Abstract
The prevalence of allergic conditions has continuously increased in the last few decades in Westernized countries. A dysbiotic gut microbiome may play an important role in the development of allergic diseases. Genetic, environmental and dietary factors may alter the commensal microbiota leading to inflammatory dysregulation of homeostasis. Murine and human studies have begun to elucidate the role of the microbiota in the pathogenesis of atopic diseases including asthma, atopic dermatitis and food allergies. However, the role of the microbiome in most eosinophilic gastrointestinal diseases (EGIDs) is not yet known. This review provides an overview of what is currently known about the development of tolerance from both molecular and clinical standpoints. We also look at the gut specific microbiome and its role in atopic conditions with the hope of applying this knowledge to the understanding, prevention and treatment of EGIDs, particularly EoE.
Keywords: asthma, atopic dermatitis, bacteria, eosinophilic esophagitis, eosinophil
I. Introduction
Atopic gastrointestinal diseases are a diverse group of antigen-mediated disorders that encompass IgE-mediated disorders related to anaphylaxis as well as delayed hypersensitivity responses that cause eosinophilic infiltration of the gut. While there is some evidence linking early gut dysbiosis to gut atopy, very little is known about the role of the microbiome in atopic and eosinophilic disorders of the gut. It is the goal of this review to examine the role of the gut microbiome in atopic disorders and attempt to explain how gut commensals influence specific gastrointestinal atopy.
II. Commensal bacterial exposure drives tolerance
The increasing incidence and prevalence of atopic diseases is most often attributed to the hygiene hypothesis. It is thought that the highly sterilized western lifestyle does not provide adequate exposure to pathogens, leading the immune system to mistake innocuous food and environmental particles for pathogens. Recent studies have found that subjects raised in rural communities with exposure to animals are less likely to develop atopic disease than subjects exposed to sterilized urban environments (1). Germ-free and antibiotic treated mouse models have also been used to test this hypothesis, and these models have supported the idea that early life exposure to diverse commensals is essential for the development of immunologic tolerance.
In murine models of allergic airway disease, failure of commensal population development causes dysregulation of the IgE-basophil axis (2). IgE is the immunoglobulin specifically linked to type 1 hypersensitivity anaphylactic reactions and in addition is responsible for binding mast cells and basophils characteristic of other atopic conditions.. Eosinophils express a number of anti-microbial products, including granule cationic proteins and defensins, and they generate DNA-containing extracellular traps (3) (4) (5). Similarly, basophils have recently been reported to kill bacteria through formation of extracellular traps (BETs) containing mitochondrial DNA and granule proteins (6). These products may change the local microbiota in atopic diseases where there is significant infiltration by these granulocytes. Hill et al. found that depletion of murine microbial populations enhances serum IgE and basophils in both antibiotic treated mice as well as germ free mice (2). These mice displayed enhanced basophil-mediated allergic inflammation of the lungs in a house dust mite model of asthma. This link between a commensal population of bacteria and IgE-basophil-mediated inflammation was found to be B cell intrinsic and MYD88 dependent, as MYD88 depleted mice had similarly enhanced levels of IgE and basophils to antibiotic-treated and germ free mice when compared to control populations. The authors postulate that the commensal population stimulates B cell MYD88 inhibition of IgE, thus decreasing basophil progenitor formation in the bone marrow.
Commensal bacteria are also important for maintaining a balance of type 1 T helper (Th1) cells and type 2 T helper (Th2) cells. Th1 cells are the effector cells against bacterial invasion, whereas Th2 cells historically deter parasitic invasion. However, in the setting of altered tolerance the Th2 response leads to the allergic responses seen in atopic disease. A careful balance of these cell types is important for tolerance. Improper exposure to commensals in infancy may be responsible for the enhanced Th2 response in atopic conditions. Oyama et al. found that treatment of mice with antibiotics in infancy resulted in an exaggerated IgE/IgG1 response with diminished IgG2 (7). Furthermore, the spleen and Peyer’s patches of these mice had decreased lymphocytes compared to untreated mice. Similar antibiotic exposure in older mice, presumably after gut microbial colonization, did not cause any of these immunologic changes, and both antibiotic treated and untreated mice had similar IgE levels. These results were supported in studies with the opposite approach, wherein exposure of germ-free mice to ova-albumin (OVA) resulted in Th2 predominant inflammation (8). Reconstitution of the microbiota with Bifidobacterium infantis in germ-free mice resulted in tolerance to OVA, though only if exposure was in the neonatal period.
