boiling method was used. First, after a passage of samples on blood agar, a single colony of each isolates was inoculated to five ml of luriabertani medium and incubated at 37°C for 20 hours on a shaker. Then, 1.5 ml of bacterial culture was transferred to the eppendorf tubes. The eppendorf tubes were centrifuged for 5 min at 14,000 rpm and sediment was placed in suspension state using thermoblock machine for 10 minutes at 95°C. The eppendorf tubes were then centrifuged again for 5 min at 14,000 rpm and subsequently the DNA in the supernatant used for the PCR.
PCR reaction for confirmation of pseudomonas aeruginosa strains
PCR was performed using DNA sample, specific primers and regents followed by gel electrophoresis analysis. Amplification was carried out using the following thermal cycling conditions: Initial DNA denaturation at 94°C for 10 min, followed by 36 cycles of denaturation at 94°C for 30 sec, annealing at 52°C for 40 sec, and extension at 72°C for 50 sec, followed by final extension at 72°C for 5 min. PCR reactions for bla-IMP and bla-VIM genes were performed containing the following.
bla-IMP. forward (GGAATAGAGTGGCTTAAYTCTC) and reverse (GGTTTAAYAAAACAACCACC) primers (designed) 232bp.
bla-VIM. forward (GATGGTGTTTGGTCGCATA) and reverse (CGAATGCGCAGCACCAG) primers (designed) 390bp.
Statistical analysis
Collected data was analysed using SPSS software version 22. Chi-square and T-student tests were applied to analyse data. P-value less than 0.05 was considered as a statistical significant level.
RESULTS
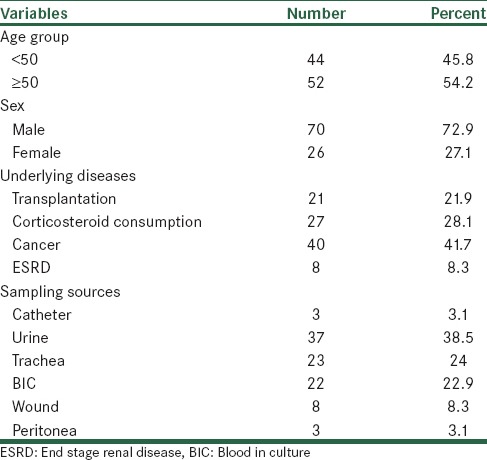
In the current study, 96 inpatients with secondary immunodeficiency in the Al-Zahra hospital participated. The mean age of the patients was 49.39 ± 16. years (range: 7-86 years).44 patients (45.8%) were less than 50 years old and 52 patients (54.2%) were aged 50 years and older. 70 (72.9%) patients were male and 26 (27.1%) were female. The underlying cause of immunodeficiency in 40 patients (41.7%) was cancer, followed by steroid use in 27 patients (28.1%), organ transplantation in 21 patients (21.9%) and ESRD in 8 patients (8.3%).
Source samples in 37 patients (38.5%) were urine, followed by trachea (24%), blood (22.9%), wound (8.3%) and peritoneal fluid (1.3%). In Table 1, the demographic characteristics of study patients have been shown.
Table 1.
Frequency distribution of demographic characteristics of study patients
PCR detection of metallo-beta-lactamase genes
Of 96 samples 27 (28.1%) were IMP positive, 5 cases (5.2%) were VIM positive and 3 samples (3.1%) were both VIM and IMP positive [Figure 1]. The source of 27 IMP positive samples were 1 catheter, 10 urine, 6 trachea, 6 blood, 3 wound and 1 peritoneal fluid. Of 5 VIM positive samples, 2 sources were urine, 2 tracheas and 1 blood. Of 3 IMP and VIM-positive samples, 2 sources were urine and 1 was trachea [Figure 2].
Figure 1.
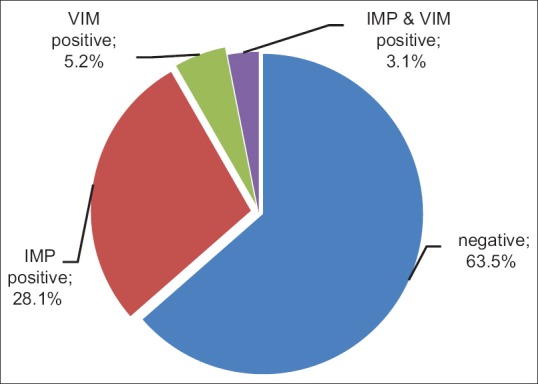
Frequency percentage of VIM and IMP expression in the studied patients
Figure 2.
