Abstract
Background:
With the advent of DNA-based culture-independent techniques, a constantly growing number of Selenomonas phylotypes have been detected in patients with destructive periodontal diseases. However, the prevalence levels that have been determined in different studies vary considerably.
Aim:
The present study was undertaken to detect and compare the presence of Selenomonas sputigena in the subgingival plaque samples from generalized aggressive periodontitis (GAP), chronic generalized periodontitis, and periodontally healthy patients using conventional polymerase chain reaction (PCR) technique.
Materials and Methods:
A total of 90 patients were categorized as periodontally healthy individuals (Group I, n = 30), chronic generalized periodontitis (Group II, n = 30), and GAP (Group III, n = 30). The clinical parameters were recorded and subgingival plaque samples were collected. These were then subjected to conventional PCR analysis.
Statistical Analysis Used:
Kruskal–Wallis ANOVA test was used for multiple group comparisons followed by Mann–Whitney U-test for pairwise comparison.
Results:
On comparison between three groups, all the clinical parameters were found to be statistically highly significant. Comparing Groups I-II and I-III, the difference in detection was found to be statistically highly significant whereas in Groups II-III, it was statistically nonsignificant. On comparison of S. sputigena detected and undetected patients to clinical parameters in various study groups, the difference was found to be nonsignificant.
Conclusion:
S. sputigena was found to be significantly associated with chronic and aggressive periodontitis. Although the difference in its detection frequency in both groups was statistically nonsignificant when compared clinically, S. sputigena was more closely associated with the GAP.
Key words: Chronic periodontitis, generalized aggressive periodontitis, polymerase chain reaction, Selenomonas sputigena
INTRODUCTION
A variety of anaerobic, Gram-negative bacteria in subgingival plaque are implicated in the initiation and progression of periodontal disease.[1] Oral members of the genus Selenomonas have repeatedly been associated with periodontitis.[2] Moreover, these Selenomonas species have been identified in significant proportions in active periodontal-diseased sites.[3] Selenomonas sputigena was first described as Spirillum sputigenum in 1886 and was reclassified as S. sputigena in 1932. Because the organism was consistently isolated from dental plaque, studies in the 1950s and 1960s investigated culture conditions, morphology, and fine structure. In the 1970s and 1980s, gingival inflammation was found to be associated with elevated numbers of S. sputigena.[4] In the 1990s, with the advent of DNA-based culture-independent techniques such as polymerase chain reaction (PCR), a constantly growing number of Selenomonas phylotypes have been detected.[1]
Most of epidemiological studies investigating the prevalence of putative periodontal pathogens include data on Selenomonas spp. These have been detected in both chronic periodontitis (CP) and generalized aggressive periodontitis (GAP) lesions.[2] However, their prevalence levels vary considerably in different studies. The picture is further complicated by the fact that most of the cited studies focus on different species of this genus such as Selenomonas noxia and S. sputigena.[2,5,6]
Although S. sputigena has been found [2,3,7,8,9,10,11] detected, there are variable results for its association with periodontally healthy, CP, and aggressive periodontitis patients. According to our extensive literature search, no study primarily aiming at establishing the prevalence of S. sputigena in periodontally healthy, CP, or aggressive periodontitis patients has been conducted so far. Thus, the present study was conducted to find the association of S. sputigena with periodontally healthy, aggressive, and CP and to compare between these groups. To the best of our knowledge, this study is first of its kind in Indian population.
MATERIALS AND METHODS
The present study is a comparative study started in February 2014 involving a total of 90 subjects from whom informed consent was obtained. The study was undertaken after ethical approval from the Institutional Ethics Committee and Review Board of College of Dental Sciences, Davangere. The sample size was determined using G*Power software, version 3.1.9 (Available at http://www.gpower.hhu.de/en.html) with values of mean and standard deviation from the literature and by calculating the effect size. Effect size (d) was estimated to be 0.8, from the values of subjects mentioned in the study by Gonclaves et al.[8] keeping the power at 0.8, Type I error at 0.05 to follow a standard protocol. A minimum sample size of 21 samples per group was estimated. Still a higher sample was taken, i.e., 30 per group, making a total sample size to be 90 in the present study.
