Summary
Advances in the development of delivery, repair, and specificity strategies for the CRISPR-Cas9 genome engineering toolbox are helping researchers understand gene function with unprecedented precision and sensitivity. CRISPR-Cas9 also holds enormous therapeutic potential for the treatment of genetic disorders by directly correcting disease-causing mutations. Although the Cas9 protein has been shown to bind and cleave DNA at off-target sites, the field of Cas9 specificity is rapidly progressing with marked improvements in guide RNA selection, protein and guide engineering, novel enzymes, and off-target detection methods. We review important challenges and breakthroughs in the field as a comprehensive practical guide to interested users of genome editing technologies, highlighting key tools and strategies for optimizing specificity. The genome editing community should now strive to standardize such methods for measuring and reporting off-target activity, while keeping in mind that the goal for specificity should be continued improvement and vigilance.
Introduction
The CRISPR-Cas9 genome engineering technology has been established as a powerful molecular tool for numerous areas of biological study where it is useful to target or modify specific DNA sequences (Hsu et al., 2014; Sander and Joung, 2014; Cox et al., 2015; Wright et al., 2016). Originally derived from bacterial adaptive immune systems known as CRISPR-Cas (Mojica et al., 2000; Jansen et al., 2002; Mojica et al., 2005; Pourcel et al., 2005; Barrangou et al., 2007), the Cas9 nuclease can localize and cleave a target DNA via a guide RNA (Brouns et al., 2008; Deltcheva et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012). The Cas9 nuclease has been engineered to edit target DNA at desired loci in eukaryotic cells (Cong et al., 2013; Mali et al., 2013c). However, in the context of large eukaryotic genomes, Cas9 is known to bind and cleave at off-target sites (Cong et al., 2013; Fu et al., 2013; Hsu et al., 2013) like its genome editing predecessors, zinc finger nucleases (ZFNs) (Bibikova et al., 2001; Urnov et al., 2005; Miller et al., 2007) and transcriptional activator-like effector nucleases (TALENs) (Boch et al., 2009; Moscou and Bogdanove, 2009; Christian et al., 2010; Miller et al., 2011). Efforts to measure, understand, and improve Cas9 specificity have accordingly been of great interest in the field. While prior review articles should be referred to for foundational background and insight into Cas9 genome editing development (Makarova and Koonin, 2015) and applications (Mali et al., 2013b; Hsu et al., 2014; Sander and Joung, 2014; Porteus, 2016; Wright et al., 2016), this article focuses on the sub-field of Cas9 specificity by comparing relevant methods for off-target improvement and detection, highlighting useful experimental and computational tools, and emphasizing further research directions for continued improvement.
Recently, several improvements have been devised for the wild-type Cas9 enzyme derived from Streptococcus pyogenes, currently the nuclease most widely used for genome editing applications. Initially, bioinformatic solutions for guide RNA selection recommended guide sequences that recognized cognate genomic DNA targets with minimal sequence homology across that given genome. Although such approaches proved reasonably effective, subsequent systematic profiles of Cas9 off-target activity both in vitro (Pattanayak et al., 2013) and in vivo (Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013a) led to the first set of data-driven guidelines for target site selection. Given a particular locus and species of interest, computational models and associated webtools typically rank guide RNAs by predicted specificity and suggest likely off-target sites that can be checked for undesired mutagenesis via targeted sequencing or enzymatic assays (see ‘Biased off-target detection’). However, the appropriate number of potential off-target sites to experimentally assay remains unclear, as the accuracy of in silico prediction can vary. Thus, improving methods of off-target detection and quantification has become a focus of a number of research groups in the CRISPR-Cas9 field.
Several protocols for the unbiased detection of genome-wide Cas9 off-target activity in cells have been recently described (Frock et al., 2015; Kim et al., 2015; Ran et al., 2015; Tsai et al., 2015; Wang et al., 2015), and are important complements to biased off-target detection methods based on in silico prediction and targeted sequencing (Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013a; Pattanayak et al., 2013) (see ‘Unbiased genome-wide off-target detection’). The adoption of these protocols marks an important departure from standardized methods of measuring an inherently biased subset of potential off-target sites. In addition to these protocols, the computational tools used for guide RNA design and off-target prediction are expanding to account for a deepening understanding of specificity-governing parameters in the cell, including detailed features of the off-target sequence and the Cas9 orthologs (see ‘Computational guide selection and predictive models’).
Furthermore, multiple methods have been developed to improve specificity by utilizing natural or engineered orthologous Cas9s (see ‘Orthologs’), engineering the Cas9 protein or guide RNA (see ‘Protein Engineering and Bespoke PAMs’ and ‘Modified Guides’), or by modulating the kinetics and regulation of the CRISPR components in the cell (see ‘Kinetics and Regulation’). Meanwhile, use of nuclease-inactivated ‘dCas9’ for targeted binding applications such as transcriptional regulation have particular considerations given the distinctions between the Cas9 target search and nucleolytic mechanisms (see ‘dCas9 specificity’). CRISPR specificity must also be considered in a broader sense to include cell type, temporal, and spatial specificity. Lastly, the standardization of off-target analysis methods and data reporting in research publications would benefit the field by enabling comparison across studies and better algorithms for guide RNA design (see ‘Broader Definitions of Specificity’ and ‘Need for Standardization’).
Biased Off-target Detection
To date, the predominant approach for identifying Cas9 nuclease off-target activity has been to: 1) computationally predict likely off-target sites based on sequence homology, and then 2) assess any potential editing activity by enzymatic assays based on mismatch-sensitive endonucleases (Guschin et al., 2010; Ran et al., 2013b), Sanger sequencing, or targeted deep sequencing (‘see The Need for Standardization’). These a priori predictions are critical as ~98% of Streptococcus pyogenes Cas9 (SpCas9) guide RNAs in human exons and promoters have at least one off-target site with three or fewer mismatches (Bolukbasi et al., 2016), and foundational Cas9 specificity studies collectively demonstrated that off-target sites with three or fewer mismatches are significantly more likely to be cleaved than more dissimilar sites (Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013a; Pattanayak et al., 2013). Selecting the most unique target site possible is thus a valuable strategy for improving specificity, since many gene editing applications have several possible guide RNAs capable of accomplishing the same experimental outcome (i.e., knocking out a gene by introducing indels in an exon of interest).
