Abstract
The new generic Total Nucleic Acid Isolation kit improved the detection limit of a cytomegalovirus (CMV) PCR from 400 to 200 copies/ml. Sensitivity and specificity in 30 CMV-positive and 100 negative samples were 100%. Analytical performance was excellent. The kit provides a reliable, standardized, and time-saving tool for DNA extraction.
PCR is widely used in all diagnostic settings and has become highly automated in routine diagnostics. However, most assays require extensive hands-on time due to labor-intensive nucleic acid isolation from the sample (4).
We report here the evaluation of a new fully automated generic extraction using the Total Nucleic Acid Isolation (TNAI) kit on the COBAS AmpliPrep instrument (both from Roche Diagnostics, Mannheim, Germany), based essentially on the method developed by Boom et al. (1). The extracted nucleic acids may thus serve for different RNA or DNA PCRs. The kit was evaluated in combination with cytomegalovirus (CMV) DNA quantification (COBAS AMPLICOR CMV MONITOR; Roche Diagnostics) (2, 3, 5, 7). For evaluation, analytical performance as well as the stability of working solutions and extracted DNA were determined. Finally, 30 CMV-positive samples were compared by using TNAI with the manual method (included in the CMV-PCR kit) as a reference (4, 6).
Analytical performance.
The lower limit of detection was determined by using serial dilution of CMV DNA (Pelispy CMV DNA monitor, 10,000 copies/ml; VQC International, Alkmaar, The Netherlands) with eight replicates by employing a probit analysis algorithm. In order to obtain reliable results, the volume of the CMV quantitation standard was adjusted to 400 μl per 4,000 μl of TNAI diluent for 48 determinations. The correlation between the sample volume and the detection limit was optimal at 200 μl with a detection limit of 200 copies/ml (95% hit rate), using an EDTA plasma matrix; when using a citrate plasma matrix the sensitivity increased slightly to 280 copies/ml with the same input volume. Increasing the sample volume to 500 μl and 850 μl further improved the sensitivity to around 110 copies/ml in EDTA plasma. However, since the lowest limit of the PCR calibration curve is at 200 copies/ml, tests with a lower concentration can hardly be quantified correctly by this method. All further analyses were performed using 200-μl samples.
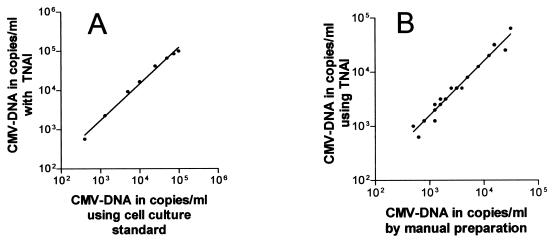
To determine linearity, EDTA and citrate plasma samples were spiked with a dilution series of the cell culture CMV stain AD 196 (VR-538; American Type Culture Collection), employing linear regression analysis. Samples were processed in three replicates/run. Linearity ranged from 400 copies/ml to 100,000 copies/ml for ETDA plasma (r2 = 0.99) (Fig. 1A) as well as for citrate plasma (r2 = 0.99). Within this range, the measured values exhibited intrasignal deviation of ≤0.3 log.
FIG. 1.
Performance of the automated generic TNAI kit. (A) Dynamic range using EDTA plasma spiked with a dilution series from cell culture virus (each dilution was tested in three replicates). (B) Correlation between the manual extraction and TNAI results for 23 clinical EDTA plasma samples that were positive by both methods.
Precision was determined with the low and high positive controls of the kit as well as with dilution series of CMV DNA from cell culture virus, using variance component analysis. Intra-assay variances determined for 21 replicates per sample were 6.9% (using 75,000-copies/ml input concentration), 9.3% (10,000 copies/ml), and 23% (1,000 copies/ml), respectively. Interassay variances were done with two replicates of each specimen in five runs with 9.6% (75,000 copies/ml), 7.0% (10,000 copies/ml), and 18.7% (1,000 copies/ml), respectively.
PCR inhibition was tested by using 100 routine blood donations (Bavarian Blood Donation Service, Munich, Germany). No PCR inhibition was detectable.