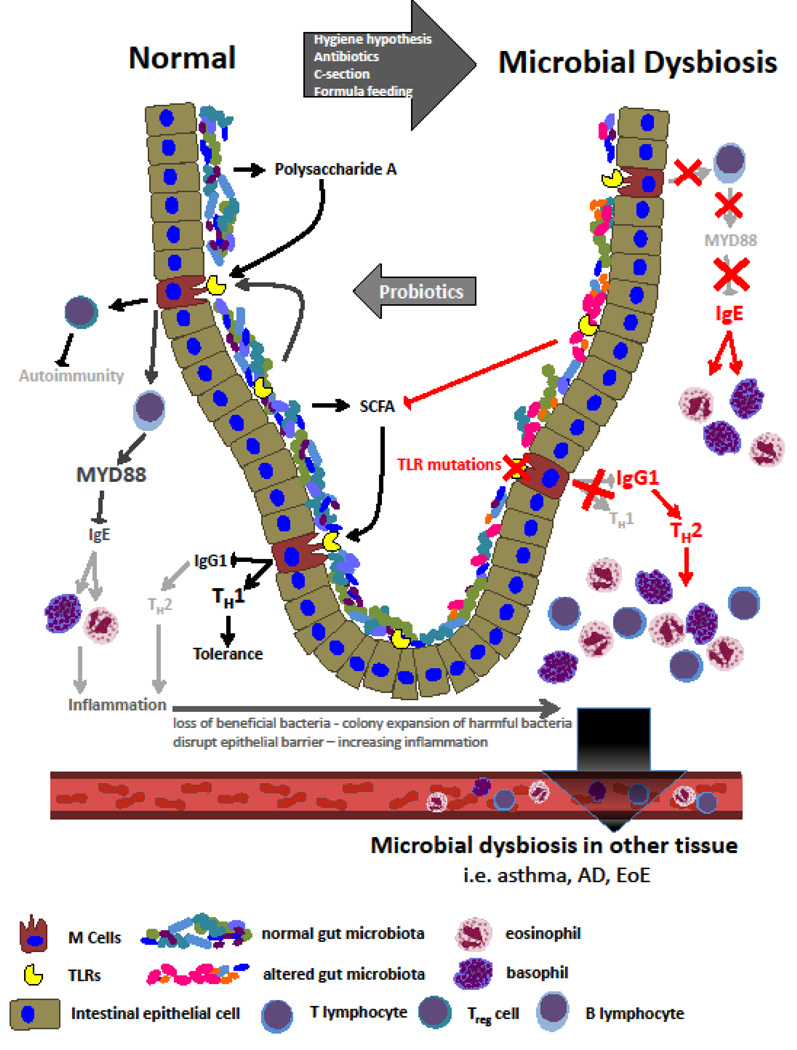
Correspondingly, an intact response to the commensal bacteria and its components is essential for the development of tolerance. The gut has evolved several systems to monitor the bacterial populations within the lumen, including signaling through toll-like receptors (TLRs). Previous studies have shown that defective TLR signaling has been implicated in development of food allergies; for example, mice with a mutation in TLR4 exposed to peanut extract undergo a systemic allergic response (9, 10). Interestingly, TLR4 wild-type mice neonatally exposed to antibiotics produce a similar response to peanut extract, suggesting that there is a TLR4-commensal response necessary for development of tolerance (11). Furthermore, exposure of the TLR4 mutant mice to the TLR9 agonist, CpG oligonucliotides (CPG-ODN), resulted in an attenuated response to the peanut extract. This CpG-ODN likely causes a shift toward the Th1 response (12), and acts to rescue the development of tolerance. Other mouse models of allergic disease have also shown that administration of CPG-ODN reduces airway inflammation and decreases IgG1 and IgE responses (12). Lastly, exposure to the commensal population’s polysaccharide A induces TLR2-dependent activation of the T regulatory population (Tregs) (13). Induction of this population of Tregs is necessary to suppress the development of autoimmunity and successfully attenuates pro-inflammatory Th17 responses that can be seen in the context of asthma and atopy (14).
In summary, exposure to commensals is necessary for proper development of tolerance through a number of mechanisms. Sterility in the murine neonatal period can lead to a shift in the IgE-basophil axis, an imbalance in Th1/Th2 activation, as well as improper activation of Tregs. Human studies to evaluate these mechanisms are limited, but some have implicated polymorphisms of TLR2 as well as TLR4 co-receptor CD14 in atopic disease (15, 16) (Figure 1). These genetic variations may provide a link between commensal bacteria and genetic disposition to atopic disease. These bacterial ligands also represent potential therapeutic targets in atopic disease.
Figure 1. Commensal bacteria and their role in the development of tolerance.
Commensal colonization decreases IgE-basophil axis and increases TLR stimulation thus promoting tolerance. Dysbiosis and ineffective TLR signaling leads to enhanced Th2 response and IgE-mediated disease.