Sequence uniqueness is thus an important initial filter for selecting guide RNAs with minimal potential off-target sites with high sequence homology. However, as the number of mismatches considered relative to the on-target site increases, the total number of potential off-target sites dramatically increases as well. For example, a benchmark guide RNA designed to target the EMX1 locus (Ran et al., 2015) with no ‘off-by-1’ mismatched off-target sites in the hg38 reference genome, has 10 ‘off-by-2’ site, 69 ‘off-by-3’ sites, and 27,480 ‘off-by-6’ sites.
As a result, even targeted sequencing – likely the most scalable of the aforementioned approaches – is biased towards a small number of sites that can reasonably be assayed. However, Cas9-mediated cleavage has been reported at off-target sites with as many as 6 mismatches to the guide sequence (Tsai et al., 2015), demonstrating the risk of missing detectable events by setting low ‘off-by’ thresholds for the sake of practical, biased off-target screening. Furthermore, most computational off-target prediction tools do not adequately consider off-target sites with gaps, bulges, or alternative protospacer adjacent motif (PAM) sequences (sequence motifs recognized by Cas9 directly 3′ of the matching guide sequence in the target DNA; typically 5′-NGG for SpCas9, but also 5′-NAG with lower efficiency) (Table 1).
Table 1.
Improvements to CRISPR-Cas9 Specificity
| Category | Improvement | Description | Advantage | Disadvantage |
|---|---|---|---|---|
| Measurement | Targeted Deep Sequencing (Ran et al., 2013b) | Targeted amplicon next-generation sequencing (NGS) of putative or known off-target sites, followed by computational analysis to quantify the proportion of reads with indels near the PAM site. | More quantitative, sensitive, and scalable than alternative assays like Surveyor. | Biased towards a subset of off-target sites that can reasonably be assessed. Best used as complement to unbiased, genome-wide off-target detection methods. |
| GUIDE-seq: Genome-wide, unbiased identification of DSBs enabled by sequencing (Tsai et al., 2015) | Captures DSBs with a double stranded oligonucleotide (dsODN), which is then used as a priming site for sequencing. | Straightforward wet-lab protocol with computational pipelines available online for data processing. | Requires efficient delivery of the dsODN, which may be toxic to some cell types at some doses, and has not been demonstrated for in vivo models. | |
| BLESS: Direct in situ breaks labeling, enrichment on streptavidin, and NGS (Ran et al., 2015; Slaymaker et al., 2016) | Biochemical ligation of NGS sequencing adapters to exposed gDNA ends. Computational filtering separates Cas9-mediated DSBs (aligned near PAM) from naturally occuring DSBs (more randomly distributed). | No exogenous bait is introduced to cells. Can be applied to tissue samples from in vivo models. | Sensitive to time of cell fixation. Current configuration requires relatively large number of cells. | |
| Digenome-seq: In vitro nuclease-digested whole genome sequencing (Kim et al., 2015; Kim et al., 2016b) | Cell-free gDNA samples are digested in vitro by Cas9 RNP with multiplexed guide RNAs before WGS. Cut sites are identified based on read alignment. | Can be applied to any cell-type as digestion performed on extracted gDNA. More sensitive than GUIDE-seq in a head-to-head. | WGS may be costly. Must be paired with one of the other methods to validate the sites discovered in vitro are truly mutagenized in the cell. | |
| Computational Guide Selection | Specificity Score (Hsu et al., 2013) and CFD (Doench et al., 2016) | An experimentally-derived function to score the likelihood of an offtarget site being edited, based on the position and nature of its mismatches. | A continuous variable to distinguish guide RNAs during design and to rank off-target sites for targeted sequencing follow-up. | Function derived from SpCas9 data with 20-mer guides, so it may not generalize to other contexts. |
| WGS of Reference Genome (Yang et al., 2014) | Whole genome sequencing (WGS) of the relevant cell line, animal model, patient, etc. (Box 1). | Identify new off-target sites created by genetic variation, which are not present in the reference genomes (ie. hg38). | Remains costly. Custom reference genomes only available with some guide design tools (Table 2). | |
| Protein Engineering | Single/Paired Nickases (Cong et al., 2013; Mali et al., 2013a; Ran et al., 2013a; Cho et al., 2014) | A point mutation at the active site of one of the Cas9 nuclease domains yields a targeted nickase. Paired nickases targeting complementary strands create a deletion. | Nicks are generally repaired with higher fidelity, so off-target edits are less frequent. Nickases can also mediate efficient homology directed repair. | Less efficient on-target editing with some guide RNAs. |
| SpCas9 PAM Variant D1135E (Kleinstiver et al., 2015b) | A single point mutation increases specificity for the 5′-NGG PAM. | Significant decrease in editing at 5′-NAG and 5′-NGA PAMs. Improved genome-wide specificity for several guides as assessed by GUIDE-seq. | On-target efficiency may be affected. It was comparable to wtCas9 for 6 guides by T7E1, but somewhat lower for 3 guides by deep sequencing. | |
| eSpCas9 (Slaymaker et al., 2016) | Three mutations within the nt-groove weaken Cas9’s non-target DNA strand stabilization and therefore increase stringency of guide RNA-DNA complementation for nuclease activation. | Off-target editing was nearly entirely avoided, as assessed by both BLESS and targeted deep sequencing. | On-target efficiency may be affected, although it was comparable to wtCas9 for most of the guide RNAs reported. | |
| SpCas9-HF (Kleinstiver et al., 2016a) | Four mutations weaken Cas9 binding to the target DNA strand and therefore increase stringency of guide RNA-DNA complementation for nuclease activation. | Off-target editing was nearly entirely avoided, as assessed by both GUIDE-seq and targeted deep sequencing. | On-target efficiency may be affected, although it was comparable to wtCas9 for most of the guide RNAs reported. | |
| RNA Modifications | Tru-guides (Fu et al., 2014b) | Truncated guides have spacers of 17 to 18-nt as opposed to the canonical 20-nt. The shorter region of RNA:DNA complementarity shifts the binding energy such that mismatches are less tolerated. | Simple to implement. Often drastically decreases activity at genomic off-target sites. | Less efficient on-target editing with some guide RNAs. May have more offtarget sites with high sequence homology. |