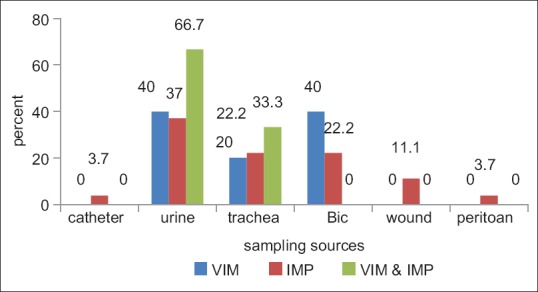
VIM and IMP expression based on source of sampling
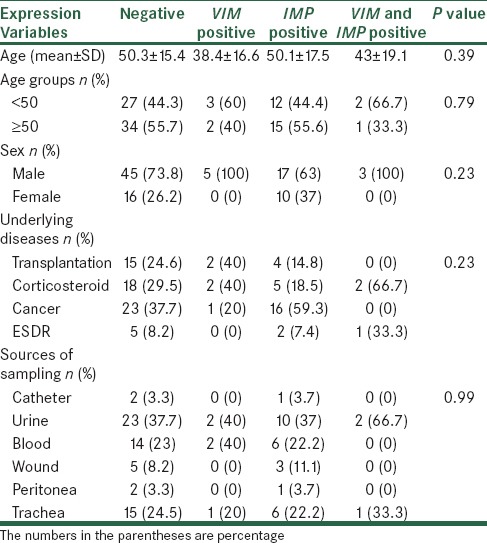
In Table 2, frequency distribution of IMP and VIM gene expression in terms of demographic and clinical characteristics has been shown. Based on the results, positive VIM patients had a lower age average, but the difference between four groups with different underlying diseases was not statistically significant. Also gene expression of studied genes in terms of other demographic and clinical characteristics of the patients was not significantly different.
Table 2.
Gene expression of IMP and VIM genes in terms of demographic and clinical characteristics
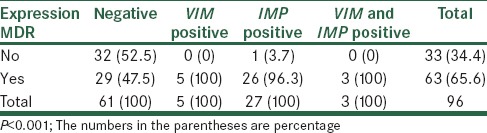
According to the results of Antibiogram test, 63 (65.6%) samples were found to have multidrug-resistant. The prevalence of multidrug resistance in the VIM and/or IMP negative samples was 47.5%, while all 5 VIM positive samples have had multidrug resistance. Also the prevalence of multi-drug resistance in IMP positive samples was 96.3% and in IMP and VIM positive samples were 100%. According to Fisher’s test, the prevalence of multi-drug resistance based on gene expression has significant difference (P < 0.001). The results are shown in Table 3.
Table 3.
Distribution of multi-drug resistance based on gene expression of IMP and VIM genes
DISCUSSION
Nosocomial infections in immunocompromised patients are highly prevalent and infection with opportunistic infections, particularly Pseudomonas aeruginosa can be associated with high morbidity and mortality rates, so this study was designed and carried out in order to determine gene expression of IMP and VIM genes and determining their relationship with antibiotic resistance.
Our finding showed that 8.3% of patients had VIM gene, 31.3% IMP gene and 3 cases had both genes.