III. Early life environment contributes to development of atopy
The quantity and quality of the population of commensals in the human gut during the neonatal period is shaped by early life exposures. Even the mode of birth can cause alterations in the gut microbiota that lead to altered immunologic responses. Jakobsson et al. showed that infants born by Cesarean section have decreased diversity of the Bacteroides phylum throughout the first two years of life compared to those born vaginally (17). Stokholm et al. showed that Cesarean section was significantly associated with colonization of the intestinal tract by Citrobacter freundii, Clostridium species, Enterobacter cloacae, Enterococcus faecalis, Klebsiella oxytoca, Klebsiella pneumoniae, and Staphylococcus aureus at 1week of life, whereas colonization by Escherichia coli was associated with vaginal birth (18). A recent pilot study by Dominguez-Bello et al. started to investigate the long-term health consequences of Cesarean section-delivered infants. They found that the microbiota of Cesarean-born infants could be partially restored via vaginal microbial transfer (19). Likewise, those born vaginally have enhanced circulating levels of Th1 cytokines compared to their Cesarean counterparts. The sterility of the surgical procedure and the lack of exposure to maternal vaginal microbiota have been blamed for this decreased colonization (20). While there is some variation in the results, there have been meta-analysis and population studies showing an increased risk of asthma and allergic rhinitis in infants born by Cesarean (21, 22), however very little is known about the relationship of delivery mode and gastrointestinal (GI) atopy.
While diet has been linked to gut microbial changes, dysbiosis and gut inflammation (23), most studies show few microbial differences between formula and human milk fed infants. There are some differences noted between the populations, with human milk fed infants having proportionally more Staphylococcus, Lactobacillus and fewer Clostridium difficile, Bacteroides, Enterococci and Enterobactericeae colonies (24). There may be some evidence that human milk is protective against early childhood wheezing, but there is no evidence that breastfeeding provides protection against food allergy (24–26). However, studies looking at breastfeeding are inherently limited due to inability to randomize and differences in duration of breastfeeding.
Antibiotic exposure is common in the neonatal period due to prematurity, maternal group B Streptococcus status, and sepsis protocols for febrile infants. It is difficult to control for this diverse population of patients who receive antibiotics early in life. There is a marginally significant increase in atopic disease in children treated with antibiotics prior to 3 months of life, but this may have been confounded by upper respiratory infections (27). In a more homogeneous population controlling for premature birth, Kummeling et al. found that antibiotic exposure in the first 6 months of life correlated with wheeze but not allergic sensitization or eczema (28). Further work must be done to understand the connections between neonatal antibiotic exposure and later development of atopic conditions.
While mode of delivery, formula feeding, and early antibiotic exposure are important factors in shaping the infant microbiome, there have been very few studies evaluating how these exposures shape the development of GI atopy. Jensen et al. performed a case-control study looking at the development of eosinophilic esophagitis (EoE) and found a 6-fold increase in EoE in children with antibiotic exposure in the first 6 months of life (29). While Cesarean delivery, preterm birth, and formula-only feeding trended toward increasing incidence of EoE, these measures were not significant. Moreover, dizygotic twins have a 10-fold higher EoE concordance rate compared with non-twin siblings, providing strong evidence for a profound impact of early-life exposures in addition to known genetic underpinnings for susceptibility to EoE (30). Sensitization to food antigen at 1 year of life was found to be associated with variations in the microbiome between 3 and 12 months of life. Specifically, infants with food sensitization by skin prick testing had increased Enterobacteria and decreased Bacteroides compared to those with negative skin prick testing (31). However, larger prospective studies looking at gastrointestinal atopy will be needed to help verify these causal relationships.
IV. Atopy and the microbiome –other organ systems
a. Asthma
The micromilieu at the site of inflammation creates a specific environmental niche where certain bacteria can proliferate and the gastrointestinal microbiota, as the central microbial habitat, can influence asthma and allergy development at other body sites. Recent evidence suggests that immune dysfunction in allergic diseases such as asthma and atopy is related to the function and composition of the gut microbiota. Arrita et al. suggest that fecal microbiota dysbiosis in the first 100 days of life, particularly reductions of the genera Faecalibacterium, Lachnospira, Veillonella and Rothia (FLVR), are linked to the development of allergic diseases (32).
Early studies using culture-based approaches have demonstrated that detection of a high abundance of either Escherichia coli or Clostridium difficile in neonatal feces is associated with significantly higher risk of developing childhood allergic disease (33) (34). Additionally, the absence of certain genera such as Bifidobacterium in neonatal stool has also been associated with an increased risk of developing childhood atopy in the first 5 years of life (35). These studies demonstrate that altered microbiota development in the neonatal gut lays the foundation for allergic disease and asthma development in childhood. Bifidobacterium adolescentis has recently been shown to be lower in fecal samples from adult allergic subjects with long term asthma and may represent a novel therapeutic target for modulation of the gut microbiota in this group of subjects (36).
b. Atopic dermatitis
Atopic dermatitis (AD) is often associated with other allergic diseases including asthma, allergic rhinitis (37) and eosinophilic gastrointestinal diseases such as EoE (38, 39) (40). With recent increase in methicillin resistant Staphylococcus aureus strains, antibiotic therapy is not the first choice for AD treatment. Corticosteroids, anti-inflammatory reagents and diluted bleach baths are more commonly used. Just as fecal microbiota therapy from healthy donors has shown success in recent years in reverting gut microbiota dysbiosis in subjects with recurrent Clostridium difficile infection, and seems promising for some other gastrointestinal diseases (41), in the future microbial skin transplant therapy may be useful for treating AD.