| 5′GGX20 guides (Cho et al., 2014) | Two additional mismatched guanines are added to the 5′ end of the guide RNA. These PAM-distal mismatches may adjust binding energetics such that Cas9 more stringently requires complementarity. | This simple modification can be rapidly assessed in all contexts. Specificity ratios at off-target sites generally improve by 10 to 100 fold or more. | This modification is only compatible with certain guide RNAs. On-target efficiency is sometimes severely decreased. | |
| Delivery | SaCas9 (Ran et al., 2015) | The Staphylococcus aureus Cas9 ortholog is 3.2 kb, uses a 20-23-nt guide RNA, and recognizes a 5′-NNGRRT PAM. | Smaller size allows AAV packaging with one or two sgRNAs in an all-in-one vector. Specificity improvements were observed with some guides by BLESS, GUIDE-seq, and targeted sequencing. | The ortholog is somewhat less characterized than SpCas9, and may be less efficient on-target in some contexts. |
| Ribonucleoprotein (RNP) (Cho et al., 2013b; Kim et al., 2014; Lin et al., 2014; Zuris et al., 2015) | Purified Cas9 protein is complexed with guide RNA before delivery to cells by lipid transfection or electroporation. | Burst-like Cas9 expression kinetics mediates efficient on-target editing without long term risk of accumulating off-target edits. | Best used ex vivo or for targeting accessible tissues (e.g. hair cells of the inner ear). |
Looking forward, assays relying on the targeted sequencing of off-target sites may become the standard due to their sensitivity and accuracy as current methodologies have a reported lower limit of detection of approximately 0.1%, or 1/1000 (Kleinstiver et al., 2016a). It is suggested that the more accessible Surveyor assay should be used only when quick, first-pass cleavage efficiency measurements are needed. The Surveyor assay is generally quantified by analyzing the band intensity of gel electrophoresis images, and is thought to have a limit of detection of ~5% indels (Fu et al., 2013), making it unsuitable for most off-target detection scenarios. Finally, targeted sequencing is perhaps best applied to verify activity at off-target sites discovered by unbiased, genome-wide methods (Kim et al., 2015; Ran et al., 2015; Tsai et al., 2015; Kleinstiver et al., 2016a; Slaymaker et al., 2016).
Unbiased Genome-wide Off-target Detection
To address the present limitations associated with in silico predictions of potential off-target sites, several unbiased methods have been recently reported for measuring Cas9 off-target activity across the genome (Table 1). We and others have recently reviewed and compared these deep sequencing-based protocols (Gabriel et al., 2015; Gori et al., 2015; Koo et al., 2015; Bolukbasi et al., 2016), including BLESS (Ran et al., 2015), HTGTS (Frock et al., 2015), GUIDE-seq (Tsai et al., 2015), IDLV capture (Wang et al., 2015), and Digenome-seq (Kim et al., 2015). These unbiased methods collectively demonstrate that sequence homology alone is not fully predictive of off-target sites. Further, although the PAM tends to be the most stringent predicate of target recognition fidelity relative to the guide sequence, off-target mutagenesis was also observed at non-canonical PAMs, confirming that biased methods may miss important Cas9 off-target sites (Frock et al., 2015; Kim et al., 2015; Ran et al., 2015; Tsai et al., 2015).
However, depending on the approach employed for enriching Cas9-induced DNA cleavage, unbiased methods tend to be less sensitive and have a lower throughput than biased targeted sequencing, in addition to typically requiring higher sequencing coverage and much more complex protocols. As a result, it is currently difficult to generate off-target data at a scale required to adequately train a predictive model for specificity. Regardless, these approaches remain valuable for researchers interested in validating individual guide RNA sequences in their experimental system.
GUIDE-seq and Digenome-seq have reported sensitivity to off-target events as low as 0.1%, or 1 in 1000 events, whereas IDLV capture reported a sensitivity of 1%. The BLESS and HTGTS authors did not explicitly report summary sensitivity metrics. It should be noted that all of these sensitivity rates vary depending on both the number of cells assayed in the experiment, the deep sequencing depth, and the bioinformatics methods used to differentiate true Cas9-induced cleavage events from background noise. As a result, differences in reported detection sensitivity between competing protocols do not necessarily reflect inherent limitations of the off-target detection method but also a need for technical optimization.
Beyond sensitivity, interested users should also compare the protocols’ applicability in their experimental context (Table 1). For example, GUIDE-seq requires optimized delivery of a defined, chemically modified double-stranded oligonucleotide (dsODN), making it potentially toxic to certain primary cell types and difficult to apply in vivo on tissue samples. However, the dsODN-specific PCR enriches for double-stranded breaks and thus should enable appropriate sequencing coverage with one sequencing run of reasonable depth. While BLESS can be applied to both ex vivo and in vivo samples after Cas9 editing alone, it requires the biochemical ligation of adapter sequences to free, unrepaired DSB ends. As a result, its sensitivity will be affected by the time-point after Cas9 editing and DNA repair kinetics. Digenome-seq, on the other hand, can immediately be applied in any Cas9-editable cell type, yet it is an in vitro method that removes genomic structural context. It originally required expensive whole genome sequencing at high coverage for four sets of genomic DNA per assay. An improved version of Digenome-seq lowers the cost by multiplexing guide RNAs and reducing the WGS requirement from four genomes to one (Kim et al., 2016b). This improved protocol was run on 10 guides that had been analyzed by GUIDE-seq and the results were compared and validated with targeted sequencing of transfected cells. While Digenome-seq generally discovered ~70 more off-target sites per guide initially, the number of validated off-target sites was similar for both methods.
