Abstract
A modified multiplex PCR method for detection of nine Staphylococcus aureus enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, and sej) and one form of immunoreactive toxic shock syndrome toxin based on a previously published method (S. R. Monday and G. A. Bohach, J. Clin. Microbiol. 37:3411-3414, 1999) has been developed. The modified PCR protocol seems robust and gives reliable results.
Staphylococcal enterotoxins (SE), toxic shock syndrome toxin (TSST), and exfoliative toxins A and B belong to a family of related pyrogenic toxins produced by Staphylococcus aureus (4, 10). By virtue of its variety of enterotoxins S. aureus is an important food-borne pathogen. In addition to the well-characterized SEA, SEB, SEC, SED, and SEE, new serological types of SEs (SEG, SEH, SEI, SEJ, SEK, SEL, SEM, SEN, SEO, SEP, SEQ, SER, and SEU) have been identified in recent years (5, 6, 7, 11, 13, 15, 16, 18). Five of the SE genes (seg, sei, sem, sen, and seo) belong to the same enterotoxin gene cluster (egc), and detection of one of these genes usually indicates the presence of all five enterotoxin genes (5, 11). For routine detection of SEs, commercially produced kits, such as reverse passive latex agglutination assays and enzyme-linked immunosorbent assays, are most commonly used. However, these methods are, to date, designed only to detect SEA, SEB, SEC, SED, and SEE. As an alternative to these more traditional methods, the PCR approach can provide detection of toxin genes and is presently designed to detect at least nine SE genes (sea, seb, sec, sed, see, seg, seh, sei, and sej) (1, 9, 12, 14).
The objective of this study was initially to establish the multiplex PCR method for detection of the toxin genes in staphylococcal isolates published by Monday and Bohach (12). According to their article, any of the nine SE genes (sea, seb, sec, sed, see, seg, seh, sei, and sej), the TSST gene, and the 16S rRNA gene can be amplified in a single multiplex reaction (results were not shown). However, while establishing the described method on two different types of thermocyclers, no detection of seb, sec, and tsst could be observed, and the amplification of sec and sei gave various results. Some modifications and adaptations were therefore considered necessary.
The following modifications were made to Monday and Bohach's method (12): (i) using another DNA isolation method; (ii) redesigning four primers (seb-sec forward, seb reverse, sei forward, and tsst reverse); (iii) splitting the multiplex into two PCRs; (iv) significantly decreasing the amount of 16S rRNA primers; and (v) adding four cycles to the PCR program.
Bacterial isolates and DNA isolation.
The S. aureus isolates used in this study are listed in Table 1. One colony of each isolate was incubated overnight in 3 ml of brain heart infusion broth medium at 37°C with agitation. Overnight culture (1.5 ml) was transferred to an Eppendorf tube and was centrifuged for 2 min at 17,500 × g. The pellet was resuspended in 1× Tris-EDTA buffer, and the cells were then lysed with 10 μl of 10-mg/ml lysostaphin (Sigma-Aldrich). This solution was incubated for 60 min at 37°C. The DNA was extracted by a standard cetyltrimethylammonium bromide procedure using chloroform followed by ethanol precipitation (17). DNA concentration was measured by the SYBR Green I method (2).
TABLE 1.
Isolates used in this study and their expected toxin genotype
| Isolate | Expected toxin genotype | Reference or source | Obtained toxin genotype in the present study bya:
|
|
|---|---|---|---|---|
| Unmodified method | Modified method | |||
| R5371/00 | sea, seg, seh, sei, and tsst | FSML,b9 | sea, seg, and seh | sea, seg, seh, sei, and tsst |
| R5460/00 | seb, seg, seh, sei, and tsst | FSML, 9 | seg and seh | seb, seg, seh, sei, and tsst |
| R5010/00 | sed, seg, sei, and sej | FSML, 9 | sed, seg, sei, and sej | sed, seg, sei, and sej |
| R4774/00 | None | FSML, 9 | None | None |
| R4571/00 | sec and tsst | FSML, 9 | sec | secc and tsst |
| R4071/00 | seb | FSML, 9 | None | seb |
| R2102/00 | sec, seg, and sei | FSML, 9 | seg and sei | sec,cseg, and sei |
| R963/00 | sed, seg, sei, and sej | FSML, 9 | sed, seg, sei, and sej | sed, seg, sei, and sej |
| FRI472 | sed, seg, sei, and sej | 12 | sed, seg, sei, and sej | sed, seg, sei, and sej |
| FRI913 | sea, sec, see, and tsst | 12 | sea, sec, and see | sea, sec,csee, and tsst |
| FRI572 | seg and sei | 12 | seg and sei | seg and sei |
| FRI445 | seg and sei | 12 | seg and sei | seg and sei |
| 3169 | secbovine, sed, sej, and tsst | 12 | sec, sed, and sej | secbovine, sed, sej, and tsst |
Our results using MJ research PTC 225 and Perkin Elmer 9700 thermocyclers.
