Abstract
Purpose
Intratumoral androgen synthesis in prostate cancer (PCa) contributes to the development of castration-resistant prostate cancer (CRPC). Several enzymes responsible for androgen biosynthesis have been shown to be overexpressed in CRPC, thus contributing to CRPC in a castrated environment. The TMPRSS2-ERG transcription factor has been shown to be present in primary PCa tumors as well as CRPC tumors. We hypothesize that TMPRSS2-ERG fusions regulate androgen biosynthetic enzyme (ABE) gene expression and the production of androgens, which contributes to the development of CRPC.
Experimental design
We used a panel of assays including lentivirus transduction, gene expression, chromatin immunoprecipitation and sequencing, Liquid chromatography-Mass spectrometric quantitation, immunocytochemistry, immunohistochemistry and bio-informatics analysis of gene microarray data bases to determine ERG regulation of androgen synthesis.
Results
We found that ERG regulated the expression of the ABE AKR1C3 in PCa cells via direct binding to the AKR1C3 gene. Knockdown of ERG resulted in reduced AKR1C3 expression, which caused a reduction in both DHT synthesis and PSA expression in VCaP PCa cells treated with 5α-androstanedione, a DHT precursor metabolite. Immunohistochemical staining revealed that ERG was co-expressed with AKR1C3 in PCa tissue samples.
Conclusions
These data suggest that AKR1C3 catalyzes the biochemical reduction of 5α-Androstanedione to DHT in PCa cells, and that ERG regulates this step through upregulation of AKR1C3 expression. Elucidation of ERG regulation of ABEs in CRPC may help to stratify TMPRSS2-ERG fusion-positive PCa patients in the clinic for anti-AR driven therapies; and AKR1C3 may serve as a valuable therapeutic target in the treatment of CRPC.
Introduction
The TMPRSS2-ERG fusion gene has been shown to be present in approximately 40% of prostate cancer (PCa) tumors, including primary and advanced PCa (1-6). The TMPRSS2-ERG fusion gene results in androgen receptor (AR) induced overexpression of the ERG transcription factor (7). The presence of TMPRSS2-ERG in PCa has been shown to be associated with poor clinical prognosis (2, 8-10). ERG fusion-positive PCa patients had significantly lower disease-free survival in watchful waiting cohorts (11, 12), and significantly greater risk of biochemical prostate-specific antigen (PSA) recurrence compared to fusion-negative PCa patients (13-15). ERG has been shown to have several biological functions that facilitate its oncogenic properties in PCa. It has been shown to regulate cellular migration and invasion, epithelial to mesenchymal transition, dedifferentiation, and AR signaling (16-19) ; however, the biological functions of ERG contributing to the development of castration-resistant PCa (CRPC) have not been fully elucidated. Interestingly, ERG fusion-positive PCa patients responded better to abiraterone treatment, a CYP17A1 enzyme inhibitor used to treat CRPC, compared to ERG fusion-negative patients (5). These data are consistent with ERG fusion-positive patients having a higher PSA recurrence, and suggests that ERG may regulate the synthesis of intratumoral androgens, and AR activation in CRPC.
Upregulation of androgen biosynthetic enzymes (ABEs) and intratumoral androgen synthesis have been shown to facilitate the development of CRPC (20-22). This can be seen with the ability of abiraterone treatment to reduce serum and intratumoral dihydrotestosterone (DHT) and testosterone levels in CRPC patients (23-25). One ABE that has recently become of particular interest is AKR1C3, because it has been shown to be highly upregulated in CRPC tumors (26). AKR1C3 enzyme functions downstream of CYP17A1 enzyme in the DHT synthesis pathway (22), and it may play a pivotal role in DHT synthesis in CRPC. Intratumoral DHT synthesis bypasses the synthesis of testosterone, and DHT is synthesized directly from androstanedione (5α-Adione) or androstanediol (22, 27, 28). Inhibitors specific for AKR1C3 are currently being developed (29-31), and may prove to be beneficial for treatment of CRPC.
This study is focused on TMPRSS2-ERG transcription factor upregulation of AKR1C3 expression and DHT synthesis in CRPC. We have evidence suggesting that AKR1C3 functions as a key ABE responsible for DHT synthesis via biochemical reduction of 5α-Adione into DHT (bypassing testosterone). Our data indicates that 5α-Adione is the primary DHT precursor metabolite, and that 5α-Adione can induce high levels of AR activation in CRPC cells. Elucidation of TMPRSS2-ERG regulation of AKR1C3 may help to stratify ERG fusion-positive PCa patients towards anti-AR targeted therapies. In addition, our data supports the notion that AKR1C3 is an attractive therapeutic target for CRPC treatment.
Material and Methods
Cell Lines and Treatments
Prostate cancer cell lines VCaP, LNCaP, and HEK293T cells were obtained from American Type Culture Collection (ATCC; Manassas, Va). BPH-1 cells were obtained from Dr. Shijie Sheng (Wayne State University). VCaP cells were maintained in ATCC DMEM media with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA), and 1% penicillin streptomycin (PS) (Gibco, Carlsbad, CA). LNCaP and BPH-1 cells were maintained in Gibco RPMI media (Gibco, Carlsbad, CA) with 10 % FBS and 1% PS. HEK293T cells were maintained in Gibco DMEM media with 10% FBS, 1% PS, and 1% sodium pyruvate. All cell lines were tested for mycoplasma contamination prior to use with Venor-GeM mycoplasma detection kit from Sigma Biochemicals (St. Louis, MO). HPLC/MS/MS, immunocytochemistry, and PSA activation experiments were performed using Gibco DMEM phenol red-free media supplemented with 1% charcoal stripped serum (C.S.S.) and 1% PS.
Lentiviral Plasmids and Transduction
Lentiviral plasmid selection and virus production were conducted as previously described with minor modifications (32). Briefly, the pGIPZ lentivector was used for shRNA ERG knockdown (5’-TGGACAGACTTCCAAGATG-3’) and shRNA AKR1C3 knockdown (5’-ACACCAGTGTGTAAAGCTA-3’), and the pLOC lentivector was used for ERG overexpression (Open BioSystems, Lafayette, CO). Truncated ERG and full length ERG were cloned into the pLOC lentivector. Cloning primers are listed in Supplementary Table 1. Lentiviral particles were generated in HEK293T cells using a Trans-Lentiviral Packaging Kit (Thermo Fisher Scientific, Waltham, MA, TLP5918). VCaP cells were infected with pGIPZ lentiviral particles and selected with (0.3 μg/ml) puromycin (Research Products International Corporation, Mount Prospect, IL). LNCaP and BPH-1 cells were infected with pLOC lentiviral particles and selected with (2-4 μg/ml) blasticidin (Research Products International Corporation, Mount Prospect, IL). A pooled lentiviral cell population was used for all lentiviral experiments. Transient transfections were performed using Lipofectamine2000 Reagent (Invitrogen, Carlsbad, CA,) in OptiMEM media (Invitrogen, Carlsbad, CA,).