The patients for this study were categorized into three groups – periodontally healthy subjects (Group I, n = 30), chronic generalized periodontitis subjects (Group II, n = 30), and GAP subjects (Group III, n = 30). This was based on the periodontal classification of the American Academy of Periodontology which included history and clinical and radiographic findings.
After selection of cases, the clinical parameters were recorded first followed by the microbiological sample collection. The clinical parameters that were assessed included plaque index (PI),[12] gingival index (GI),[13] and probing pocket depth (PPD). The radiographic assessment was performed using orthopantomogram.
Sample collection
Subgingival plaque samples were collected from the selected teeth (three sites with probing depth of ≥5 mm in periodontitis patients and <3 mm in periodontally healthy patients), using sterile paper points, i.e., 3 paper points-International Organization for Standardization (ISO) #45, each kept for 10s in each periodontal pocket [14] [Figure 1]. The sample sites were selected randomly in each quadrant and isolated with cotton rolls; supragingival plaque was removed using sterile cotton pellets and the sites were air dried then. Using a tweezers, a paper point was inserted into the base of the pocket/sulcus. After 10 s, the paper point was removed from the selected site, and paper point end with collected plaque sample was first directed into the vial containing transport medium. The same procedure was repeated for 2 more paper points at the same site. Thus, a total of 9 paper points from 3 sites from each patient were together transported to Eppendorf vials containing transport media, viz., TE buffer (10 mM Tris-HCL, 1 mM EDTA pH 8; Hi-Media, Mumbai, Maharashtra), which were sent to the microbiological laboratory within 24 h.
Figure 1.
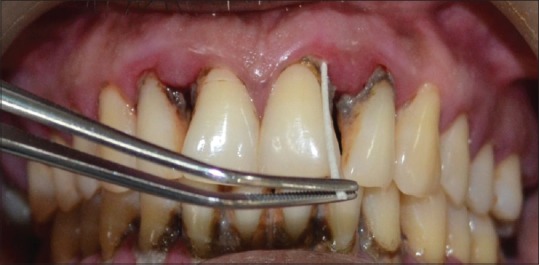
Collection of subgingival plaque samples with paper points
Polymerase chain reaction procedure for detection
In the laboratory, DNA extraction was done using modified proteinase-K method [15] which was then stored at −20°C until processing. It was followed by PCR amplification (conventional technique)[1] was then done. PCR master mix (Chromous Biotech Master mix-Chromous Biotech Pvt. Ltd., Bengaluru, India) was used containing Taq polymerase buffer containing 1.5 mM MgCl2, dNTP mix, and Taq polymerase enzyme. A volume of 25 µl of above master mix was taken to thermostable PCR tubes, and 1.2 µl each of forward and reverse primers specific for S. sputigena was added (30 pmole). A 5 µl template DNA (<100 ng) was added and finally the total volume was made to 50 µl by adding water.
The primer sets (Bioserve Biotechnology India Pvt Ltd., Hyderabad, India) were used for specifically amplifying S. sputigena-R8F: 5'-AGAGTTTGATCCTGGCTCAG-3' and SS-R: 5'-CTCAATATTCTCAA GCTCGGTT-3' [Figure 2]. The tubes were kept in a thermal cycler (Applied Biosystem, Waltham, Massachusetts, USA) to carry out the temperature cycles for amplification [Figure 3].
Figure 2.
Primers for Selenomonas sputigena
Figure 3.