FSML, Jim McLauchlin, Food Safety Microbiology Laboratory, Public Health Laboratory Service, London, United Kingdom.
This isolate generates both the seb-sec and the sec amplification products and should therefore be analyzed further with unique primers for seb to ensure that there is no seb hidden in the seb-sec product.
Amplification of selected staphylococcal genes.
Monday and Bohach (12) compared and evaluated the sequences of the SE genes. Unique primer sequences were identified for each gene with the exception of seb and sec. The seb-sec primer pair produces an identical product for both seb and sec. As a solution to the lack of specificity of the seb-sec primers, Monday and Bohach (12) designed a separate sec forward primer that works in combination with the seb-sec primers and that produces an amplification product unique to sec. The sec primers detect all three subclasses of enterotoxin SEC (C1, C2, and C3) (3, 8). All samples were tested for the presence of the 16S rRNA gene in order to ensure correct interpretation of toxin-negative isolates (12). However, due to the existence of multiple copies of the 16S rRNA gene, the concentration of the corresponding primers was reduced in the modified method to avoid 16S rRNA amplification products outcompeting other amplification products.
Monday and Bohach used 22 primers in the same PCR (12). Many primer sets in a single reaction may reduce robustness. Furthermore, some of the PCR products are similar in size and the bands were therefore found to be difficult to differentiate, particularly when analyzing unknown isolates. To avoid these problems we investigated several alternative primer combinations before the 11 primer sets were divided into two halves (reaction mixtures 1 and 2). Primers for sed, see, seg, sei, and tsst (Table 2) were combined in reaction mixture 1, and primers for sea, seb-sec, sec, seh, sej, and 16S rRNA (Table 2) were combined in reaction mixture 2. Monday and Bohach (12) used Gibco BRL Taq DNA polymerase (Life Technologies, Inc., Rockville, Md.) and an Amplitron II thermocycler (Barnstead Thermolyne Co., Dubuque, Iowa) for PCR amplification, whereas we used the following protocol: 5 μl of DNA (10 ng/μl) was added to 45 μl of reaction mixture containing final concentrations of 1× AmpliTaq buffer, 4 mM MgCl2, 2 U of AmpliTaq Gold polymerase (all from Applied Biosystems), 400 μM each deoxynucleoside triphosphate (ABgene, Epsom, United Kingdom), 300 nM each SE primer, and 60 nM 16S rRNA primers. DNA was amplified on an MJ Research thermocycler PTC 225 by initial denaturation for 10 min at 95°C followed by 15 cycles of 95°C for 1 min, 68°C for 45 s, 72° for 1 min, 20 cycles of 95°C for 1 min, 64°C for 45 s, 72° for 1 min, and a final extension at 72°C for 10 min. PCR products were separated by electrophoresis of 10 μl of reaction product in a 2.5% agarose gel (0.5× Tris-borate-EDTA buffer at 100 V for 100 min) and visualized on a UV transilluminator (Gel Documentation System; Bio-Rad, Hercules, Calif.). Product size was determined by comparison with a pUC-mix molecular weight ladder (Fermentas, Vilnius, Lithuania). Both the original and the modified protocol were tested on two different thermocyclers: MJ Research PTC 225 and Perkin-Elmer GeneAmp PCR System 9700.
TABLE 2.