Western Blotting and Antibodies
Western blotting was performed using SDS-PAGE with gel transfer to a nitrocellulose membrane. Membranes were blocked with 5% nonfat milk and probed with primary antibody in 5% milk. Membranes were probed with HRP-linked secondary antibody in 5% milk. Protein bands were detected using enhanced chemiluminescence substrate and autoradiography film. Primary and secondary antibodies are listed in Supplementary Table 2. Densitometry was performed using ImageJ software (NIH).
RT-PCR and RT-qPCR
RNA was extracted from VCaP and LNCaP cells using the Trizol (Invitrogen, Carlsbad, CA,) method. For RT-PCR experiments, cDNA was amplified using an Eppendorf Mastercycler Realplex2 qPCR machine. PCR products were loaded onto a 0.8% agarose gel containing ethidium bromide. For RT-qPCR experiments, cDNA was amplified with SYBR green from the SensiFAST SYBR kit obtained from Bioline (Taunton, MA, BIO-98002). Gene expression was quantified with previously described qPCR method (33). Fold changes in message levels were quantified using comparative Ct method (34) . PCR primers are listed in Supplementary Table 2.
Chromatin Immunoprecipitation (ChIP) PCR
ChIP-PCR was carried out using the ChIP-IT Express kit obtained from Active Motif (Carlsbad, CA) as previously described with following modification (17). Briefly, five million VCaP cells were fixed with formaldehyde and chromatin was extracted according to the manufacturer`s instructions. For immunoprecipication, 15-25 μg of chromatin was incubated with 3-5 μg of anti-ERG primary antibody ERG1/2/3 (H-95)X or normal IgG antibody (Santa Cruz Biotechnology, Dallas, TX) and protein G agarose beads. Immunoprecipitated chromatin was reverse cross-linked with proteinase K. PCR was performed using 750-1000 ng DNA template per sample and ran at 30-40 cycles using an Eppendorf Mastercycler PCR machine. ChIP-PCR primers are listed in Supplementary Table 1. PCR products were analyzed onto a 0.8% agarose gel containing ethidium bromide. ChIP seq methodology and data analysis was performed as previously described (35).
Mass Spectrometry (HPLC/MS/MS)
Androgen metabolites obtained from Steraloids (Newport, RI) and Sigma-Aldrich (St. Louis, MO) (Supplementary Table 3.) were used for MS standards and in vitro steroid treatments. VCaP shScrambled and shERG cells were plated at a cell density of 500,000 cells/well in a 6-well plate. Cells were serum starved for 24 hours using 1% C.S.S. phenol red-free DMEM media. Cells were treated with androgen metabolites or 0.1% ethanol (vehicle control) for 24 hours in 1% C.S.S. phenol red-free media. Cells were treated with (100 nM) androgen metabolite or a concentration gradient of (10 nM), (50 nM), or (100 nM) androgen metabolite. In order to serve as an internal control, 1 ng of DHEA-D2 and DHT-D3 was added to the media directly before cell lysate preparation. Cells and media were collected and probe sonicated on ice. Lipids were purified using a Sep-pak cartridge obtained from Waters Corporation (Milford, MA, WAT023590). Lipid samples were eluted with acetonitrile into HPLC/MS/MS vials obtained from MicroSOLV Technology (Eatontown, NJ, 9512S-0CV vials, 9502S-10C-B tops). A cell count control plate was plated and cell number was determined on the day of sample collection. HPLC/MS/MS experiments and data analysis were performed at the Wayne State University Lipidomics Core Facility.
Immunocytochemistry
VCaP cells were plated on cover slips at a cell density of 400,000 cells per cover slip. Cells were serum starved for 24 hours using 1% C.S.S. phenol red-free DMEM media. Cells were treated with (100 nM) 5α-Androstanedione or 0.1% ethanol for 24 hours in 1% C.S.S. phenol red-free DMEM media. Cell coverslips were washed with 1X PBS, fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, blocked with 2% horse serum, and incubated overnight at 4 °C with either 1.) AR antibody (rabbit) or 2.) AR antibody (mouse), 1:1000 in PBS (Santa Cruz Biotechnology, Dallas, TX). Cover slips were washed with PBS and incubated at room temperature in a dark box for 1 hour with either 1.) Texas Red antibody (rabbit) (Vector Labs, Burlingame, CA) or 2.) Alexa Fluor 633 antibody (mouse), 1:200 in PBS (Molecular Probes, Carlsbad, CA). Cover slips were mounted onto slides using Vectashield mounting medium with DAPI (Vector Labs, H-1200). Slides were subjected to confocal imaging at the Wayne State University School of Medicine Microscopy, Imaging, and Cytometry Resources Core Facility using the Ziess LSM 780 confocal microscope at a magnification of 40X.
Immunohistochemistry
Human prostate cancer tissues from patients undergoing radical prostectomy were used for immunohistochemical analysis. Formalin-fixed, paraffin-embedded serial tissue sections from human prostate tumor tissue specimens and LuCaP xenograft tumors were deparaffinized with xylene and rehydrated in graded EtOH. Endogenous peroxidase activity was blocked by incubating in 3% H2O2 for 20 min. Antigen retrieval was performed with Antigen Retrieval Citra Plus Solution (BioGenex, Freemont, CA) in a steamer. Slides were then blocked with Blocking Serum from ABC Vectastain Kit (Vector Labs, Burlingame, CA). Slides were incubated at 4°C overnight in a humidified chamber with antibodies directed against ERG (1:100; Epitomics, Burlingame, CA) or AKR1C3 (1:5000; Sigma-Aldrich, St. Louis, MO). After washing, sections were incubated with ABC Vectastain Kit, according to manufacturer's protocol, followed by incubation with 3,3-diaminobenzidine tetrahydrochloride (Vector Labs). Nuclei were counterstained with Mayer's hematoxylin (Sigma-Aldrich). Sections were then dehydrated with graded EtOH, washed with xylene, and mounted with Permount (Sigma-Aldrich).
Gene Expression Omnibus Database
Gene expression omnibus (GEO) publicly available microarray data was used to analyze correlation between ERG expression and AKR1C3 expression. The 25 metastatic prostate cancer expression profiles were extracted from database GDS2545 using ERG accession ID 914_g_at and AKR1C3 accession ID 37399_at, and analyzed using “GEO2R.” The patient tissues were procured from warm autopsy program. The metastatic samples were obtained from warm autopsy program. The in vitro cell line expression profiles were extracted from databases GSE22010, GSE39353, and GSE39354 using ERG accession ID's 8070297, 211626_x_at, and 211626_x_at respectively, and AKR1C3 accession ID's 7925929, 209160_at, and 209160_at respectively.