Thermal cycler
PCR cycling conditions for amplification using primers specific for S. sputigena included an initial denaturation step at 95°C for 5 min, followed by 36 cycles of denaturation at 95°C for 1 min, annealing at 52°C for 30 s, extension at 72°C for 1 min, and final elongation step at 72°C for 5 min. The samples were kept at 4°C following PCR. Amplified products were subjected to electrophoresis through 1.5% agarose gel containing 1X TAE; 20 µl of each amplified product was mixed with 3 µl of bromophenol blue loading dye [Figure 4]. Electrophoresis was performed at 25V for 2 h. The gel was visualized under ultraviolet light illuminator after staining with ethidium bromide (0.5 µg/ml) in a dark room, and the image was captured with a digital camera for further analysis The microorganism (S. sputigena) was identified based on the formation of bands in the form of ladders in the captured image [Figures 5 and 6].
Figure 4.
Agarose gel with loaded amplification products to be tested
Figure 5.
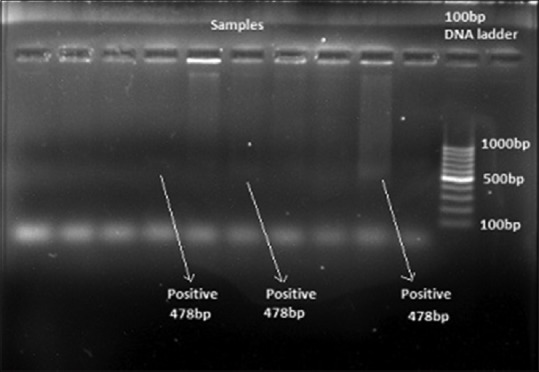
Bands showing detection of Selenomonas sputigena
Figure 6.
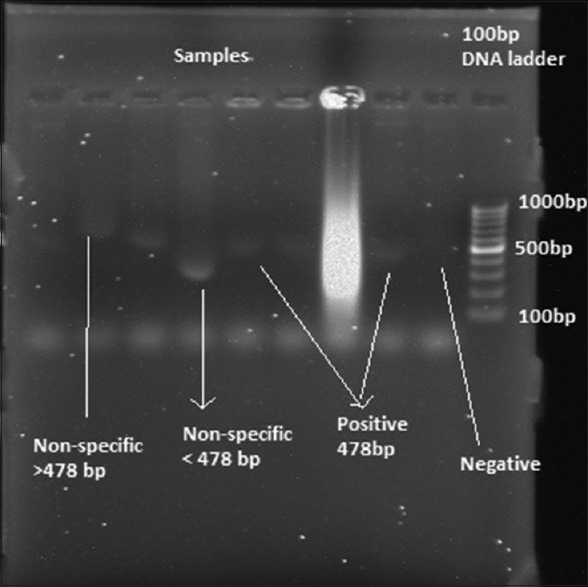
Bands showing positive, negative, and nonspecific bands
Statistical analysis
Descriptive data that included mean and standard deviations were determined for each clinical parameter in each group and were used for analysis. As the data were categorical, nonparametric tests were applied. Kruskal–Wallis ANOVA test was used for multiple group comparisons followed by Mann–Whitney U-test for pairwise comparison.
For all the tests, a P ≤ 0.05 was set for statistical significance.
RESULTS
The comparison of clinical parameters between various study groups is shown in Table 1 [Graph 1]. There was statistical significance for mean PI, GI, PD (P < 0.001) between the various groups except the mean PI which was statistically nonsignificant (P = 0.268) on comparison between Group I and III.
Table 1.
Comparison of clinical parameters between various study groups
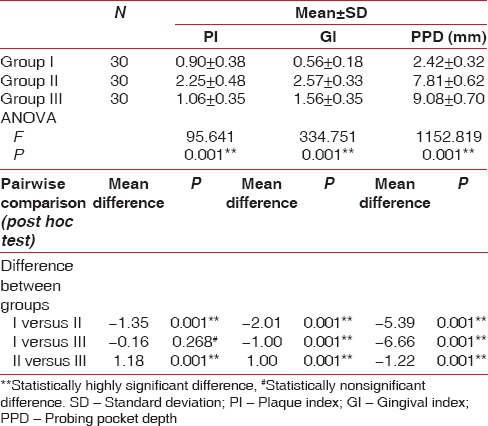
Graph 1.