The gut microbiota in AD has not yet been as deeply investigated as the skin microbiota, but it has been recently reported by Song et al. that the gut microbiota in 90 AD subjects was enriched in Faecalibacterium prausnitzii, a species previously described to be deficient in the gut of Crohn’s disease subjects. Additionally, the gastrointestinal microbiota was enriched in genes encoding the use of nutrients that could be released from damaged gut epithelium, indicating a bloom of auxotrophic bacteria. Furthermore, anti-inflammatory bacterial metabolites such as butyrate and propionate had decreased levels in fecal samples from AD subjects (42). Specifically, studies have shown that disease improvement in infants was correlated with an increased abundance of fecal butyrate-producing bacteria Coprococcus eutactus (including Clostridial families Lachnospiracea and Ruminococeae) (43). These results suggest that the dysbiotic gut microbiota in AD and dysregulation of the gut inflammation and epithelial barrier could lead to an aberrant Th2-type immune response to allergens in the skin.
b. Food allergies
The increasing use of antibiotics in humans (particularly in infancy) and in agriculture for growth-promoting properties of livestock, and the increasing consumption of a high-fat/low fiber diet, have had a major impact on the gut microbiota, and have been associated with an increased allergic response to food in industrialized countries in the last few decades (44) (45). Animal models suggest that the gut microbiota, particularly in early life, play a crucial role in susceptibility to food sensitization and food allergy. Murine models of neonatal exposure to antibiotics have shown reduced gut microbiota diversity, and are associated with aberrant immunity to respiratory and dietary antigens and enhanced food allergen sensitization (46).
Atarashi et al. have identified Clostridia species in both mice and humans as being potent inducers of colonic Tregs capable of suppressing food allergy (47, 48). Stefka et al. reported that Clostridia colonization increased colonic Tregs and IL-22 expression in the small and large intestine. This alteration led to enhanced epithelial barrier function and prevented uptake of food allergens (46). Interestingly, Noval Rivas et al. have shown that the intestinal microbiota regulates susceptibility to food allergy. They identified a unique microbial signature in genetically food-allergy susceptible mice that can transmit food allergy susceptibility when transferred to germ free mice (49). Additionally, that same group recently identified a Th2-like phenotype in Tregs of food allergic mice; these specific Tregs play a key role in controlling food allergy (50).
Co-administration of bacterial adjuvants with oral immunotherapy (OIT) has been suggested as a potential treatment for food allergy. In the first double-blind, placebo-controlled randomized trial with 62 children on the probiotic Lactobacillus rhamnosus and peanut OIT (probiotic and peanut oral immunotherapy [PPOIT]) in children with peanut allergy, Tang et al. found that sustained unresponsiveness was achieved in 82.1% receiving PPOIT and 3.6% receiving placebo. Of the subjects receiving PPOIT 89.7% were desensitized. Although these results are very promising, further work is required to confirm sustained unresponsiveness after a longer period of secondary peanut elimination, and to clarify the relative contributions of probiotics versus OIT. In addition, the active group had more reactions suggesting an imbalance in treatment, which would affect efficacy (51).
As previously demonstrated in murine models, human studies also link the use of antibiotics with food allergies; maternal use of antibiotics during pregnancy and in the first month of life was associated with increased risk of cow’s milk allergy in infants (52). Infants with cow's milk allergy have a fecal microbial community dominated by Lachnospiraceae and Ruminococcaceae. Dietary intervention with extensively hydrolyzed casein formula supplemented with Lactobacillus rhamnosus GG was shown to expand butyrate-producing bacterial strains and accelerated tolerance acquisition in infants with cow's milk allergy (53).
The fecal microbiota of food allergic infants is characterized by increased relative abundance of Clostridium I and Anaerobacter and a decrease of Bacteroides and Clostridium XVIII (54) (Table 1). Chen et al. have recently shown that children with food sensitization in early life have an altered fecal microbiota compared to healthy controls, with lower microbiota diversity. Children with food sensitization had a significantly decreased number of Bacteroidetes and a significantly increased number of Firmicutes compared with healthy children. The most differentially abundant taxa in children with food sensitization was characterized by increased abundances of Clostridium IV and Subdoligranulum and decreased abundances of Bacteroides and Veillonella (55) (Table 1). Exploring novel Clostridia-based biotherapeutics or butyrate administration as adjunct therapy to promote tolerance to food allergens may be useful in the future.
Table 1.