Oncomine Database
The Oncomine database was used to extract survival data for 363 prostate tumor specimens from Setlur et al. “prostate cancer” (2008) (36). Survival data were given as dead or alive at 1 year, 3 years, 5 years, and endpoint. For survival data past 5 years the last time to follow up was used as the endpoint. Prostate tumor expression profiles were extracted from the GEO database GSE8402 using AKR1C3 accession ID DAP4_3222. AKR1C3 expression was determined as high or low by taking the median expression value of 363 tumor specimens, and adding or subtracting the standard deviation. This gave a sample size of 46 high tumors and 51 low tumors, with their respective available survival data taken from the Oncomine database. For ERG analysis the presence or absence of the TMPRSS2-ERG fusion gene was determined in the Setlur et al. study by FISH or PCR analysis, and this data was made available in the Oncomine database as well as the GEO database. Survival data and TMPRSS2-ERG fusion status data were available for analysis from 354 of the prostate tumor specimens.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism Version 5.0.
Results
TMPRSS2-ERG regulates androgen biosynthetic enzyme expression in prostate cancer cells
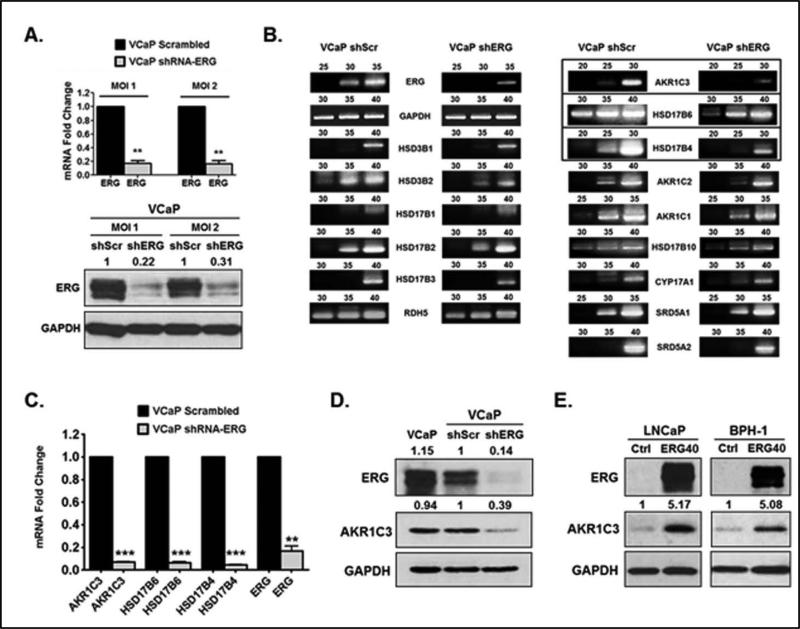
To determine ERG regulation of androgen biosynthetic enzymes (ABEs) in prostate cancer (PCa) we performed shRNA knockdown of ERG inTMPRSS2-ERG fusion-positive VCaP cells using lentivirus transduction. Lentiviral shRNA knockdown of ERG resulted in decreased ERG expression at the mRNA and protein levels compared to VCaP shScrambled control (Figure 1A). Using VCaP shScrambled and shERG cells, we performed an screening of ABE gene expression. Among the tested genes, expression of AKR1C3, HSD17B4, and HSD17B6 was highly reduced in VCaP shERG cells, and expression of HSD3B2 and HSD17B10 was also reduced to some extent (Figure 1B). Using quantitative PCR we showed that AKR1C3, HSD17B4, and HSD17B6 expression was significantly reduced in shERG knockdown cells (Figure 1C). ERG knockdown in VCaP cells resulted in reduced protein expression of AKR1C3 (Figure 1D) and HSD17B4 enzymes (Supplementary Figure 1A), whereas HSD17B6 protein expression was undetectable (data not shown).
Figure 1.
TMPRSS2-ERG regulates androgen biosynthetic enzyme (ABE) expression in PCa cells. A.) RT-qPCR analysis of ERG in VCaP shScrambled (shScr) and VCaP shERG lentiviral cells. Western blot analysis of ERG in VCaP shScr and shERG cells. MOI = multiplicity of infection for lentiviral transduction. B.) RT-PCR analysis of ABE expression in VCaP shScr and shERG cells. PCR cycle numbers are indicated above each DNA gel. C.) RT-qPCR analysis of HSD17B6, HSD17B4, and AKR1C3 in VCaP shScr and VCaP shERG cells. D.) Western blot analysis of AKR1C3 in VCaP, VCaP shScr and shERG cells. E.) Western blot analysis of AKR1C3 in LNCaP ERG40 and BPH-1 ERG40 lentiviral cells. LNCaP Ctrl and BPH-1 Ctrl cells represent empty vector controls. For statistical analysis a two-tailed, paired, student's t-test was performed (N=3) **P<0.01, ***P<0.001. Numbers above western blots represent fold changes determined by densitometry. Representative of three independent experiments.
Next, we overexpressed the truncated form of ERG that is most commonly found in PCa patients, TMPRSS2 exon 1 fused to ERG exon 4 (T1-E4) (37). T1-E4 is also the truncated form of ERG expressed in VCaP cells (Supplementary Figure 1B). We termed this truncated form “ERG40” because it has a deletion of the first 39 amino acids from the N-terminus (38). Lentivirus was used to overexpress the truncated ERG40 in LNCaP PCa cells, and in BPH-1 cells (benign prostatic hyperplasia). Overexpression of ERG40 resulted in a significant increase in AKR1C3 mRNA compared to empty vector control (Supplementary Figure 1C). Overexpression of ERG40 caused an increase in AKR1C3 protein expression in both LNCaP cells and BPH-1 cells (Figure 1E). Interestingly, similar results were seen in LNCaP cells overexpressing full length (wild type) ERG. LNCaP ERGwt cells showed an increase in AKR1C3 mRNA compared to empty vector control (Supplementary Figure 1D). These data demonstrate that ERG positively regulates the expression of DHT synthesis enzyme, AKR1C3, in PCa cells.