Importantly, as all unbiased methods require manipulation of the genome, they will each introduce different and subtle artifacts to the set of identified off-target sites and likely will not report the exact same sites with identical frequencies. The techniques have been compared for some ‘benchmark’ guide RNAs, but a comprehensive head-to-head comparison is lacking at this time and may be needed (Gori et al., 2015). The improved Digenome-seq protocol can be rapidly transferred to new laboratories, as it consists of in vitro Cas9 cleavage and WGS. BLESS and GUIDE-seq, as currently configured, may be the most accessible protocols, as they enrich for DSBs and require fewer sequencing reads per sample. Future generations of these methods should collectively look to improve accuracy and throughput as well as reduce the input cell number required.
In sum, these biased and unbiased methods can and perhaps should be applied in a complementary manner. For example, off-target sites can be discovered in vitro (e.g. Digenomeseq), in easily cultured cell types (e.g. HTGTS, GUIDE-seq), or even in vivo (e.g. BLESS) prior to validation in vivo or in cell lines of interest via targeted sequencing. Although combinations of biased and unbiased methods have been applied to a small set of guide RNAs, larger data sets will be required to determine how well in vitro off-target activity corresponds with Cas9 targeting in vivo and across different cell types. These types of datasets will, in turn, drive continued improvement of bioinformatics tools for target site selection.
Computational guide RNA selection and predictive models
Many such online tools and applications have been developed for the in silico design of guide RNAs and prediction of their off-target sites (Table 2). Some, such as Cas-OFFinder, simply identify off-target sites via sequence similarity and rank them based on relative orthogonality, while others use a specificity score calculation we have previously described (Hsu et al., 2013). This formula scores guides based on a weighted sum of the mismatches in their off-target sites, with weights experimentally determined to reflect the effect of the mismatch position on cleavage efficiency.
Table 2.
Computational tools for designing guide RNAs.
| Name | Website | Specificity Score | Genomes Supported |
Single / Paired Design |
Cas9 Orthologs Supported |
Custo m Guide Length |
Lists Off Targets |
Considers Off Targets with Indels |
|---|---|---|---|---|---|---|---|---|
| Benchling | http://benchling.com | (Hsu et al., 2013) | 22 | Both | 5 | Yes | Yes | No |
| BLAST (on predesigned guide) | http://blast.ncbi.nlm.nih.gov/Blast.cgi | Sequence similarity | 1000+ | N/A | N/A | Yes | Yes | Yes |
| Cas-OFFinder | http://www.rgenome.net/cas-offinder/ | No | 34 | Single | 4 | Yes | Yes | Yes |
| MIT CRISPR Design Tool | CRISPR.mit.edu | (Hsu et al., 2013) | 16 | Both | 1 | No | Yes | No |
| Deskgen | http://deskgen.com | (Hsu et al., 2013) | 5 | Both | 4 | Yes | Yes | No |
| DNA 2.0 CRISPR gRNA Design Tool | https://www.dna20.com/eCommerce/cas9/input | Relative ranking based on orthogonality | 5 | Both | 1 | No | No | No |
| E-CRISP | http://www.e-crisp.org/E-CRISP/designcrispr.html | Relative ranking based on orthogonality | 34 | Both | 1 | Yes | Yes | No |
| EuPaGDT | https://grna.ctegd.uga.edu | Relative ranking based on orthogonality | 28 | Single | Custom PAM | Yes | Yes | No |
| GenScript gRNA Design Tool | http://www.genscript.com/gRNA-design-tool.html | No | 2 | Single | 1 | Yes | No | No |
| ZiFiT | http://zifit.partners.org/ZiFiT/Disclaimer.aspx | Orthogonality of Offtargets | 9 | Both | 1 | Yes | Yes | No |
| Broad GPP Portal | http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design | Cutting Frequency Determination (CFD) (Doench et al., 2016) | 2 | Single | 1 | No | No | Yes |
| CROP-IT | http://cheetah.bioch.virginia.edu/AdliLab/CROP-IT/ | (Singh et al., 2015) | 2 | Both | 1 | No | Yes | No |
Other studies have similarly established that mismatches at the 5′ end of the spacer are more tolerated than PAM-proximal mismatches (Cong et al., 2013; Fu et al., 2013; Pattanayak et al., 2013). This ‘seed sequence’ effect has been corroborated by biochemical experiments demonstrating that Cas9 unwinds and binds DNA in a directional manner moving away, or 3′ to 5′, relative to the PAM (Sternberg et al., 2014), and that proper matching of the 5′ end of the guide sequence is important for proper nuclease domain positioning and successful Cas9 cleavage (Jiang et al., 2015). Beyond weighting for PAM-proximal mismatches, the specificity score also reflects the finding that consecutive mismatches attenuate Cas9 activity more than an equivalent number of interspaced mismatches. Importantly, as these target site selection guidelines were derived from studies on SpCas9, their general applicability to less commonly used Cas9 orthologs remains unclear.
Overall, these bioinformatic tools generally perform well in ranking guide RNAs such that, for a given locus, users can easily select guide sequences that are optimized for high genomic specificity. However, as our mechanistic and empirical understanding of Cas9 activity has progressed, several caveats as well as avenues for the improvement of in silico prediction have emerged.
For example, there is evidence suggesting off-target sites with small insertions or deletions in the targeted sequence or alternative PAMs can indeed be cleaved by Cas9, albeit with low frequency (Hsu et al., 2013; Jiang et al., 2013; Kleinstiver et al., 2015b; Ran et al., 2015). Cas9 specificity is similarly known to vary due to other variables such as cell type, species, delivery modality, and dosage (see ‘Kinetics and Regulation’). Although combining existing Cas9 binding and cleavage data sets with whole-genome chromatin state information has been suggested to provide more accurate off-target prediction (Singh et al., 2015), generally such data has been collected for a limited number of guides. It is difficult to integrate all of these observations in a systematic, data-driven way, so these parameters are not taken into account by the majority of available off-target prediction tools.