Antibiogram test showed 65.6% of patients had multi-drug resistance (MDR) and it depends entirely on these gene expressions. The relationship between the presence of these genes and multidrug resistance has been demonstrated in several studies. In a study conducted by Doosty et al. in 2013 in the Valiasr hospital, 70 isolates of Pseudomonas aeruginosa from patients hospitalized in intensive care unit were studied. Among 44 isolates resistant to imipenem, 36 isolates were producing MBL enzyme. by performing PCR, 33 strains MBLs producing P. aeruginosa were identified as a positive phenotype, that 23 isolated genes (52.2%) were VIM positive and 10 isolated genes (22.3%) were IMP positive.[16] other study which was done by Yazdani et al. in 2014 in the Al-Zahra hospital on 46 samples of P. aeruginosa isolated from patients with primary immunodeficiency reported that 7 patients (14.6%) had VIM gene and 8 patients (17.4%) had IMP gene. Also, MDR samples with these genes were significantly higher.[17] A study in Zahedan, Iran showed that the rate of resistance against imipenem was 5.7% and after carrying out the phenotypic experiments, nine species were identified as of MBL producer. Seven species were confirmed by Polymerase Chain Reaction (PCR) method. Gene VIM-1 was the predominant gene among the positive (antibiotic resistant) species.[18] Samira Aghamiri and et al. in Tehran reported that in the DDST phenotypic method, among the 100 imipenem resistant isolates, 75 strains were MBLs positive. The PCR test indicated that 70 strains (33%) carried bla-VIM gene and 20 strains (9%) harbored bla-IMP. In addition, the extent of antibiotic resistance among Pseudomonas aeruginosa is on the rise.[19] In a study carried out by Mai et al. In Egypt, blaVIM and blaOXA genes were the most prevalent genes in P. aeruginosa.[20] Other study conducted by Hassan et al. detected that the first plasmid-mediated quinolone-resistance determinants qnrB1 and qnrS1 among clinical strains obtained from patients in Turkey.[21] In a study by esther et al. between 2007 and 2010 in Spain, 175 pseudomonans aeruginosa samples were studied. They found a significant difference between multidrug resistance and presence of VIM-2 gene.[22] Therefore, there can be different genes, especially two common genes VIM and IMP, which may eventually contribute to producing Carbapenemase enzymes and development of MDR Pseudomonans aeroginosa. As a result, identification of these genes in patients with secondary immune deficiency could be a solution to deal with multi-drug resistance in these patients and prevention of deaths caused by nosocomial infection. Today metalloproteinase carbapenemase-producing organisms have been expanding and are endemic in several European countries, the Middle East, South America and Asia that causes a serious challenge in the diagnosis and treatment of nosocomial infections. On the one hand, choosing the drug for treatment of these infections are inherently limited. On the other hand due to the pharmacological properties or toxicity of these drugs, their clinical use is limited.[23] Considering the results, the MBL-producing genes of IMP and VIM, probably play a significant role in the incidence of multi-drug resistance in patients with secondary immunodeficiency. Therefore, further studies are strongly recommended in patients with secondary immunodeficiency, with clinical signs of infection. In addition to bacteriological cares, Multiplex PCR for the detection of beta-lactamase-producing gene and the Metallo-Beta-lactamase-producing gene should be performed, although these tests are faced with significant limitation in term of funding and financial support.
CONCLUSION
Based on the results of this study it can be concluded that, a significant percentage of patients with secondary immunodeficiency that suffer nosocomial infections with multidrug resistance, especially Pseudomonas aeruginosa, are probably MBL-producing gene positive. according to importance of hospital infections and high mortality rate of infections in immunocompromised patients, It is recommended that the hospital care system in infection control, particularly in ICUs be further strengthened and by observing clinical signs of infection, immediate action to identify the cause of infection and their features, the presence of genes involved in the development of multi-drug resistance and antibiotic therapy should be evaluated followed by antibiotic therapy for these patients according to strict and systematically protocol.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Rodrigues AC, Chang MR, Nobrega GD, Rodrigues MS, Carvalho NC, Gomes BG, et al. Metallo-beta-lactamase and genetic diversity of Pseudomonas aeruginosa in intensive care units in Campo Grande, MS, Brazil. Braz J Infect Dis. 2011;15:195–9. doi: 10.1016/s1413-8670(11)70174-x. [DOI] [PubMed] [Google Scholar]