ERG regulates AKR1C3 expression via direct DNA binding to the AKR1C3 gene
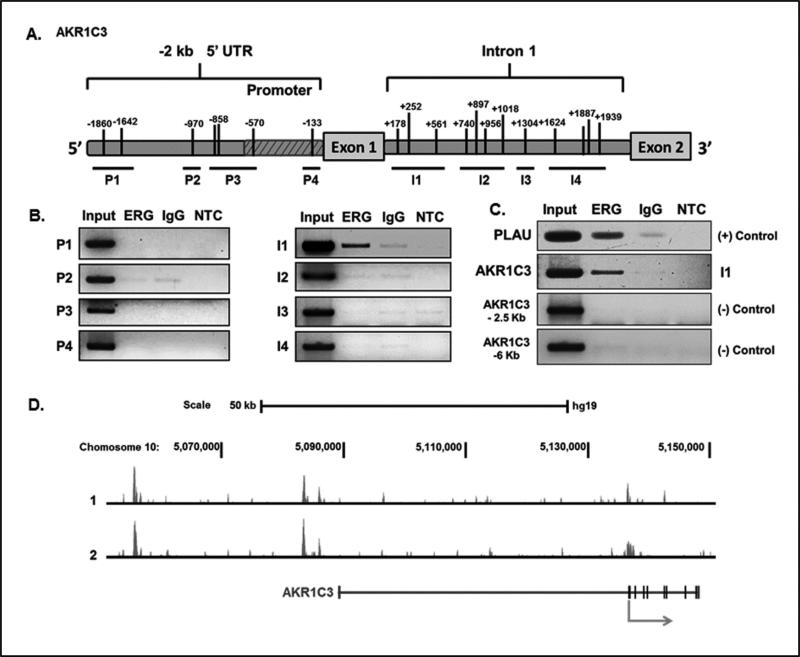
To investigate whether ERG regulation of AKR1C3 expression in PCa occurs through direct DNA binding, we performed ChIP-PCR in VCaP cells. Primers were used to access ERG binding to the AKR1C3 gene region, spanning from -2 kb upstream of the transcription start site (TSS) to +2 kb downstream of the TSS. Several ERG consensus binding sequence sites (5’-GGAA/T-3’) were identified in the 2 kb 5’untranslated region and in the first intron of the AKR1C3 gene (Figure 2A). Four primer sets were designed to span the −2 kb promoter region (P1-P4) and the intron 1 region (I1-I4) (Figure 2A). ERG was shown to bind directly to the intron 1 region of the AKR1C3 gene, at a site located +154 to +670 downstream of the TSS (Figure 2B). The known ERG target gene Plasminogen Activator Urokinase (PLAU) (39) was also enriched in anti-ERG antibody immunoprecipitates, suggesting the validity of the assay. To determine the specificity of ERG's interaction with the AKR1C3 gene, we designed negative control primers that span regions of the AKR1C3 gene that do not contain putative ERG consensus binding sites. As expected, there was no ERG binding to these regions of the AKR1C3 gene (Figure 2C).
Figure 2.
ERG binds directly to the AKR1C3 gene. A.) Diagram illustrating the AKR1C3 gene located on chromosome 10. Black tick marks indicate ERG binding sites (5’-GGAA/T-3’) located in the region of −2 Kb to +2 Kb relative to the transcription start site. B.) ChIP-PCR analysis of putative ERG binding sites in the AKR1C3 gene. ChIP was performed in VCaP cells using anti-ERG antibody; IgG antibody was used as a control for non-specific binding. NTC is non-template control. C.) ChIP-PCR analysis of a known ERG binding site in PLAU promoter region using VCaP cells. Negative control primers were used in VCaP cells to demonstrate specificity of the ChIP-PCR technique. D.) ChIP-Seq analysis performed in duplicate using anti-ERG antibody in VCaP cells.
ChIP-Seq was performed in VCaP cells with two independent experiments using anti-ERG antibody. ERG binding enrichment peaks were observed at three different sites along the AKR1C3 gene. Two of the binding peaks were located far upstream of the TSS in the 5’ UTR of AKR1C3; one at approximately −50 kb, and another at −75 kb, relative to the TSS. The third peak was shown to be located near the TSS and intron 1 region (Figure 2D), consistent with the ChIP-PCR data. These data demonstrate that ERG directly binds to the AKR1C3 gene and regulates AKR1C3 gene expression in fusion-positive PCa cells.
Androstanedione is a precursor metabolite for DHT in TMPRSS2-ERG fusion-positive cells
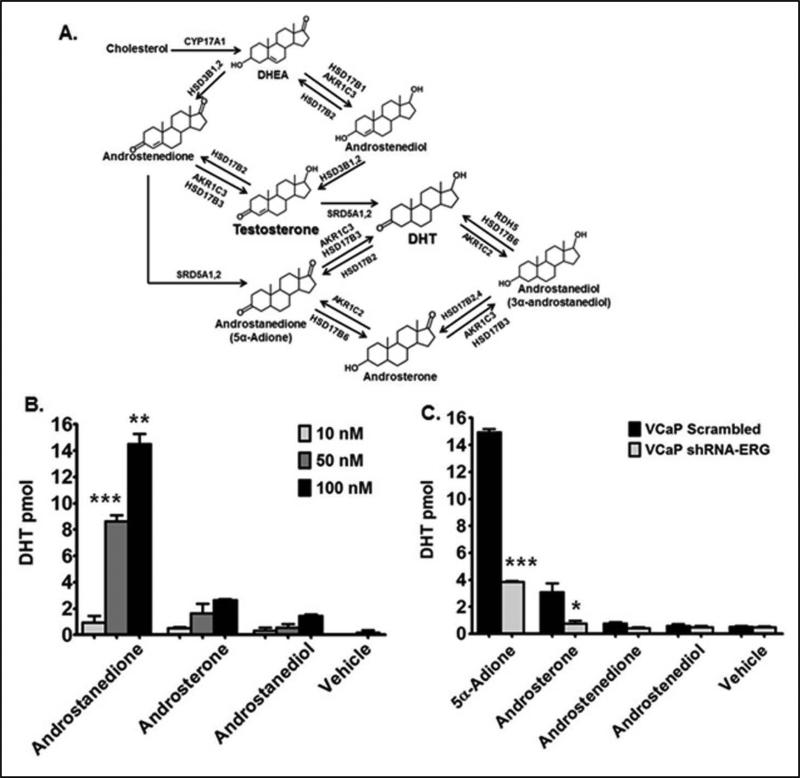
AKR1C3 participates in testosterone and DHT synthesis in the androgen biosynthetic pathway (Figure 3A). DHT can be produced directly from testosterone and indirectly through androstenedione without involving testosterone (testosterone bypass pathway). Using HPLC/MS/MS analysis we investigated the production of DHT from its immediate precursors’ as shown in Figure 3A. Previous studies support the notion that androstendione to 5α-Adione is the preferred pathway in PCa cells for the production of DHT (27). VCaP shScrambled cells were treated with (10 nM), (50 nM), and (100 nM) of 5α-Adione, androsterone and androstanediol, and cells and media were collected for MS analysis. A significant dose-dependent increase in DHT synthesis was observed following treatment with 5α-Adione (Figure 3B). These data show that in VCaP cells DHT is produced from 5α-Adione pathway .
Figure 3.
Androstanedione is a precursor metabolite for DHT in TMPRSS2-ERG fusion-positive cells. A.) Diagram illustrating the DHT biosynthesis pathway and the enzymes that synthesize DHT. B.) HPLC/MS/MS analysis in VCaP shSrc cells; DHT levels were quantified as pmol per one million cells. C.) HPLC/MS/MS analysis in VCaP shScr and shERG cells; DHT levels were quantified as pmol per one million cells. Vehicle controls represent 0.1% EtOH treated samples. For statistical analysis a two-tailed, unpaired, student's t-test was performed (N=3) *P<0.05, **P<0.01, ***P< 0.001.