Microbiota associated with human gut atopy
| Disease | Microbiota | Behavior of microbiota | Site | References and Authors |
|---|---|---|---|---|
|
Cow Milk Allergy |
Lachnospiraceae and Ruminococcaceae |
increased relative abundance | fecal | (53) Berni Canani et al. |
|
Food Allergies |
Clostridium I and Anaerobacter |
increased relative abundance | fecal | (54) Ling et al. |
|
Bacteroides and Clostridium XVIII |
decreased relative abundance | fecal | (54) Ling et al. | |
|
Sphingomonas, Sutterella, Bifidobacterium, Collinsella, Clostridium sensu stricto, Clostridium IV, Enterococcus, Lactobacillus, Roseburia, Faecalibacterium, Ruminococcus, Subdoligranulum, and Akkermansia |
increases in the numbers | fecal | (55) Chen et al. | |
|
Bacteroides, Parabacteroides, Prevotella, Alistipes, Streptococcus, and Veillonella |
decreases in the numbers | fecal | (55) Chen et al. | |
| EoE |
Prevotella, Streptococcus and Neisseria |
shared lineages | oral and esophageal |
(56) Benitez et al. |
|
Neisseria and Corynebacterium |
enrichment in active EoE | esophageal | (56) Benitez et al. | |
|
Granulicatella and Campylobacter |
enrichment with reintroduction of highly allergenic foods |
esophageal | (56) Benitez et al. | |
| Proteobacteria (Haemophilus) |
enriched in untreated EoE | esophageal | (58) Harris et al |
V. Microbiome in EoE
a. Philadelphia Experience
Once thought sterile or composed of few cultivable microbes, the esophagus has been shown to have a composition of around 300 bacteria species. New culture independent techniques available today have allowed scientists to identify the microbial composition of the esophagus in children and adults. Investigators at Children’s Hospital of Philadelphia (CHOP) sought to characterize the oral and esophageal microbiota in 33 children diagnosed with EoE as well as 35 controls. Oral samples were obtained through a swab method prior to endoscopy and esophageal samples from esophageal biopsies. The majority of subjects included in the study were on proton pump inhibitor therapy and on a food elimination or reintroduction diet. Subjects on antibiotics or presenting with any GI inflammatory process were excluded from the study.
Benitez et al. reported marked differences between the oral and esophageal microbiota with strains exclusively present in esophageal samples (56). However, some lineages were shared between the oral and esophageal samples, mostly members of the genus Prevotella, Streptococcus and Neisseria. Further analysis of the microbial composition of the esophagus revealed significant differences between EoE and control subjects. These differences were driven by EoE subjects with active esophageal inflammation when compared to controls, and only when taking into account relative abundance of species. An enrichment of the genus Neisseria and Corynebacterium was reported in esophageal samples of subjects with EoE and an enrichment of Streptococcus and Atopobium in the non-EoE control group.
Changes in the fecal microbiota have been largely associated with dietary interventions. Benitez et al. found that dietary changes affected the esophageal microbiota. Even though there were no significant differences in bacterial composition between active and control groups, an enrichment of the genus Granulicatella and Campylobacter was detected in subjects who reintroduced a highly allergenic food from the six-food elimination diet (Table 1).
This study in many ways is in agreement with previous reports in adults where the genus Streptococcus dominated the microbial composition of the healthy esophagus, while the genus Neisseria dominated the inflamed esophagus. Importantly, it would be of future interest to determine if specific bacterial lineages drive the inflammatory response or can be used to ameliorate the inflammation arising from the condition itself.
b. Denver and Chicago experience
Using the minimally invasive esophageal string test (EST), it was determined that the esophageal microbiota in 15 normal individuals was similar to the microbiota detected on esophageal biopsies, and that the esophageal bacterial genera were different from the bacterial contents of samples collected from the nasal and oral cavity (57).
More recently Harris et al. have shown that in 70 EST samples examined from children and adults, the bacterial load was increased with inflammation in the esophagus of subjects with EoE and gastroesophageal reflux disease (GERD) compared to controls (58). This increase in bacteria was independent of the degree of esophageal mucosal eosinophilia and treatment status. Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria were identified as the four predominant phyla in the esophagus of EoE, GERD and control subjects.
In about half of the subjects on proton-pump-inhibitor treatments in both Denver and Chicago, it was found that the phylum Proteobacteria was enriched in untreated (no steroids or diet) EoE subjects, similar to what has been observed in the Philadelphia study in active EoE subjects on PPI by Benitez et al. (56). The results from both of these studies demonstrate that active esophageal inflammation in EoE is associated with Proteobacteria enrichment on mucosal biopsies and EST samples (59).