To realize the full potential of in silico prediction, comprehensive datasets addressing such limitations must be generated. Large-scale guide RNA efficiency datasets have been used to create increasingly accurate computational models that predict the knockout efficiency of new guide RNAs (Doench et al., 2014; Chari et al., 2015; Xu et al., 2015). Most recently, a pooled screen analysis of 9,914 guide RNA variants covering all single mismatch and single RNA indels for 65 DNA targets was used to derive a cutting frequency determination (CFD) score capable of predicting the likelihood of off-target activity (Doench et al., 2016).
Overall, this data underscores the complexity of predicting Cas9 specificity and the importance of considering both position and mismatch identity in metrics like the CFD. For example, 256 of 289 (89%) rG:dT mismatched guides retained high activity, whereas only 37% of rC:dC mismatches were active. Looking just at position 16 in the guide sequence, all 46 purine-purine mismatches tested attenuated Cas9 activity, whereas 97% of rN:dT mismatches were tolerated. Furthermore, in a validation study, modest correlation was reported between the CFD and observed off-target activity at 60 sites with 1–2 mismatches. Generally, well-designed guide RNAs avoid off-by-1 and off-by-2 mismatch sites, so these models should be improved to predict activity at off-by-3 and off-by-4 mismatch sites. Future refinements and insights will likely be derived from large training datasets that consider off-target sites with >1 mismatch or indel.
Lastly, the online tools compared in Table 2 will be of practical utility for most users, but bioinformaticians may benefit from the performance and flexibility of offline tools, such as CRISPRseek (Zhu et al., 2014). A comprehensive comparison of these computational tools can be found with the ‘CRISPR Software Matchmaker’ tool (MacPherson, 2015).
Cas9 orthologs and new CRISPR proteins
Recently, a broader array of Cas9 ortholog proteins have been engineered as genome editing tools, such as the Staphylococcus aureus Cas9 (SaCas9), which is over 1 kb smaller than the more commonly used SpCas9 and thus easily packaged into AAV paired with guide RNA expression cassettes for in vivo genome editing (Friedland et al., 2015; Nishimasu et al., 2015; Ran et al., 2015). Interestingly, SaCas9 demonstrates highly efficient nuclease activity with target sequences of 21–24-nt when compared to the 20-nt targets used with SpCas9, while employing a 5′-NNGRRT PAM as opposed to the 5′-NGG of SpCas9. SaCas9 also demonstrates substantial cleavage efficiency at 5′-NNGRR PAM sites. It remains to be determined how a longer target sequence and longer PAM preference affects the genome-wide off-target activity of SaCas9, although initial results with the unbiased BLESS and GUIDE-seq protocols appear promising (Friedland et al., 2015; Ran et al., 2015). Given identical guides targeting sites with nested 5′-(NGG)RRT PAMs, editing with SaCas9 as opposed to SpCas9 resulted in lower indel frequencies at fewer off-target sites (Ran et al., 2015).
Cas9s derived from Neisseria meningitides (NmCas9) and Streptococcus thermophilus (StCas9) CRISPR1 and CRISPR3 have also been engineered for specific genome editing in mammalian cells (Mali et al., 2013a; Muller et al., 2016). Similar to SaCas9, NmCas9 requires a more complex 5′-NNNNGATT PAM and uses a longer crRNA spacer (21–24-nt) than SpCas9 (Hou et al., 2013; Mali et al., 2013a; Lee et al., 2016). Importantly, while the longer PAM limits the targeting range, it also limits the number of off target sites for any given guide. For example, NGG motifs occur roughly every 8 bp, while NNNNGATT is found every 128 bp (Lee et al., 2016). A systematic study of NmCas9 mismatch and indel tolerance found an overall improvement over SpCas9, and a promising absence of any off-target editing at all sites with ≤3 mismatches (Lee et al., 2016). However, the on-target efficiency is consistently below that of SpCas9, possibly due to weaker DNA unwinding activity (Ma et al., 2015). The two StCas9 orthologs similarly might be more specific than the wild-type SpCas9, but they also suffer from weaker efficiency (Muller et al., 2016).
Overall, Cas9 orthologs with improved specificity profiles can likely be discovered and engineered. By screening available genome databases for type II CRISPR loci, over 56 bacterial species have been identified with predicted novel tracrRNAs (Chylinski et al., 2013). Furthermore, Cas9 proteins phylogenetically clustered within the same subgroup can interchangeably use their crRNA-tracrRNA (dual-RNA) duplexes to cleave DNA, suggesting that Cas9-guide RNA pairings could be optimized by mining the natural diversity or engineering improved guide architectures (Fonfara et al., 2014).
Furthermore, the large natural diversity of bacterial CRISPR systems extends beyond type II systems featuring Cas9 as the sole effector nuclease. Recently, a newly discovered CRISPR protein named Cpf1 has also been used for human genome editing (Zetsche et al., 2015). Also targeted by a single guide RNA, Cpf1 recognizes a 5′-TTN PAM upstream (rather than downstream) of the target site and creates a 5-nt staggered overhang DSB distal to the PAM (Fonfara et al., 2016; Yamano et al., 2016). It is important to note that 8 Cpf1 orthologs were screened in human cells and only 2 mediated efficient cleavage. Cpf1 genome-wide specificity was recently assayed with Digenome-seq (Kim et al., 2016a) and GUIDE-seq (Kleinstiver et al., 2016b) and can be comparable or greater than SpCas9 for many guide RNAs. Other putative CRISPR effector proteins, known as c2c1, c2c2, and c2c3, have also been bioinformatically mined from metagenomic datasets (Shmakov et al., 2015). C2c2 has since been characterized as a programmable RNA-guided RNA-targeting nuclease in bacterial cells (Abudayyeh et al., 2016). Interestingly, once c2c2 binds its complementary target RNA, it is active as a non-specific RNase and significantly impedes cell growth. It remains largely unclear how these new CRISPR proteins’ different characteristics (i.e. guide RNAs, PAMs, nuclease domains) will affect their specificity, but their discovery reflects great potential for gene editing beyond SpCas9.