To investigate whether ERG activation of AKR1C3 gene expression drives androgen synthesis in fusion-positive PCa cells, we used HPLC/MS/MS analysis and performed a substrate feeding experiment comparing VCaP shScrambled and shERG cells. ERG knockdown did not significantly change the production of testosterone from androstenedione (Supplementary Figure 2C). However, in ERG knockdown cells DHT production from 5α-Adione and androsterone was significantly reduced compared to shScrambled controls (Figure 3C). Interestingly, testosterone precursors androstenedione and androstenediol were converted to DHT at very low levels in VCaP shScrambled or shERG cells (Figure 3C), suggesting that 5α-Adione pathway has a more profound effect on DHT synthesis. AKR1C3 and HSD17B3 are the enzymes responsible for catalyzing the biochemical reduction of 5α-Adione to DHT. However, in VCaP cells PCR data showed that HSD17B3 was expressed at very low levels, and AKR1C3 was shown to be highly expressed. This suggests that AKR1C3 may be the enzyme responsible for the conversion of 5α-Adione to DHT in VCaP cells. Taken together these data suggest that AKR1C3 enzyme catalyzes the biochemical reduction of 5α-Adione to DHT, and that ERG regulates this step through upregulation of AKR1C3 expression in fusion-positive PCa cells.
Androstanedione induces AR activation through ERG regulated AKR1C3 expression
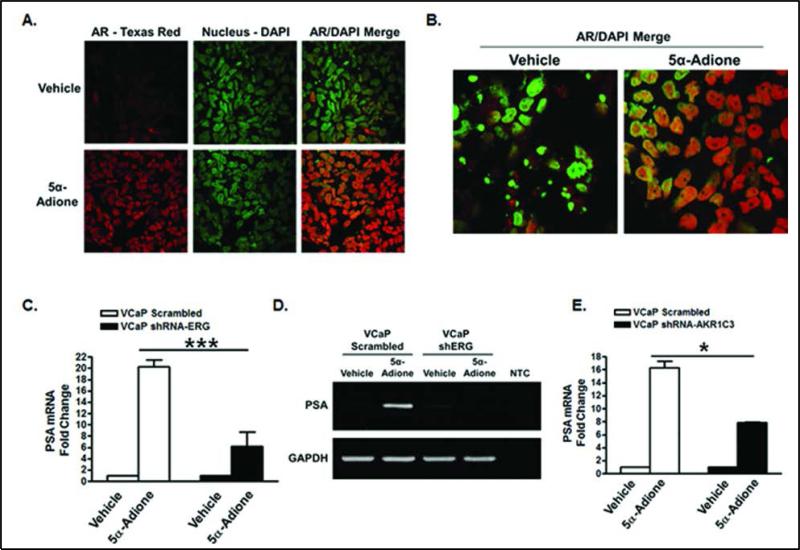
The data above show that androstanedione (5α-Adione) treatment of VCaP cells caused an increase in DHT synthesis; therefore, we investigated whether this had any functional consequence on AR activation. AR activation can be determined by AR translocation into the nucleus, and by AR target gene expression. Immunocytochemistry in VCaP cells demonstrated that (100 nM) 5α-Adione treatment induced a marked increase in AR translocation into the nucleus compared to vehicle controls (Figure 4A and Figure 4B). AR co-localization with the nucleus was quantified using Velocity software (PerkinElmer) analysis and represented as a global Pearson's correlation coefficient. In vehicle treated cells AR had a weak to moderate correlation with the nucleus (r = 0.38), however in 5α-Adione treated cells AR had a strong correlation with the nucleus (r = 0.72) (Supplementary Figure 3A). RT-qPCR analysis showed a significant increase in PSA expression in VCaP shScrambled cells following treatment with (100 nM) 5α-Adione compared to VCaP shERG cells (Figure 4C). In addition, RT-PCR data showed that activation of PSA expression was abrogated in VCaP shERG cells treated with 5α-Adione (Figure 4D); this further supports the notion that AKR1C3 is the enzyme responsible for the conversion of 5α-Adione to DHT. In order to study the direct role of 5α-Adione synthesis to DHT in AR activation, we performed shRNA knockdown of AKR1C3 in VCaP cells using shAKR1C3 lentivirus (Supplementary Figure 3C). RT-qPCR analysis showed a significant increase in PSA expression in VCaP shScrambled cells following treatment with (100 nM) 5α-Adione compared to VCaP shAKR1C3 cells (Figure 4E). Taken together, these data show that ERG enhances 5α-Adione induced AR activation through direct upregulation of AKR1C3 expression.
Figure 4.
Androstanedione induces AR activation through ERG regulated AKR1C3 expression. Immunocytochemistry in VCaP cells treated with vehicle or (100 nM) 5α-Adione for 24 hours. Confocal images were shown in panels A and B; cells were stained with anti-AR, Texas Red and DAPI as a green pseudo color. PCR amplification of PSA expression is shown in panel C-D. C.) RT-qPCR of PSA expression in VCaP shScr and VCaP shERG lentivirus infected cells, D.) RTPCR analysis of PSA expression in VCaP shScr and shERG lentivirus infected cells. NTC is non-template control. E.) RT-qPCR analysis of PSA expression in VCaP shScr and shAKR1C3 lentivirus infected cells. For statistical analysis a two-tailed, unpaired, student's t-test was performed C.) (N=5) and E.) (N=2) *P<0.05, ***P<0.001.