The stability of the working solutions and of the extracted DNA was tested with the low and the high positive controls. The working solution was stable for at least 3 days on board the instrument (28 to 32°C). CMV DNA in the eluate was stable for 24 h on board (28 to 32°C), allowing overnight operations, and for at least 7 days in the fridge (2 to 8°C). In all stability analyses the target recovery deviated by less than 0.15 log compared to the unstressed sample.
Specificity and sensitivity in clinical samples.
Specificity was determined in 100 samples from healthy blood donors. All samples were CMV DNA negative; specificity was thus 100% (confidence interval, 96.4 to 100%).
For sensitivity, results from 30 EDTA plasma samples from transplant recipients with a proven CMV infection (established by other methods: pp65 antigen detection and CMV PCR from leukocytes) (7) were compared with TNAI and with the manual extraction method (Fig. 1B). Four samples were negative with both extraction methods. This is in line with the well-known fact that detection of CMV DNA in plasma during CMV replication is limited to a shorter period of time than the detection of CMV antigen or CMV DNA in leukocytes (7). However, three samples were repeatedly CMV DNA positive by TNAI but repeatedly negative by the manual method. Thus, as suggested by the analytical data, the TNAI method confirmed its higher sensitivity in clinical samples. Moreover, a strong correlation between the two methods was observed when comparing the viral loads of the 23 clinical samples positive with both methods (r2 = 0.98).
As major advantages, the TNAI kit extracts DNA and RNA simultaneously, thus enabling different PCRs from one extraction, and it reduces the overall processing time for 24 samples from 150 min with the manual method to 90 min. Moreover, it includes an in-process control, since the internal quantification standard is added at the first step of extraction, resulting in high reliability and precision.
Taken together, the TNAI method meets the general requirements for nucleic acid extraction systems (3, 5), showing stability of solutions and the eluated DNA under routine conditions, high dynamic range in quantification, and high precision in intra-assay and interassay variances. Moreover, viral load correlated excellently between TNAI and the manual method.
Importantly, the sensitivity of the PCR was improved by using TNAI in the analytical performance as well as with clinical samples.
In this study the performance of the extraction kit was evaluated with the help of CMV DNA detection. The data may apply to DNA extraction in general. However, future evaluation should be extended to RNA as the target.
Taken together, the TNAI kit combines an accurate, sensitive, and standardized delivery system of nucleic acids for PCR in an easy-to-handle automated open nucleic acid extraction kit.
Acknowledgments
This work was supported by Roche Diagnostics, Penzberg, Germany.
We are indebted to Martina Walter, Sabine Boehm, Annedore Bender, and Michael Wiedmann for analyzing the samples and for critical review of the manuscript.
REFERENCES
- 1.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiDomenico, N., H. Link, R. Knobel, T. Caratsch, W. Weschler, Z. G. Loewy, and M. Rosenstraus. 1996. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin. Chem. 42:1915-1923. [PubMed] [Google Scholar]
- 3.Hiyoshi, M., S. Tagawa, T. Takubo, K. Tanaka, T. Nakao, Y. Higeno, K. Tamura, M. Shimaoka, A. Fujii, M. Higashihata, Y. Yasui, T. Kim, A. Hiraoka, and N. Tatsumi. 1997. Evaluation of the AMPLICOR CMV test for direct detection of cytomegalovirus in plasma specimens. J. Clin. Microbiol. 35:2692-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction—our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Sia, I. G., J. A. Wilson, M. J. Espy, C. V. Paya, and T. F. Smith. 2000. Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J. Clin. Microbiol. 38:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelzl, E., A. Kormann-Klement, J. Haas, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Von Muller, L., W. Hampl, J. Hinz, H. Meisel, A. Reip, E. Engelmann, R. Heilbronn, B. Gartner, O. Kramer, H. Einsele, H. Hebart, T. Ljubicic, J. Loffler, and T. Mertens. 2002. High variability between results of different in-house tests for cytomegalovirus (CMV) monitoring and a standardized quantitative plasma CMV PCR assay. J. Clin. Microbiol. 40:2285-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]