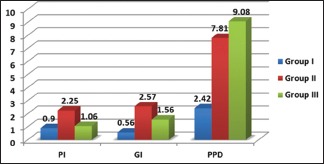
Comparison of clinical parameters between various study groups
Table 2 clearly depicts the detection frequency of S. sputigena in various study groups [Graph 2]. S. sputigena was present in plaque samples from all the three groups. The maximum and least number of samples in which S. sputigena was detected belonged to Group III and I, respectively. However, on intergroup analysis, the presence of S. sputigena was found to be statistically highly significant between Group I and II (P = 0.003 and P = 0.001, respectively). The striking feature to notice is that it was statistically nonsignificant (P = 0.072) between Group II and III.
Table 2.
Detection frequency of Selenomonas sputigena in various study groups
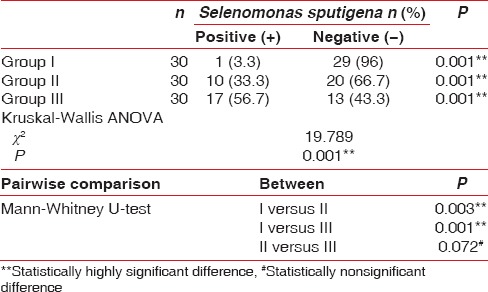
Graph 2.
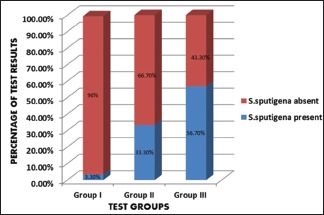
Detection frequency of Selenomonas sputigena in various study groups
On comparing the clinical parameters between S. sputigena detected (+) and undetected (−) patients in various study groups, none of the parameters (PI, GI, PPD) were found to be statistically significant [Table 3 and Graph 3].
Table 3.
Comparison of clinical parameters between Selenomonas sputigena detected (+) and undetected (-) patients in various study groups using Mann-Whitney U-test
Graph 3.
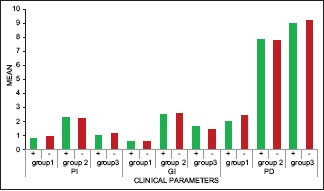
Comparison of clinical parameters between Selenomonas sputigena detected (+) and undetected (-) patients in various study groups
DISCUSSION
There is abundant evidence to implicate microorganisms as the primary etiologic agents of various forms of periodontal disease.[16] Tanner et al. (1979)[17] isolated an anaerobic, Gram-negative, motile curved rod bacterium, named S. sputigena from the periodontal pockets of patients with advanced destructive periodontal disease. This bacterium has since been identified in patients as a dominant member of the microbiota of the periodontal pocket. The production of antibody with high titer in patients against S. sputigena and the bone resorption activity of this microorganism seen in germ-free rats suggest that this bacterium is a periodontopathic agent in humans.[18] A variety of epidemiological studies investigating the prevalence of putative periodontal pathogens include data on Selenomonas spp. They have been detected in both CP and GAP lesions using culture- and DNA-based techniques.[2] Thus, the present study was conducted to find the association of S. sputigena with periodontally healthy, aggressive, and CP and compare between these groups. To the best of our knowledge, this study is first of its kind in Indian population. In this study, subgingival plaque samples were collected for analysis as the organism of interest here (S. sputigena) was known to be found in periodontal pockets. The subgingival plaque sample can be collected by using either curettes or paper points. As studied by Jervoe-Storm et al., 2007,[19] both techniques seem suitable for microbiological diagnostics. In this study, sterile paper points ISO #45 were used.[10] For detection of concerned organism in this study, conventional PCR has been used. High sensitivity and specificity and rapid identification of the microorganism are undeniable advantages of conventional PCR over culture method. The prime objective of this study was only the detection of S. sputigena in subgingival dental plaque biofilms and not the quantification justifying the use of conventional PCR over real-time PCR as the latter is known to quantify microorganisms' DNA and has a higher cost.