ERG and AKR1C3 are co-expressed in human prostate tumor tissue specimens, and predict lower survival probability
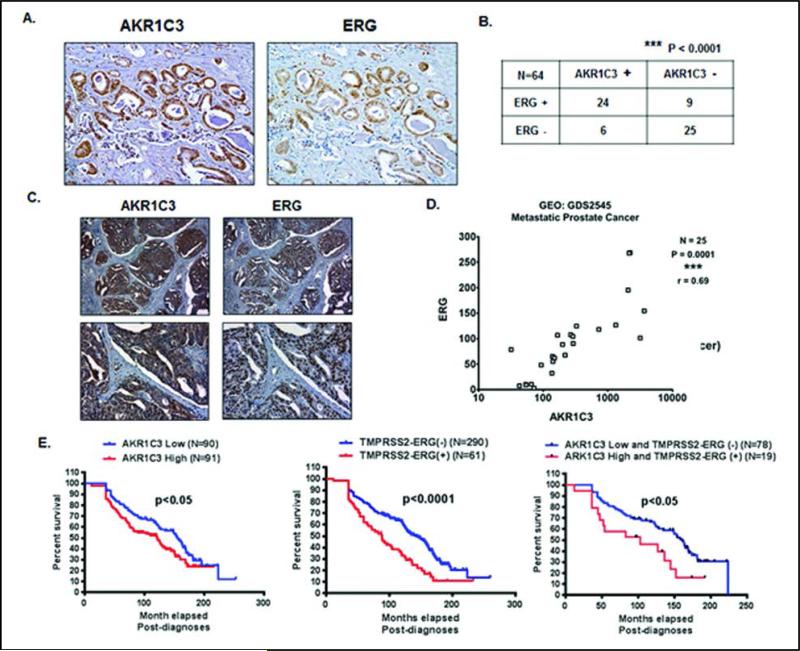
To determine the expression of ERG and AKR1C3 in human tumor tissues, immunohistochemical (IHC) staining of ERG and AKR1C3 in 64 human prostate tumor tissues was performed. IHC staining revealed that ERG and AKR1C3 were co-expressed in human prostate tumor tissue (Figure 5A). The distribution of 64 patient tumor tissue samples for the presence or absence of ERG and AKR1C3 showed that there was significant co-expression of the two genes (Figure 5B). ERG and AKR1C3 were also co-expressed in TMPRSS2-ERG fusion-positive LuCaP xenograft tissue specimens (Figure 5C). Analysis of 25 metastatic prostate tumor specimens extracted from Gene Expression Omnibus publicly available microarray data revealed a significant correlation (***P = 0.0001; r = 0.69) between ERG and AKR1C3 expression (Figure 5D). ERG and AKR1C3 expression also correlated in GEO microarray data from in vitro prostate cell line experiments (Supplementary Figures 4B-4D). Data extracted from the Oncomine database containing 363 prostate tumor tissue specimens was used to perform Kaplan-Meier Survival plots of AKR1C3 low vs. high expression tumors (Supplementary Figure 4A), and TMPRSS2-ERG fusion-negative vs. fusion-positive tumors. Analysis of 90 patients by low or high expression of AKR1C3 and ERG in prostate tumor tissues, showed that there was a significant decrease (P = 0.022 for AKR1C3 and P<0.0001 for ERG) in survival for AKR1C3 high expression tumors, and for TMPRSS2-ERG fusion-positive tumors (Figure 5E). The median survival for AKR1C3 low tumors or TMPRSS2-ERG fusion-negative tumors was 12-13 years, whereas the median survival for AKR1C3 high tumors or TMPRSS2-ERG fusion-positive tumors was 7-10 years. Analysis of these patients by AKR1C3 low and ERG negative vs AKR1C3 high and ERG positive showed that there was a significant decrease (P = 0.017) in survival for AKR1C3 high and ERG positive tumors. The median survival for AKR1C3 low and ERG negative tumors was 13 years, whereas the median survival for AKR1C3 high and ERG positive tumors was 8.5 years.
Figure 5.
ERG and AKR1C3 are co-expressed in human prostate tumor tissue specimens, and predict lower survival probability. A.) Immunohistochemical analysis of AKR1C3 and ERG in human prostate tumor tissue specimens. B.) Distribution of 64 patients by presence or absence of AKR1C3 and ERG in prostate tumor tissues, P<0.0001. C.) Immunohistochemical analysis of AKR1C3 and ERG in LuCaP xenograft specimens. D.) Correlation plot of AKR1C3 and ERG expression using microarray data from 25 metastatic PCa samples. Data was extracted from Gene Expression Omnibus database E.) Kaplan-Meier survival plot of (Left) AKR1C3 low tumors vs. AKR1C3 high tumors,(Middle) ERG fusion-negative tumors vs. ERG fusion-positive tumors and (Right) AKR1C3 low and TMPRSS2-ERG negative vs. AKR1C3 high and TMPRSS2-ERG positive tumors. Data was extracted from Oncomine database. For statistical analyses of Kaplan-Meier Survival a Mantel-Cox Test and a Gehan-Breslow-Wilcoxon Test were performed..
Discussion
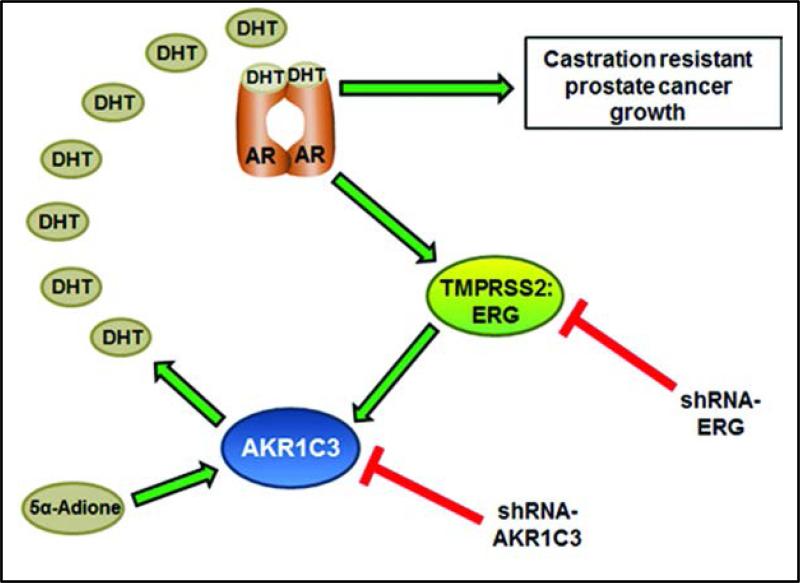
The TMPRSS2-ERG fusions persist in castrate resistant prostate cancer (low androgen state) and fuel tumor growth through oncogenic ERG mechanisms. This implies that in order for ERG to be expressed in tumors, there should be at least some active androgen synthesis and AR signaling. As advanced tumors often have low systemic androgen exposure, it is well accepted that tumors gain the capability of androgen synthesis for tumor growth, but the transcriptional mechanisms contributing to the androgen biosynthetic machinery are not known. Here we describe a model in which AR activates expression of the TMPRSS2-ERG transcription factor, which then activates androgen synthesis via regulation of androgen biosynthetic enzyme (ABE), AKR1C3. Under this model, AKR1C3 can utilize 5α-Adione as a substrate and synthesize intratumoral DHT in order to provide continual AR activation in a castrate environment. This can fuel AR signaling, as well as provide a feed-forward activation loop of TMPRSS2-ERG and AKR1C3 expression in prostate tumors (Figure 6).
Figure 6.
Schematic representation of proposed role of TMPRSS2-ERG fusions in androgen biosynthesis and AR activation via feed-forward activation.