Protein Engineering and Bespoke PAMs
Meanwhile, biological engineers have rapidly optimized and successfully matured SpCas9’s utility as a tool, with increasing precision as understanding of its molecular structure and mechanism has deepened (Table 1). The first generation of improvements leveraged knowledge of the two nuclease domains of Cas9, initially identified based on sequence homology (Sapranauskas et al., 2011). For example, a single point mutation (D10A) can inactivate the RuvC domain and change the SpCas9 nuclease into a nickase capable of cleaving only the DNA strand complementary to the guide RNA (Cong et al., 2013). Using paired guide RNAs to create two appropriately offset nicks results in efficient indel formation while improving specificity by as much as 1500-fold, likely because single DNA nicks are repaired with high fidelity compared to double strand breaks (Mali et al., 2013a; Ran et al., 2013a; Cho et al., 2014; Friedland et al., 2015). Paired nickases have been applied to efficiently generate mouse models with no detectable off-target edits, whereas wild type Cas9 can generate off-target edits that are inherited by founders’ progeny(Shen et al., 2014). Beyond nickases, a second point mutation in the HNH catalytic domain creates a fully nuclease-inactive ‘dCas9’ (Nishimasu et al., 2014). By fusing dCas9 to the FokI nuclease domain and again expressing paired guide RNAs, one can create a dimerization-dependent system that also improves specificity (Guilinger et al., 2014; Tsai et al., 2014).
Another recent protein engineering approach, known as SpCas9MT-pDBD fusions, involves linking a mutated Cas9 with attenuated PAM-binding (SpCas9MT) to a programmable DNA-binding domain (pDBD) such as a ZF or TALE that targets nearby genomic DNA (Bolukbasi et al., 2015). SpCas9MT-pDBD provided specificity improvements up to 150-fold, likely by leveraging cooperativity between two independent DNA-binding events to improve specificity, much like the aforementioned nickase and FokI strategies.
However, these strategies have the overall disadvantage of requiring additional components and larger transgenes. Recently, a crystal structure-based, rational engineering strategy generated SpCas9 and SaCas9 nucleases with ‘enhanced specificity’ (eSpCas9 and eSaCas9, respectively). These Cas9s differ by only 3 and 4 codon substitutions, respectively, within the non-targeted DNA strand groove (nt-groove) of Cas9 (Slaymaker et al., 2016). These mutations are thought to weaken non-target strand binding by the protein, encouraging DNA rehybridization in competition with guide RNA invasion of the target DNA strand (Figure 1). With increased stringency of RNA-DNA matching, these Cas9s demonstrate dramatic reduction in the number of genome-wide off-target sites detectable by BLESS or follow-up targeted sequencing. Moreover, eSpCas9’s on-target efficiency appeared to be comparable to wild-type SpCas9 for the 24 guide RNAs tested, suggesting a surprising de-coupling of efficiency and specificity without discretely modifying the mechanism of action (as accomplished by the cooperativity strategies).
Figure 1. Model of Cas9 DNA targeting and cleavage.
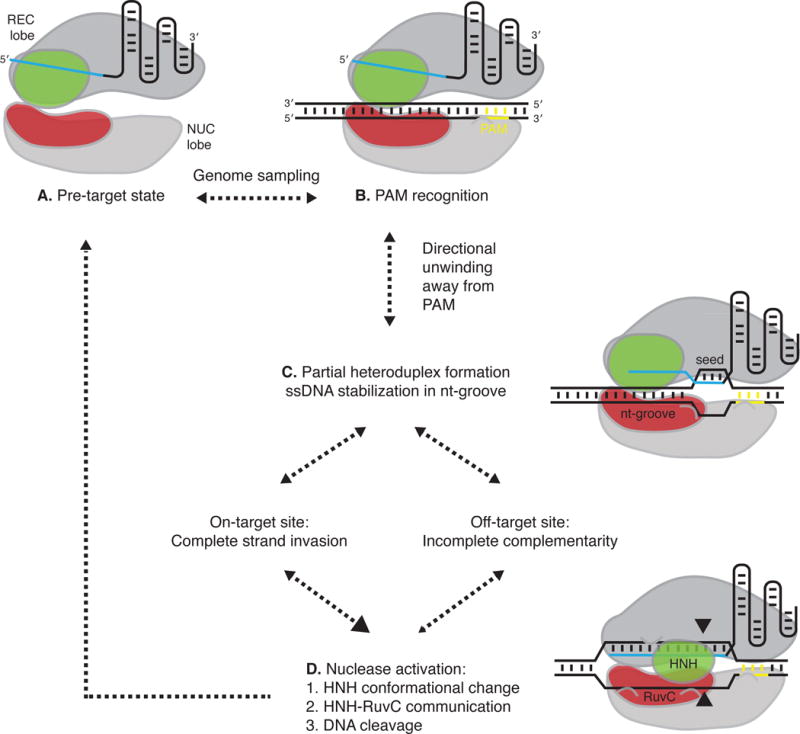
A) Cas9 in complex with guide RNA samples the genome for PAM matches by diffusion, and undersamples heterochromatin regions under certain conditions (Knight et al., 2015). B) Upon PAM recognition and dsDNA binding, the DNA is directionally unwound as the guide RNA interrogates the target strand for complementary sequences. C) The non-target ssDNA strand is stabilized in a non-sequence specific manner in Cas9’s nt-groove. Incomplete complementarity or a short 15-nt PAM-proximal region of complementarity is sufficient for Cas9 binding, as demonstrated by dCas9 or ‘dead-guides’ with wtCas9, respectively. Near-complete complementarity of >17-nt of RNA:DNA heteroduplex is required for nuclease activation. D) After this RNA strand invasion, the HNH domain undergoes a conformational change and communicates through a linker to the RuvC domain. The HNH and RuvC catalytic domains simultaneously cleave the target and non-target ssDNA strand, respectively. Cas9 may remain bound to the cut site for an extended period before returning to the pre-target state (Richardson et al., 2016). The model shown is adapted from several references (Anders et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014; Sternberg et al., 2015; Jiang et al., 2016; Slaymaker et al., 2016).