Harris et al. showed that Haemophilus genus from the Proteobacteria phylum was significantly increased in the esophagus of untreated EoE subjects (Table 1). Streptococcus, the predominant genus in the esophagus, was decreased in PPI treated GERD subjects, and Aggregatibacter was enriched in the esophagus of PPI treated GERD subjects. Results from Benitez et al. and Harris et al. indicate that not only the degree of inflammation but also the treatment regimen has a strong impact on the esophageal microbiota in EoE. These results offer a baseline of knowledge of bacterial communities in the esophagus in EoE subjects, and provide important findings for the design of future studies, longitudinal analyses and the impact of specific treatments on the microbiota in EoE and other EGIDs. Further studies in different countries with larger numbers of subjects will help delineate the effect of alterations in the microbiota and treatment on esophageal inflammation, and potentially help to identify novel therapeutic targets for these diseases.
c. Pros and Cons and future needs
According to Hippocrates (460-377 BC), all diseases begin in the gut. The mucosal surface of the gastrointestinal tract and its microbiota act as an important organ for host defense. In the EoE microbiome studies performed in Philadelphia, Chicago and Denver it was observed that independent of the sites of collection, subjects with active EoE have an enrichment in Proteobacteria. However, each center has different treatment approaches, and we know that treatment has an impact on the esophageal microbiota. In the future it will be necessary to examine the microbiome of EoE patients on dietary therapy and compare it to the steroid treated population. Further complicating the microbiome of the EoE population, a new population of patients with PPI responsive esophageal eosinophilia has been recently identified. These patients have esophageal eosinophilia and inflammation, but show clinical and histologic improvement in response to PPI therapy. Further investigation of the microbiota in these individuals may help determine which type of bacteria are associated with inflammation and which are associated with specific treatments (60).
Additionally, to determine the functional consequences of specific alterations in microbiome composition the field is in need of models development (animal, germ-free, in vitro and ex vivo) to better understand the impact of an altered microbiota or specific bacteria to demonstrate causality of the microbiota in allergic diseases and inflammation. There are multiple mouse models of EoE (61–68). Some of these models are transgenic combined with antigen exposure while others involve inducing an immune response with cutaneous or inhaled antigen exposure. The inflammation in these models varies in location (whole gut vs. esophagus alone), the type of granulocytes infiltrating, as well as position of inflammation within the esophagus (lamina propria vs. epithelia). Because of these differences, teasing the functional role of the microbiota in EoE will have additional obstacles to overcome before the data translates to the human population and will likely need to be performed in multiple murine and germ-free models.
Future multicenter studies will be necessary to determine if the gastrointestinal microbiome in subjects with atopy (asthma, AD, EGIDs) have some similar bacterial communities characteristic of allergic disease that may have the potential to be targeted and altered to reconstitute a physiologically “normal microbiota”.
VI. Conclusion
We are only at the beginning of untangling the complex network and relationships between the human gut microbiota, environmental factors and the genetics in health and atopic disease. However, the number of publications describing an altered microbiota in allergic disease has significantly increased in recent years. Further work including longitudinal studies and multicenter projects are still needed to better understand if the dysbiotic microbiota identified in various atopic diseases is a cause or consequence of the disease. Changes in lifestyle such as increase in Cesarean sections, early life formula feeding and increased use of antibiotics in infancy have all been linked to disturbed microbial homeostasis and appear to significantly influence childhood allergic disease susceptibility. However, the use of supplementation with probiotics to stabilize microbial homeostasis seems promising in some studies but conflicting in others, indicating that large multicenter trials, with well characterized populations and long term follow up will be necessary in the future to better understand the potential beneficial impact from probiotics, prebiotics and bacterial produced metabolites in the treatment of allergic disease.
Acknowledgments
We thank Drs. Glenn T. Furuta, Dan Atkins, Marc Rothenberg, Sandeep K. Gupta and Steven J. Ackerman, for their insights, critical review and editing of the manuscript.
Funding: JMS, ABM and SAF are funded in part by the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). CEGIR (U54 AI117804) is part of the Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), NCATS, and is funded through collaboration between NIAID, NIDDK, and NCATS. ABM is supported by NIH 1K08DK106444-01 and the AGA Donald and June Castell Award; JMS and AJB are funded The Children’s Hospital of Philadelphia Eosinophilic Esophagitis Research Fund.
Abbreviations
- AD
atopic dermatitis
- BETs
basophil extracellular traps
- EETs
eosinophil extracellular traps
- EGIDs
eosinophilic gastrointestinal diseases
- EoE
eosinophilic esophagitis
- EST
esophageal string test
- GERD
gastroesophageal reflux disease
- IG
Immunoglobulin
- GI
gastrointestinal
- CPG-ODN
CpG oligonucliotides
- OVA
ova-albumin
- PPI
proton pump inhibitor
- PPOIT
probiotic and peanut oral immunotherapy
- TLR
toll-like receptor
Footnotes
Conflict of interest: no potential conflict of interest.
Author contributions: ABM, AJB, JMS and SAF wrote and revised the manuscript, KD designed the figure.