It has been shown previously that androgens can regulate AKR1C3 expression. Specifically, high androgen levels suppress activation of AKR1C3 expression, whereas androgen-starved conditions activate expression of AKR1C3 (22, 40). This provides negative and positive regulation where in the presence of high and low androgen, respectively, AKR1C3 expression is regulated as a means to balance intracellular androgen levels. Androgen inhibition of AKR1C3 is thought to be regulated, in part, by AR-induced recruitment of lysine-specific demethylase 1 (LSD1) to the AKR1C3 gene (40). Whether TMPRSS2-ERG plays a role in androgen-induced regulation of AKR1C3 remains unclear; although, microarray data from supplementary figure 4B suggests that ERG positively regulates AKR1C3 expression independent of androgen status. The ChIP and ChIP-seq data from our study (Figure 2) show a novel transcription mechanism, whereby ERG transcription factor directly binds to the AKR1C3 gene locus and activates gene expression. We cannot rule out direct ERG factor regulation of AR activity, as Yu et al. demonstrated that ERG can occupy AR targets and regulate their expression (19). In such cases, ERG can override AR repressive activity and directly regulate AKR1C3 expression. AKR1C3 genome analysis (Figure 2A and B) supports this idea as shown by direct binding of ERG to AKR1C3 intron and 5’`-UTR. Another implication of Yu`s study is that ERG inhibits AR signaling involving pro-differentiation function and shifts cells to stem cell dedifferentiation and oncogenesis. As our data support the notion that ERG activates androgen synthesis through AKR1C3 expression, the tumor-expressed androgens can mediate growth function through AR activation. As ERG depends on AR for its own expression, ERG-induced androgens are a link for sustained expression of ERG in advanced tumors via a feed-forward loop (Figure 6). Sustained expression of ERG, restored AR activity, and expression of androgen biosynthetic enzymes exist together in castrate-resistant prostate cancer (CRPC) (41); our study provides a mechanistic link among these entities, which in concert promote tumor growth.
Intracrine androgen synthesis has been charged with activation of AR and tumor growth despite gonadal depletion of androgens. Recent reports demonstrate that DHT production in tumors sustains AR-mediated tumor growth, and strongly implicate the involvement of ABEs in DHT production (27). Specifically, this study demonstrates that DHT synthesis bypasses testosterone production and that 5α-Adione is readily converted to DHT. Our data show that, among various DHT precursors, 5α-Adione is more potent in making DHT in ERG-positive cells (Figure 3). In addition, AKR1C3 has been shown to be highly expressed in VCaP cells (41) and our data demonstrate that ERG regulates AKR1C3 expression in fusion-positive cells. Even though it can participate in production of both testosterone and DHT, our data suggest that ERG- knockdown cells have severe impairment in production of DHT while testosterone production is maintained. This suggests that tumor cells have preference for DHT production over testosterone and that the ERG fusion-gene product regulates the key enzymes in the pathway. Consistent with this idea, AKR1C3 expression is low or absent in fusion-negative cells and higher in fusion-positive cells (26). This gene is highly expressed in CRPC patients, thus, fusion status may dictate expression of AKR1C3 and DHT production.
Based on our data fusion-positive cells have higher propensity to synthesize DHT from DHEA → androstendione → 5α-Adione → DHT. In this pathway 3β−hydroxysteroid dehydratase (HSD3B) catalyzes the rate-limiting step in conversion of adrenal steroids such as DHEA to androstenedione. Tumors have been shown to express a gain-of-function mutation in HSD3B1 which increases the stability of the protein and provides a higher flux of DHT production from adrenal steroid precursors (42). Our studies also show that, in VCaP cells, ERG regulates HSD3B2 (Figure 1B) gene expression; thus, not only enzyme stability but overexpression of this enzyme may contribute to DHT production. Taken together, our data demonstrate that ERG participates in the expression of multiple ABEs in the DHT biosynthetic pathway.
Our data support the hypothesis that, in TMPRSS2-ERG fusion-positive cells, DHT is produced from the 5α-Adione pathway, and this drives AR activation. However, the finding that AKR1C3 may catalyze 5α-Adione into DHT is a novel finding that lends further support for therapeutically targeting AKR1C3 in PCa patients. It may be beneficial in the future to screen PCa patients for levels of 5α-Adione and AKR1C3 in their tumor biopsies. This study suggests that TMPRSS2-ERG fusion-positive PCa patients may have favorable response rates to anti-AR targeted therapies, such as enzalutamide, abiraterone, or AKR1C3-specific inhibitors.
Supplementary Material
Translational relevance.
Intracrine androgen synthesis is a viable target for clinical management of patients undergoing traditional androgen deprivation therapies. Despite newer therapies such as abiraterone acetate, a drug which targets androgen biosynthetic enzyme Cyp17, the disease eventually relapses (abiraterone-resistance). Elucidating molecular mechanisms contributing to the expression of biosynthetic enzymes in androgen production helps to understand the biological mechanisms contributing to the development of castration resistant prostate cancer (CRPC). ERG factor regulates key enzymes that participate in DHT production in prostate cancer cells. ERG specifically binds to AKR1C3 gene, regulates its expression, promotes DHT production via 5-α androstanedione and results in AR activation. Further, both ERG factor and AKR1C3 are co-expressed in human prostate tumors and predict lower survival in patients. As TMPRSS2-ERG fusions are prevalent in prostate cancer including CRPC and our data present a link between ERG expression, androgen biosynthesis and AR activation, this study suggests that TMPRSS-ERG fusion-positive patients may have favorable responses to anti-AR therapies.
Acknowledgements
The work was supported by U.S. Department of Defense, W81XWH-09-1-0250, and NIH-NCI Grant CA151557. Katelyn Powell was supported by T32 CA009531 pre-doctoral training grant. We would like to thank lipidomics core facility for performing MS analysis androgens and androgen precursors. Lipidomics core was supported by National Center for Research Resources, National Institutes of Health Grant S10RR027926. We would like to thank Dr. Arul Chinnaiyan and Dr. Xuhong Cao for sharing ChiP Seq data. We would like to thank Dr. Daniel Bonfil for providing LuCaP xenograft tumor slides for IHC analysis. The authors thank Dr. Michael Tainsky and Dr. Raymond Mattingly of Wayne State University for critically reading the manuscript.
¶ The Fund for Cancer Research, and NIH-NCI Cancer Center Support Grant CA-22453.
Footnotes
Conflict of interest: None.