- 2.Woods D E, Iglewski B. HToxins of Pseudomonas aeruginosa: New perspectives. Reviews of Infectious Disease. 1983;5:714–22. doi: 10.1093/clinids/5.supplement_4.s715. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–60. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 4.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 5.Khosravi AD, Mihani F. Detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn Microbiol Infect Dis. 2008;60:125–8. doi: 10.1016/j.diagmicrobio.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–35. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng X, Wang P, Wang Y, Zhang H, Tao C, Yang W, et al. Identification and distribution of the clinical isolates of imipenem-resistant Pseudomonas aeruginosa carrying metallo-beta-lactamase and/or class 1 integron genes. J Huazhong Univ Sci Technolog Med Sci. 2008;28:235–8. doi: 10.1007/s11596-008-0301-8. [DOI] [PubMed] [Google Scholar]
- 8.Bahar MA, Jamali S, Samadi kuchaksaraei A. Imipenem-resistant Pseudomonas aeruginosa strains carry metallo-beta-lactamase gene bla (VIM) in a level I Iranian burn hospital. Burns. 2010;36:826–30. doi: 10.1016/j.burns.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Khan AA, Cerniglia CE. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of exotoxin A gene using PCR. Appl Environ Microbiol. 1994;60:3739–45. doi: 10.1128/aem.60.10.3739-3745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vos D, Lim A, Jr, Pirnay JP, Struelens M, Vandenvelde C, Duinslaeger L, et al. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J ClinMicrobiol. 1997;35:1295–9. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borer A, Saidel-Odes L, Riesenberg K, et al. Attributable mortality rate for carbapenem-resistant Klebsiella pneumonia bacteremia. Infect Control Hosp Epidemiol. 2009;30:972–6. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 12.Schwaber MJ, Klarfeld-Lidji S, Navon-VeneziaS, et al. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother. 2008;52:1028–33. doi: 10.1128/AAC.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnin RA, Nordmann P, Potron A, et al. Carbapenem-hydrolyzing GES-type extended-spectrum beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:349–54. doi: 10.1128/AAC.00773-10. Epub 2010 Oct 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuzon G, Ouanich J, Gondret R, et al. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumonia isolates in France. Anti microb Agents Chemother. 2011;55:2420–3. doi: 10.1128/AAC.01452-10. Epub 2011 Feb 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miriagou V, Cornaglia G, Edelstein M, et al. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. ClinMicrobiol Infect. 2010;16:112–22. doi: 10.1111/j.1469-0691.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- 16.Doosti M, Ramezani A, faghihi M. Metallo-beta-lactamase and genetic diversity of Pseudomonas aeruginosa in intensive care units in Valiasr Hospital in Zanjan. J Kordestan University Med Sci. 2013;18:97–105. [Google Scholar]
- 17.Yazdani M, Shabani SH. Detection of different types of Metallo β Lactamases among Pseudomonas aeruginosa isolates clinical from Intensive Care Units patients. Thesis for obtaining of special degree in infectious diseses, medical school of Isfahan University of Medical sciences, 2014 [Google Scholar]
- 18.Ghamgosha M, Shahreki zahedani S, Kafilzadeh F, Bameri Z, Taheri RA, Farnoosh G. Metallo-beta-Lactamase VIM-1, SPM-1, and IMP-1 Genes Among Clinical Pseudomonas aeruginosa Species Isolated in Zahedan, Iran. Jundishapur J Microbiology. 2015;8:e17489. doi: 10.5812/jjm.8(4)2015.17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samira Aghamiri, Nour Amirmozafari, Jalil Fallah Mehrabadi, Babak Fouladtan, Hossein Samadi Kafil. Antibiotic Resistance Pattern and Evaluation of Metallo-Beta Lactamase Genes Including bla-IMP and bla-VIM Types in Pseudomonas aeruginosa Isolated from Patients in Tehran Hospitals. ISRN Microbiology 2014. 2014:1–6. doi: 10.1155/2014/941507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai M, Zafer, Mohamed H, Al-Agamy, Hadir A. El-Mahallawy, Magdy A, Amin, Mohammed Seif El-Din Ashour. Antimicrobial Resistance Pattern and Their Beta-Lactamase Encoding Genes among Pseudomonas aeruginosa Strains Isolated from Cancer Patients. BioMed Research International Volume 2014. doi: 10.1155/2014/101635. Article ID 101635:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasan Nazik. Betigül Öngen and Nuray Kuvat. nvestigation of Plasmid-Mediated Quinolone Resistance among Isolates Obtained in a Turkish Intensive Care Unit. Jpn J Infect Dis. 2008;61:310–2. [PubMed] [Google Scholar]
- 22.Esther V, Carlos J, Joaquín R, Antonio O, Fernando Ch. Draft Genome Sequence of VIM-2-Producing Multidrug-Resistant Pseudomonas aeruginosa ST175, an Epidemic High-Risk Clone. Genome Announc. 2013;1:e00112–13. doi: 10.1128/genomeA.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beirão EM, Furtado JJ, Girardello R, et al. Clinical and microbiological characterization of KPC-producing Klebsiella pneumoniae infections in Brazil. Braz J Infect Dis. 2011;15:69–73. doi: 10.1016/s1413-8670(11)70143-x. [DOI] [PubMed] [Google Scholar]