Primers used in this study for detection of SE genes, TSST gene (tsst), and 16S rRNA gene
| Primera | Primer sequence (5′-3′) | Amplified product size (bp) | GeneBank accession no. | Reference or source | Multiplex PCR reaction mixture no. |
|---|---|---|---|---|---|
| sea forw. | GCA GGG AAC AGC TTT AGG C | 521 | M18970 | 12 | 2 |
| sea rev. | GTT CTG TAG AAG TAT GAA ACA CG | ||||
| seb-sec forw. | ACA TGT AAT TTT GAT ATT CGC ACT G | 667 | M11118 (seb) | This study | 2 |
| seb rev. | TGC AGG CAT CAT GTC ATA CCA | ||||
| sec forw. | CTT GTA TGT ATG GAG GAA TAA CAA | 284 | X05815 (sec1) | 12 | 2 |
| sec rev. | TGC AGG CAT CAT ATC ATA CCA | AY450554 (sec2) | |||
| X51661 (sec3) | |||||
| sed forw. | GTG GTG AAA TAG ATA GGA CTG C | 385 | M28521 | 12 | 1 |
| sed rev. | ATA TGA AGG TGC TCT GTG G | ||||
| see forw. | TAC CAA TTA ACT TGT GGA TAG AC | 171 | M21319 | 12 | 1 |
| see rev. | CTC TTT GCA CCT TAC CGC | ||||
| seg forw. | CGT CTC CAC CTG TTG AAG G | 328 | AF064773 | 12 | 1 |
| seg rev. | CCA AGT GAT TGT CTA TTG TCG | ||||
| seh forw. | CAA CTG CTG ATT TAG CTC AG | 359 | U11702 | 12 | 2 |
| seh rev. | GTC GAA TGA GTA ATC TCT AGG | ||||
| sei forw. | CAA CTC GAA TTT TCA ACA GGT ACC | 466 | AF064774 | This study, 12 | 1 |
| sei rev. | CAG GCA GTC CAT CTC CTG | ||||
| sej forw. | CAT CAG AAC TGT TGT TCC GCT AG | 142 | AF053140 | 12 | 2 |
| sej rev. | CTG AAT TTT ACC ATC AAA GGT AC | ||||
| tsst forw. | GCT TGC GAC AAC TGC TAC AG | 559 | J02615 | This study, 12 | 1 |
| tsst rev. | TGG ATC CGT CAT TCA TTG TTA T | ||||
| 16S rRNA forw. | GTA GGT GGC AAG CGT TAT CC | 228 | X68417 | 12 | 2 |
| 16S rRNA rev. | CGC ACA TCA GCG TCA G |
forw., forward; rev., reverse.
Analysis of S. aureus isolates using both the unmodified and modified multiplex PCR methods.
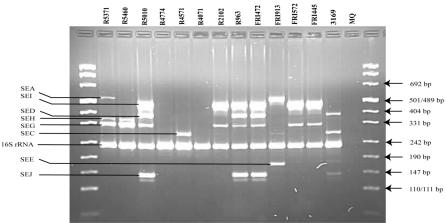
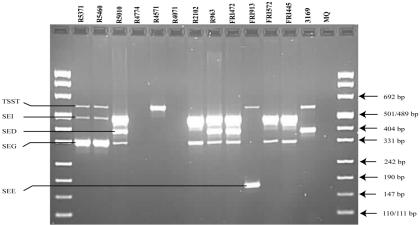
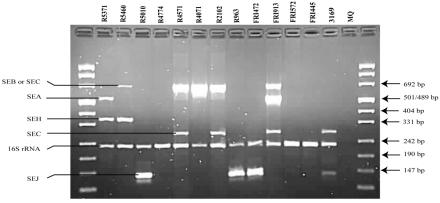
Figure 1 shows PCR results with the unmodified multiplex PCR protocol as described by Monday and Bohach (12) performed on the MJ Research PTC 225 thermocycler. No detection of seb-sec and tsst was observed, and the amplification of sec and sei gave various results. The modified multiplex PCR protocol gave correct identification of all expected toxins (Table 1). Figures 2 and 3 show the PCR results with reaction mixtures 1 and 2 and detection of the SED, SEE, SEG, SEI, and TSST genes and the SEA, SEB-SEC, SEC, SEH, SEJ, and 16S rRNA genes, respectively. When DNA from several isolates was combined (R5460 plus R5010 plus FRI913 or FRI472 plus FRI913 plus R5460), the modified method was able to detect all enterotoxin genes simultaneously (results not shown). The modified protocol, including redesigned seb-sec forward and seb reverse primers (Table 2), resulted in the expected amplification of the SEB-SEC gene in known isolates (Table 1, Fig. 3). However, it has been suggested (9, 10, 14) that isolates that give both the seb-sec and sec products should be analyzed further with unique primers for seb to ensure that there is no seb hidden in the seb-sec product. It is well known that some isolates express SECbovine and thereby generate only the sec product instead of both the seb-sec and sec products (12). This was confirmed in this study by isolate 3169 (Table 1, Fig. 3). The TSST gene could not be detected in the isolates previously reported as TSST producers with the unmodified protocol (Table 1, Fig. 1). After redesigning the tsst reverse primer (Table 2), the expected tsst amplification product was detectable by the modified protocol (Table 1, Fig. 2). Similarly, after redesigning the sei forward primer (Table 2) we were able to detect the expected sei amplification product in two isolates (R5371/00 and R5460/00) that were negative by the unmodified protocol (Table 1, Fig. 1 and 2). In a personal communication, Monday and Bohach suggested we try different brands of thermocyclers, because they also observed that their original protocol did not work on an Eppendorf thermocycler while it worked on a Perkin-Elmer GeneAmp model 2400 (Applied Biosystems), possibly due to differences in ramping time and block controls. In the present study we did not observe differences in performance between the MJ Research PTC 225 and Perkin Elmer GeneAmp 9700 in either the unmodified or modified method.