A similar strategy by a different research group resulted in a ‘high fidelity’ SpCas9-HF (Kleinstiver et al., 2016a). In contrast with eSpCas9, SpCas9-HF contains 4 alanine substitutions at residues that interact with the phosphate backbone of the targeted DNA strand. While on-target efficiency appeared to be slightly more variable than eSpCas9 for the guides tested, it remained comparable (>70%) to wild-type efficiency for 86% of 37 guide RNAs. Altogether, these engineered Cas9s greatly improve specificity without much sacrifice in efficiency. Interestingly, these improved variants present a new challenge to improve the limit of detection of biased and unbiased assays beyond 0.1%, in order to capture remaining rare off-target events that may be important in clinical applications.
Another successful approach has been to alter the PAM recognition sequence of Cas9 itself. Since the PAM is perhaps the most stringent determinant of Cas9 specificity, Cas9s with alternative PAMs could not only increase the number of targetable sites across a genome of interest, but also potentially demonstrate improved genome-wide specificity by requiring a longer PAM or a PAM with less abundance in the target genome. One strategy is to swap the putative PAM-interacting domain (PID) of the protein for the PID from an ortholog recognizing a different PAM. This has been previously demonstrated with SpCas9 and Streptococcus thermophilus CRISPR3 Cas9 (St3Cas9), which share 60% sequence identity and can swap dual guide RNA sequences whilst retaining function (Fonfara et al., 2014; Nishimasu et al., 2014).
More powerful yet is leveraging directed evolution to alter the PAM specificity, as shown recently for SpCas9 (Kleinstiver et al., 2015b) and SaCas9 (Kleinstiver et al., 2015a). Notably, just 4 mutations resulted in a modified ‘VRER’ SpCas9 with high specificity for NCGC PAMs, which are roughly 23 times less abundant in the human genome than the canonical NGG PAM for standard 5′-G-N19 guide RNAs. Directed evolution also yielded a single point mutation (D1135E) SpCas9 that increases specificity for NGG PAMs over the alternative NAG PAM (Hsu et al., 2013; Jiang et al., 2013; Kleinstiver et al., 2015b). In the case of SaCas9, directed evolution was applied to modify the PAM from NNGRRT to NNNRRT, in order to increase the targeting range by two to four-fold (Kleinstiver et al., 2015a).
One advantage of this unbiased directed evolution approach is that it does not require structural data and can be rapidly applied to diverse orthologs. It could prove important for therapeutic targets requiring a very specific edit and lacking nearby canonical PAMs. Finally, bespoke PAMs are likely compatible with the enhanced specificity and high fidelity mutations. Chimeras, directed evolution, and other protein engineering approaches may also be fruitful for generating Cas9s that recognize AT-rich PAMs, which could be useful for targeting intronic regions or AT-rich genomes.
dCas9 Specificity
Aside from DNA cleavage, a large collection of Cas9 applications employ catalytically inactivated Cas9 (dCas9), which can be fused to various effector domains for applications ranging from programmable chromosomal labeling to targeted transcriptional and epigenetic control (Hsu et al., 2014). However, the specificity profile of dCas9 binding vs. wild type (wtCas9) cleavage applications needs to be carefully considered. Recent work studying the target search mechanisms (Figure 1) of the active Cas9 enzyme indicates that Cas9 initially scans the genome for PAM sites (Sternberg et al., 2014; Jiang et al.), resulting in a transient binding state stabilized by a 5-bp seed sequence of the guide RNA (Wu et al., 2014). Additional bases are interrogated for guide RNA-target DNA complementarity until the nuclease achieves an active state upon full guide RNA-target hybridization (Jiang et al., 2013; Jiang et al., 2015).
In accordance with this model, dCas9 immunoprecipitation studies demonstrate a high promiscuity of Cas9 binding that correlates poorly with Cas9 cleavage (Kuscu et al., 2014; Wu et al., 2014; Ran et al., 2015): across nearly 300 off-target binding loci identified via dCas9 ChIP-seq, only one of these sites was mutated slightly above background level by wtCas9 (Wu et al., 2014). In a GUIDE-seq genome-wide study of DNA cleavage, only 2% of off-target sites intersected with a set of 585 dCas9 ChIP-seq off-target binding sites (Tsai et al., 2015). Moreover, dCas9 ChIP-seq identified sites with ~7 mismatches on average, whereas cleavage sites harbored ~3 mismatches on average (Tsai et al., 2015). Although a useful unbiased genome-wide assay for specificity of dCas9 applications, one should not consider dCas9 ChIP-seq as a proxy for genome-wide detection of ‘true’ cleavage events.
Just as for its nuclease counterpart, a combination of biased and unbiased approaches provides the most reliable assessment of dCas9 off-target activity. One method is a mismatch reporter assay in which a library of mismatched target sites is cloned upstream of a promoter driving expression of barcoded RNAs (Mali et al., 2013a). RNA-seq is then used to determine the extent to which that mismatched target site enabled dCas9-mediated activation or repression of the promoter. This method revealed that dCas9-effector fusions are largely insensitive to point mutations and can repress or activate transcription while tolerating up to 3 mismatches, especially near the 5′ end of the guide RNA. That mismatch tolerance may be less than that measured by ChIP-seq due to the longer duration of promoter binding required to make the transcriptional regulatory function detectable.