In this study, S. sputigena was found in all three groups. However, there was a difference in frequency of detection of S. sputigena between the three groups. It was highest in Group III, GAP subjects (56.7%) followed by Group II, chronic generalized periodontitis (33.3%) where as it was least in Group I, periodontally healthy subjects (3.33%) of the subjects. The findings in this study are consistent with findings by Moore, 1984,[20] where they found S. sputigena in subgingival plaque samples from three patients, i.e., three men 35–52 years of age with generalized moderate (chronic) periodontitis and by Dzink et al., 1985,[5] who isolated S. sputigena in higher proportions from active sites as compared to inactive sites in patients with active destructive periodontal lesions. Faveri, 2008,[7] found S. sputigena in nine out of ten subgingival plaque samples from patients with GAP and Kamma et al., 1994,[9] conducted a study where S. sputigena was detected only at bleeding sites in patients with rapidly progressive periodontitis aged 25–35 years. From this study results, it can be inferred that S. sputigena is associated with both chronic and aggressive periodontitis patients in contrast to periodontally healthy subjects.
On comparing the detection frequency of S. sputigena in Group I (periodontally healthy subjects) and Group II (chronic generalized periodontitis), the difference was found to be statistically significant. These findings were consistent with findings of Paster, 2001,[21] who found that S. sputigena was among the many species or phylotypes that were commonly detected in a variety of periodontal diseases but not (or rarely) in health. These also correlate with study of Sawada et al., 2000,[1] who conducted a study where subgingival plaque samples were collected from periodontal patients with adult periodontitis and periodontally healthy volunteers and concluded that the populations of centipeda periodontitis and S. sputigena may increase in colonized periodontal sites as the disease progresses.
The detection frequency of S. sputigena in Group I (periodontally healthy subjects) when compared with Group III (generalized aggressive periodontitis) was found to be highly significant. These findings correlate with study conducted by Gonclaves, 2012,[8] where GAP subjects showed significantly higher mean counts of Porphyromonas gingivalis, S. Sputigena, and Selenomonas oral clone CS002 as compared to periodontally healthy. On the contrary, there was statistically nonsignificant difference in detection frequency of S. sputigena between Group II (chronic generalized periodontitis) and Group III (GAP). However, to correlate this finding, there is lack of evidence.
In this study, statistically non-significant difference was found when all the clinical parameters were compared between S.sputigena positive and negative groups. This lack of significant relationship between S. sputigena and clinical parameters may be attributed to the identical probing depths between sites, small sample size, and low number of active sites. It is in contrast to study by Kamma et al., who have shown the relationship between S. sputigena, and PD in GAP was higher in sites with increased probing depths.[9] Many studies have proven that S. sputigena is found in active periodontal lesions and bleeding sites.[22,23]
CONCLUSION
The findings of our study indicate that S. sputigena is a part of the subgingival microbiota associated with chronic and aggressive periodontitis and may play a role in disease onset and progression. It can be concluded that S. sputigena is more closely associated with GAP than chronic generalized periodontitis. The present investigation has shown that uncultivated micro-organism, S. sputigena, recently identified could be added to the list of periodontal microbiota. However, future studies that provide quantitative information on proportions of this microorganism at the sites of disease activity are needed. Its role in inducing periodontal destruction cannot be drawn unless longitudinal studies are performed.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Sawada S, Kokeguchi S, Takashiba S, Murayama Y. Development of 16S rDNA-based PCR assay for detecting centipeda periodontii and Selenomonas sputigena. Lett Appl Microbiol. 2000;30:423–6. doi: 10.1046/j.1472-765x.2000.00735.x. [DOI] [PubMed] [Google Scholar]