FIG. 1.
Agarose gel electrophoresis of PCR products amplified with the multiplex PCR method described by Monday and Bohach (12). Primers detecting genes encoding all the nine SEs, the TSST, and the 16S rRNA are represented in this PCR mixture. S. aureus isolates, MQ (MilliQ water) negative control, amplification products, and DNA fragment sizes are indicated.
FIG. 2.
Agarose gel electrophoresis of PCR products amplified with the modified multiplex PCR protocol—reaction mixture 1. Primers detecting genes encoding SED, SEE, SEG, SEI, and TSST are represented in this PCR mixture. S. aureus isolates, MQ (MilliQ water) negative control, amplification products, and DNA fragment sizes are indicated.
FIG. 3.
Agarose gel electrophoresis of PCR products amplified with the modified multiplex PCR protocol—reaction mixture 2. Primers detecting genes encoding SEA, SEB, SEC, SEH, SEJ, and 16S rRNA are represented in this PCR mixture. S. aureus isolates, MQ (MilliQ water) negative control, amplification products, and DNA fragment sizes are indicated.
Splitting the multiplex PCR into two halves increases the workload. However, this may be justified by the observation that this approach yielded more reliable results. Furthermore, the modified protocol appeared to be more robust, as observed by the absence of differences in performance on two different brands of thermocyclers.
Acknowledgments
We are grateful to Jim McLauchlin for the isolates from the Food Safety Microbiology Laboratory (London, United Kingdom) and to Claudia Deobald and Greg Bohach for the isolates from the Department of Microbiology, Molecular Biology and Biochemistry, University of Idaho (Moscow).
REFERENCES
- 1.Akineden, O., C. Annemuller, A. A. Hassan, C. Lammler, W. Wolter, and M. Zschock. 2001. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin. Diagn. Lab. Immunol. 8:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berdal, K. G., and A. Holst-Jensen. 2001. Roundup Ready (R) soybean event-specific real-time quantitative PCR assay and estimation of the practical detection and quantification limits in GMO analyses. Eur. Food Res. Technol. 213:432-438. [Google Scholar]
- 3.Hsiao, M. H., T. R. Chen, and H. Y. Tsen. 2003. Novel PCR primers for specific detection of C1, C2 and C3 enterotoxin genes in Staphylococcus aureus. J. Food Drug Anal. 11:338-343. [Google Scholar]
- 4.Iandolo, J. J. 1989. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu. Rev. Microbiol. 43:375-402. [DOI] [PubMed] [Google Scholar]
- 5.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. A highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:4260. [DOI] [PubMed] [Google Scholar]
- 6.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Z. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Q. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 7.Letertre, C., S. Perelle, F. Dilasser, and P. Fach. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol. 95:38-43. [DOI] [PubMed] [Google Scholar]
- 8.Marr, J. C., J. D. Lyon, J. R. Roberson, M. Lupher, W. C. Davis, and G. A. Bohach. 1993. Characterization of novel type C staphyloccal enterotoxins: biological and evolutionary implications. Infect. Immun. 61:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLauchlin, J., G. L. Narayanan, V. Mithani, and G. O'Neill. 2000. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 63:479-488. [DOI] [PubMed] [Google Scholar]
- 10.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mempel, M., G. Lina, M. Hojka, C. Schnopp, H. P. Seidl, T. Schafer, J. Ring, F. Vandenesch, and D. Abeck. 2003. High prevalence of superantigens associated with the egc locus in Staphylococcus aureus isolates from patients with atopic eczema. Eur. J. Clin. Microbiol. Infect. Dis. 22:306-309. [DOI] [PubMed] [Google Scholar]
- 12.Monday, S. R., and G. A. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37:3411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omoe, K., D. L. Hu, H. Takahashi-Omoe, A. Nakane, and K. Shinagawa. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun. 71:6088-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omoe, K., M. Ishikawa, Y. Shimoda, D. L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orwin, P. M., J. R. Fitzgerald, D. Y. M. Leung, J. A. Gutierrez, G. A. Bohach, and P. M. Schlievert. 2003. Characterization of Staphylococcus aureus enterotoxin L. Infect. Immun. 71:2916-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orwin, P. M., D. Y. M. Leung, H. L. Donahue, R. P. Novick, and P. M. Schlievert. 2001. Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willems, R. J., J. Top, B. N. van den, A. van Belkum, D. J. Mevius, G. Hendriks, M. Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarwood, J. M., J. K. McCormick, M. L. Paustian, P. M. Orwin, V. Kapur, and P. M. Schlievert. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3—implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138-13147. [DOI] [PubMed] [Google Scholar]