Citations
- 1.Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20(5):538–44. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 2.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer research. 2006;66(17):8337–41. doi: 10.1158/0008-5472.CAN-06-1482. Epub 2006/09/05. [DOI] [PubMed] [Google Scholar]
- 3.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22(8):1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rickman DS, Chen YB, Banerjee S, Pan Y, Yu J, Vuong T, et al. ERG cooperates with androgen receptor in regulating trefoil factor 3 in prostate cancer disease progression. Neoplasia. 2010;12(12):1031–40. doi: 10.1593/neo.10866. Epub 2010/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer research. 2009;69(7):2912–8. doi: 10.1158/0008-5472.CAN-08-3667. Epub 2009/04/03. [DOI] [PubMed] [Google Scholar]
- 6.Baweja R, Calhoun S, Singareddy R. Sleep problems in children. Minerva pediatrica. 2013;65(5):457–72. Epub 2013/09/24. [PubMed] [Google Scholar]
- 7.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.FitzGerald L, Agalliu I, Johnson K, Miller M, Kwon E, Hurtado-Coll A, et al. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: results from a population-based study of prostate cancer. BMC Cancer. 2008;8(1):230. doi: 10.1186/1471-2407-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2007;27(3):253–63. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21(12):1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 11.Hagglof C, Hammarsten P, Stromvall K, Egevad L, Josefsson A, Stattin P, et al. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PloS one. 2014;9(2):e86824. doi: 10.1371/journal.pone.0086824. Epub 2014/02/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 13.Barwick BG, Abramovitz M, Kodani M, Moreno CS, Nam R, Tang W, et al. Prostate cancer genes associated with TMPRSS2-ERG gene fusion and prognostic of biochemical recurrence in multiple cohorts. British journal of cancer. 2010;102(3):570–6. doi: 10.1038/sj.bjc.6605519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. British journal of cancer. 2007;97(12):1690–5. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonaccorsi L, Nesi G, Nuti F, Paglierani M, Krausz C, Masieri L, et al. Persistence of expression of the TMPRSS2:ERG fusion gene after pre-surgery androgen ablation may be associated with early prostate specific antigen relapse of prostate cancer: preliminary results. J Endocrinol Invest. 2009;32(7):590–6. doi: 10.1007/BF03346514. Epub 2009/06/06. [DOI] [PubMed] [Google Scholar]
- 16.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai JPK, Singareddy Rajareddy, Kropinski Anthony, Sheng Shijie, Cher Michael L, Chinni Sreenivasa R. Androgens Induce Functional CXCR4 via ERG Factor Expression in TMPRSS2-ERG Fusion Positive Prostate Cancer Cells. Translational Oncology. 2010;3(3) doi: 10.1593/tlo.09328. Epub (Not yet in print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leshem O, Madar S, Kogan-Sakin I, Kamer I, Goldstein I, Brosh R, et al. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PloS one. 2011;6(7):e21650. doi: 10.1371/journal.pone.0021650. Epub 2011/07/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17(5):443–54. doi: 10.1016/j.ccr.2010.03.018. Epub 2010/05/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer MJ, Smit FP, Sedelaar JP, Schalken JA. Steroidogenic enzymes and stem cell markers are upregulated during androgen deprivation in prostate cancer. Mol Med. 2011;17(7-8):657–64. doi: 10.2119/molmed.2010.00143. Epub 2011/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasiah KK, Gardiner-Garden M, Padilla EJ, Moller G, Kench JG, Alles MC, et al. HSD17B4 overexpression, an independent biomarker of poor patient outcome in prostate cancer. Molecular and cellular endocrinology. 2009;301(1-2):89–96. doi: 10.1016/j.mce.2008.11.021. Epub 2008/12/23. [DOI] [PubMed] [Google Scholar]
- 22.Mitsiades N, Sung CC, Schultz N, Danila DC, He B, Eedunuri VK, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer research. 2012;72(23):6142–52. doi: 10.1158/0008-5472.CAN-12-1335. Epub 2012/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efstathiou E, Titus M, Tsavachidou D, Tzelepi V, Wen S, Hoang A, et al. Effects of abiraterone acetate on androgen signaling in castrate-resistant prostate cancer in bone. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(6):637–43. doi: 10.1200/JCO.2010.33.7675. Epub 2011/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. The New England journal of medicine. 2013;368(2):138–48. doi: 10.1056/NEJMoa1209096. Epub 2012/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. The New England journal of medicine. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. Epub 2011/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamid AR, Pfeiffer MJ, Verhaegh GW, Schaafsma E, Brandt A, Sweep FC, et al. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol Med. 2012;18:1449–55. doi: 10.2119/molmed.2012.00296. Epub 2012/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang KH, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(33):13728–33. doi: 10.1073/pnas.1107898108. Epub 2011/07/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohler JL, Titus MA, Wilson EM. Potential prostate cancer drug target: bioactivation of androstanediol by conversion to dihydrotestosterone. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(18):5844–9. doi: 10.1158/1078-0432.CCR-11-0644. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liedtke AJ, Adeniji AO, Chen M, Byrns MC, Jin Y, Christianson DW, et al. Development of potent and selective indomethacin analogues for the inhibition of AKR1C3 (Type 5 17beta-hydroxysteroid dehydrogenase/prostaglandin F synthase) in castrate-resistant prostate cancer. Journal of medicinal chemistry. 2013;56(6):2429–46. doi: 10.1021/jm3017656. Epub 2013/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamieson SM, Brooke DG, Heinrich D, Atwell GJ, Silva S, Hamilton EJ, et al. 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic Acids: highly potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase AKR1C3. Journal of medicinal chemistry. 2012;55(17):7746–58. doi: 10.1021/jm3007867. Epub 2012/08/11. [DOI] [PubMed] [Google Scholar]
- 31.Adeniji AO, Chen M, Penning TM. AKR1C3 as a target in castrate resistant prostate cancer. The Journal of steroid biochemistry and molecular biology. 2013;137:136–49. doi: 10.1016/j.jsbmb.2013.05.012. Epub 2013/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singareddy R, Semaan L, Conley-Lacomb MK, St John J, Powell K, Iyer M, et al. Transcriptional regulation of CXCR4 in prostate cancer: significance of TMPRSS2-ERG fusions. Molecular cancer research : MCR. 2013;11(11):1349–61. doi: 10.1158/1541-7786.MCR-12-0705. Epub 2013/08/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chinni SR, Sivalogan S, Dong Z, Filho JC, Deng X, Bonfil RD, et al. CXCL12/CXCR4 signaling activates Akt-1 and MMP-9 expression in prostate cancer cells: The role of bone microenvironment-associated CXCL12. Prostate. 2006;66(1):32–48. doi: 10.1002/pros.20318. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82. doi: 10.1038/nature13229. Epub 2014/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, et al. Estrogen-Dependent Signaling in a Molecularly Distinct Subclass of Aggressive Prostate Cancer. J Natl Cancer Inst. 2008;100(11):815–25. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Cai Y, Ren C, Ittmann M. Expression of Variant TMPRSS2/ERG Fusion Messenger RNAs Is Associated with Aggressive Prostate Cancer. Cancer research. 2006;66(17):8347–51. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 38.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41(5):524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10(2):177–88. doi: 10.1593/neo.07822. Epub 2008/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer cell. 2011;20(4):457–71. doi: 10.1016/j.ccr.2011.09.001. Epub 2011/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai C, Wang H, Xu Y, Chen S, Balk SP. Reactivation of Androgen Receptor-Regulated TMPRSS2:ERG Gene Expression in Castration-Resistant Prostate Cancer. Cancer research. 2009;69(15):6027–32. doi: 10.1158/0008-5472.CAN-09-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang KH, Li R, Kuri B, Lotan Y, Roehrborn CG, Liu J, et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell. 2013;154(5):1074–84. doi: 10.1016/j.cell.2013.07.029. Epub 2013/09/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.