References
- 1.Ege MJ, Bieli C, Frei R, van Strien RT, Riedler J, Ublagger E, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006;117(4):817–823. doi: 10.1016/j.jaci.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- 2.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18(4):538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J Immunol. 1989;142(12):4428–4434. [PubMed] [Google Scholar]
- 4.Driss V, Legrand F, Hermann E, Loiseau S, Guerardel Y, Kremer L, et al. TLR2-dependent eosinophil interactions with mycobacteria: role of alpha-defensins. Blood. 2009;113(14):3235–3244. doi: 10.1182/blood-2008-07-166595. [DOI] [PubMed] [Google Scholar]
- 5.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14(9):949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 6.Yousefi S, Morshed M, Amini P, Stojkov D, Simon D, von Gunten S, et al. Basophils exhibit antibacterial activity through extracellular trap formation. Allergy. 2015;70(9):1184–1188. doi: 10.1111/all.12662. [DOI] [PubMed] [Google Scholar]
- 7.Oyama N, Sudo N, Sogawa H, Kubo C. Antibiotic use during infancy promotes a shift in the T(H)1/T(H)2 balance toward T(H)2-dominant immunity in mice. J Allergy Clin Immunol. 2001;107(1):153–159. doi: 10.1067/mai.2001.111142. [DOI] [PubMed] [Google Scholar]
- 8.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159(4):1739–1745. [PubMed] [Google Scholar]
- 9.Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, et al. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J Allergy Clin Immunol. 2000;106(1 Pt 1):150–158. doi: 10.1067/mai.2000.107395. [DOI] [PubMed] [Google Scholar]
- 10.Bashir ME, Andersen P, Fuss IJ, Shi HN, Nagler-Anderson C. An enteric helminth infection protects against an allergic response to dietary antigen. J Immunol. 2002;169(6):3284–3292. doi: 10.4049/jimmunol.169.6.3284. [DOI] [PubMed] [Google Scholar]
- 11.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172(11):6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 12.Liu N, Ohnishi N, Ni L, Akira S, Bacon KB. CpG directly induces T-bet expression and inhibits IgG1 and IgE switching in B cells. Nat Immunol. 2003;4(7):687–693. doi: 10.1038/ni941. [DOI] [PubMed] [Google Scholar]
- 13.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo JG, Assa'ad A, Heizer AB, Bernstein JA, Hershey GK. The-159 C-->T polymorphism of CD14 is associated with nonatopic asthma and food allergy. J Allergy Clin Immunol. 2003;112(2):438–444. doi: 10.1067/mai.2003.1634. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Radler D, Illi S, Klucker E, Turan E, von Mutius E, et al. TLR2 polymorphisms influence neonatal regulatory T cells depending on maternal atopy. Allergy. 2011;66(8):1020–1029. doi: 10.1111/j.1398-9995.2011.02573.x. [DOI] [PubMed] [Google Scholar]
- 17.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63(4):559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 18.Stokholm J, Thorsen J, Chawes BL, Schjorring S, Krogfelt KA, Bonnelykke K, et al. Cesarean section changes neonatal gut colonization. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22(3):250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016 doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clin Exp Allergy. 2008;38(4):629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 22.Pistiner M, Gold DR, Abdulkerim H, Hoffman E, Celedon JC. Birth by cesarean section, allergic rhinitis, and allergic sensitization among children with a parental history of atopy. J Allergy Clin Immunol. 2008;122(2):274–279. doi: 10.1016/j.jaci.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146(6):1564–1572. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prince BT, Mandel MJ, Nadeau K, Singh AM. Gut Microbiome and the Development of Food Allergy and Allergic Disease. Pediatr Clin North Am. 2015;62(6):1479–1492. doi: 10.1016/j.pcl.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matheson MC, Allen KJ, Tang ML. Understanding the evidence for and against the role of breastfeeding in allergy prevention. Clin Exp Allergy. 2012;42(6):827–851. doi: 10.1111/j.1365-2222.2011.03925.x. [DOI] [PubMed] [Google Scholar]
- 26.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 27.Wickens K, Ingham T, Epton M, Pattemore P, Town I, Fishwick D, et al. The association of early life exposure to antibiotics and the development of asthma, eczema and atopy in a birth cohort: confounding or causality? Clin Exp Allergy. 2008;38(8):1318–1324. doi: 10.1111/j.1365-2222.2008.03024.x. [DOI] [PubMed] [Google Scholar]
- 28.Kummeling I, Stelma FF, Dagnelie PC, Snijders BE, Penders J, Huber M, et al. Early life exposure to antibiotics and the subsequent development of eczema, wheeze, and allergic sensitization in the first 2 years of life: the KOALA Birth Cohort Study. Pediatrics. 2007;119(1):e225–e231. doi: 10.1542/peds.2006-0896. [DOI] [PubMed] [Google Scholar]
- 29.Jensen ET, Kappelman MD, Kim HP, Ringel-Kulka T, Dellon ES. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013;57(1):67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 30.Alexander ES, Martin LJ, Collins MH, Kottyan LC, Sucharew H, He H, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134(5):1084–1092. e1081. doi: 10.1016/j.jaci.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. 2015;45(3):632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 32.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 33.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107(1):129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 34.Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, et al. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy. 2006;36(12):1602–1608. doi: 10.1111/j.1365-2222.2006.02599.x. [DOI] [PubMed] [Google Scholar]