- 2.Drescher J, Schlafer S, Schaudinn C, Riep B, Neumann K, Friedmann A, et al. Molecular epidemiology and spatial distribution of Selenomonas spp. in subgingival biofilms. Eur J Oral Sci. 2010;118:466–74. doi: 10.1111/j.1600-0722.2010.00765.x. [DOI] [PubMed] [Google Scholar]
- 3.Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun. 1989;57:3194–203. doi: 10.1128/iai.57.10.3194-3203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kingsley VV, Hoeniger JF. Growth, structure, and classification of Selenomonas. Bacteriol Rev. 1973;37:479–521. doi: 10.1128/br.37.4.479-521.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dzink JL, Tanner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648–59. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 6.Ximénez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27:648–57. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- 7.Faveri M, Mayer MP, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol. 2008;23:112–8. doi: 10.1111/j.1399-302X.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- 8.Gonçalves LF, Fermiano D, Feres M, Figueiredo LC, Teles FR, Mayer MP, et al. Levels of Selenomonas species in generalized aggressive periodontitis. J Periodontal Res. 2012;47:711–8. doi: 10.1111/j.1600-0765.2012.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamma JJ, Nakou M, Manti FA. Microbiota of rapidly progressive periodontitis lesions in association with clinical parameters. J Periodontol. 1994;65:1073–8. doi: 10.1902/jop.1994.65.11.1073. [DOI] [PubMed] [Google Scholar]
- 10.Habashneh RA, Karasneh JA, Khader YS. Predominant microflora in chronic and generalized aggressive periodontitis in a Jordanian population. Dentistry. 2014;4:2–6. [Google Scholar]
- 11.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 12.Silness J, Loe H. Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condtion. Acta Odontol Scand. 1964;22:121–35. doi: 10.3109/00016356408993968. [DOI] [PubMed] [Google Scholar]
- 13.Loe H, Silness J. Periodontal disease in pregnancy. I. Prevalence and severity. Acta Odontol Scand. 1963;21:533–51. doi: 10.3109/00016356309011240. [DOI] [PubMed] [Google Scholar]
- 14.Slots J. Bacterial specificity in adult periodontitis. A summary of recent work. J Clin Periodontol. 1986;13:912–7. doi: 10.1111/j.1600-051x.1986.tb01426.x. [DOI] [PubMed] [Google Scholar]
- 15.van Pelt-Verkuil E, van Belkum A, Hays JP. Principles and Technical Aspects of PCR Amplification. 1st ed. Netherlands: Springer Netherlands; 2008. Deoxynucleotide triphosphates and buffer components; pp. 91–101. [Google Scholar]
- 16.Socransky SS. Microbiology of periodontal disease – Present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 17.Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. Journal of Clinical Periodontology. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 18.Kumada H, Watanabe K, Nakamu A, Haishima Y, Kondo S, Hisatsune K, et al. Chemical and biological properties of lipopolysaccharide from Selenomonas sputigena ATCC 33150. Oral Microbiol Immunol. 1997;12:162–7. doi: 10.1111/j.1399-302x.1997.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 19.Jervoe-Sorm PM, AlAhdab H, Koltzcher Max, Fimmers R, Jepsen S. Comparison of Curet and Paper point Sampling of subgingival bacteria as analysed by Real Time Polymerase Chain Reaction. J Periodontol. 2007;78:909–17. doi: 10.1902/jop.2007.060218. [DOI] [PubMed] [Google Scholar]
- 20.Moore WE, Holdeman LV, Cato EP, Good IJ, Smith EP, Ranney RR, et al. Variation in periodontal floras. Infect Immun. 1984;46:720–6. doi: 10.1128/iai.46.3.720-726.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–23. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 23.Socransky SS, Haffajee AD, Dzink JL, Hillman JD. Associations between microbial species in subgingival plaque samples. Oral Microbiol Immunol. 1988;3:1–7. doi: 10.1111/j.1399-302x.1988.tb00596.x. [DOI] [PubMed] [Google Scholar]