- 35.Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39(4):518–526. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 36.Hevia A, Milani C, Lopez P, Donado CD, Cuervo A, Gonzalez S, et al. Allergic Patients with Long-Term Asthma Display Low Levels of Bifidobacterium adolescentis. PLoS One. 2016;11(2):e0147809. doi: 10.1371/journal.pone.0147809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapoor R, Menon C, Hoffstad O, Bilker W, Leclerc P, Margolis DJ. The prevalence of atopic triad in children with physician-confirmed atopic dermatitis. J Am Acad Dermatol. 2008;58(1):68–73. doi: 10.1016/j.jaad.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 38.Jyonouchi S, Brown-Whitehorn TA, Spergel JM. Association of eosinophilic gastrointestinal disorders with other atopic disorders. Immunol Allergy Clin North Am. 2009;29(1):85–97. x. doi: 10.1016/j.iac.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Brown-Whitehorn TF, Spergel JM. The link between allergies and eosinophilic esophagitis: implications for management strategies. Expert Rev Clin Immunol. 2010;6(1):101–109. doi: 10.1586/eci.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spergel JM. An allergist's perspective to the evaluation of Eosinophilic Esophagitis. Best Pract Res Clin Gastroenterol. 2015;29(5):771–781. doi: 10.1016/j.bpg.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aroniadis OC, Brandt LJ. Intestinal microbiota and the efficacy of fecal microbiota transplantation in gastrointestinal disease. Gastroenterol Hepatol (N Y) 2014;10(4):230–237. [PMC free article] [PubMed] [Google Scholar]
- 42.Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 43.Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70(2):241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 44.Branum AM, Lukacs SL. Food allergy among children in the United States. Pediatrics. 2009;124(6):1549–1555. doi: 10.1542/peds.2009-1210. [DOI] [PubMed] [Google Scholar]
- 45.Berni Canani R, Gilbert JA, Nagler CR. The role of the commensal microbiota in the regulation of tolerance to dietary allergens. Curr Opin Allergy Clin Immunol. 2015;15(3):243–249. doi: 10.1097/ACI.0000000000000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111(36):13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 48.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131(1):201–212. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noval Rivas M, Burton OT, Wise P, Charbonnier LM, Georgiev P, Oettgen HC, et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42(3):512–523. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, et al. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J Allergy Clin Immunol. 2015;135(3):737–744. e738. doi: 10.1016/j.jaci.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 52.Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother's and offspring's use of antibiotics and infant allergy to cow's milk. Epidemiology. 2013;24(2):303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 53.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016;10(3):742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80(8):2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. 2016;27(3):254–262. doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 56.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3:23. doi: 10.1186/s40168-015-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fillon SA, Harris JK, Wagner BD, Kelly CJ, Stevens MJ, Moore W, et al. Novel device to sample the esophageal microbiome--the esophageal string test. PLoS One. 2012;7(9):e42938. doi: 10.1371/journal.pone.0042938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS One. 2015;10(5):e0128346. doi: 10.1371/journal.pone.0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang R, Wagner BD, Harris JK, Fillon SA. Zero-inflated negative binomial mixed model: an application to two microbial organisms important in oesophagitis. Epidemiol Infect. 2016:1–9. doi: 10.1017/S0950268816000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parashette KRSV, Toh E, Hon EC, Janga SC, Nelson D, Gupta SK. Esophageal Microbiome in Healthy Children and Eosinophilic Esophagitis: A Prospective Study. Gastroenterology. 2015;148:S637–S638. [Google Scholar]
- 61.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013;19(8):1005–1013. doi: 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masterson JC, McNamee EN, Hosford L, Capocelli KE, Ruybal J, Fillon SA, et al. Local hypersensitivity reaction in transgenic mice with squamous epithelial IL-5 overexpression provides a novel model of eosinophilic oesophagitis. Gut. 2014;63(1):43–53. doi: 10.1136/gutjnl-2012-303631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dutt P, Shukla JS, Ventateshaiah SU, Mariswamy SJ, Mattner J, Shukla A, et al. Allergen-induced interleukin-18 promotes experimental eosinophilic oesophagitis in mice. Immunol Cell Biol. 2015;93(10):849–857. doi: 10.1038/icb.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129(3):985–994. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 65.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107(1):83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168(5):2464–2469. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 67.Rajavelu P, Rayapudi M, Moffitt M, Mishra A, Mishra A. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302(7):G645–G654. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubinstein E, Cho JY, Rosenthal P, Chao J, Miller M, Pham A, et al. Siglec-F inhibition reduces esophageal eosinophilia and angiogenesis in a mouse model of eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2011;53(4):409–416. doi: 10.1097/MPG.0b013e3182182ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]