Whole transcriptome RNA-seq also works as an unbiased method for dCas9-effector off-target analysis (Perez-Pinera et al., 2013; Konermann et al., 2015; Polstein et al., 2015). Generally, this assay confirms the highly specific upregulation of the genes targeted by dCas9 activators. This specificity is unsurprising given dCas9-activators are only efficient within a limited window of the transcription start site – binding events >200-nt upstream of a start site will have little effect on transcription (Gilbert et al., 2014; Konermann et al., 2015). Notably, one RNA-seq study identified a moderate yet significant downregulation of IL32 mRNA – a proinflammatory cytokine – in response to dCas9-VP64 transcriptional activators even without a guide RNA (Perez-Pinera et al., 2013). This result emphasizes the importance of considering off-target effects of Cas9 beyond the first order risk of binding or cutting activity at a degenerate target site, such as unintended biological activity in response to the introduced genome editing components (see ‘Broader Definitions of Specificity’).
Modified Guides
While the aforementioned strategies engineer the Cas9 protein, specificity improvements may also be achieved by modifying the guide RNA (Table 1). Recent characterization of single and dual-guide RNA systems for type II CRISPR systems identified 6 functional modules within the guide (Briner et al., 2014). These modules can be mutated, extended, and deleted to modulate on-target efficiency. For example, extending the single guide RNA (sgRNA) duplex region by 5-nt and/or mutating the string of 4 T’s shortly downstream of the spacer can improve efficiency by changing the RNA’s structure and transcription rate, respectively (Chen et al., 2013; Dang et al., 2015). This approach also improves efficiency for SaCas9 sgRNAs, indicating the general applicability of guide modifications across multiple Cas9 orthologs (Chen et al., 2016; Tabebordbar et al., 2016). Further investigation into guide RNA features will provide a basis for the optimization of guide RNAs for specificity, functionalization, and other applications.
The length of the guide sequence itself is also an important mediator of specificity. Although extending the 20-nt guide sequence of SpCas9 to 30-nt appeared to result in processing back to the natural 20-nt length (Ran et al., 2013a), shortening the guide sequence proved to be a successful strategy. Spacer sequences of 17-nt or 18-nt have been reported to mediate more precise genome editing, potentially by increasing the sensitivity of Cas9 binding to mismatches within the shorter complementary sequence (Fu et al., 2014b). While truncated guides can eliminate activity at many off-target sites, they can also create new off-target sites due to their shorter length (Fu et al., 2014b; Tsai et al., 2015; Wyvekens et al., 2015; Slaymaker et al., 2016). Interestingly, truncated guides are not compatible with the ‘enhanced specificity’ nt-groove mutants or SpCas9-HF, as the combination results in very low on-target efficiency (Kleinstiver et al., 2016a; Slaymaker et al., 2016). It remains to be seen if this simple guide design modification could improve the specificity of other Cas9 orthologs while maintaining editing efficiency, which also varies by guide length (Friedland et al., 2015; Ran et al., 2015).
Notably, shortening the guide further to 14-nt or 15-nt creates a ‘dead guide’ that can mediate efficient wild-type Cas9 binding without nuclease activation, enabling multiplexed applications of DNA cleavage and targeted effector activity (Dahlman et al., 2015; Kiani et al., 2015). This engineered guide approach benefited from insight into the mechanism of Cas9 cleavage, which has been shown to require near-complete strand invasion and complementarity (Figure 1). Whole transcriptome RNA-seq revealed highly similar specificity profiles for both 15-nt and 20-nt guides applied for transcriptional activation (Dahlman et al., 2015). Although ‘dead guides’ may have more predicted off-target binding sites, they likely exhibit acceptable specificity (<3 off-target genes significantly activated) because off-target sites must fall within a limited window of a transcription start site in order to perturb transcription.
Another simple modification, known as ‘GGX20’, adds two additional mismatched guanine nucleotides to the 5′ end of the guide RNA (Cho et al., 2014). Several studies have demonstrated specificity improvements of 10 to 100-fold or greater, although the on-target efficiency varies from comparable to severely reduced (Cho et al., 2013a; Cho et al., 2014; Sung et al., 2014; Kim et al., 2016b). Despite such variability, ‘GGX20’ and truncated guides are easily generated and assessed, although they should still be compared with standard-length guides on a case-by-case basis to optimize on-target efficiency.
Finally, chemical modification of guide RNAs can also impact Cas9 specificity. Synthesis of guide RNAs with 2′-O-methyl (M), 2′-O-methyl 3′ phosphorothioate (MS), or 2′-O-methyl 3′ thioPACE (MSP) at the 5 and 3 ends significantly improves editing efficiency in human primary T cells and CD34+ hematopoetic stem and progenitor cells (Hendel et al., 2015). However, these RNA modifications increased off-target activity at some off-target sites. In a contrasting study, synthetic guide RNAs with 10 M-modified 5′ bases and interspersed 2′-fluoro modifications towards the 3′ end were more stable and specific, but less efficient on-target (Rahdar et al., 2015). Given these mixed early findings, the ability to modulate RNA stability and mismatch binding kinetics through alternative nucleotide chemistries of guide RNAs warrants further exploration.
Kinetics and Regulation
A critical and underexplored determinant of Cas9 specificity is the pharmacokinetic profile of the delivered components across cell and tissue types, especially the form factor of the genome editing components (DNA, RNA, or protein) and the delivery vehicle (viral or non-viral).
Lentiviral-mediated integration of Cas9 and guide RNA into melanoma cell lines has been reported to achieve saturating indels up to 100% at on-target sites (Shalem et al., 2014). While both on- and off-target activity generally appeared stable between days 7 and 14, indels at one off-target site with 3 mismatches rose from ~40% to ~50%. Because off-target levels can increase with sustained exposure to Cas9, controlling or limiting the time of Cas9 expression in the cell or nucleus may improve specificity.
As such, the Cas9 delivery modality is an important mediator of specificity (Table 1). For example, cationic lipid-mediated RNP delivery improved the specificity of 3 SpCas9 guides at 11 assessed off-target sites (typically by 10-fold) compared to plasmid DNA transfection, after normalizing for equivalent on-target efficiency (Zuris et al., 2015). In a similar study, RNP-Cas9 improved the on/off-target ratio by approximately 3-fold for two guides tested (Kim et al., 2014). Delivering MS-modified synthetic guide RNAs as RNPs significantly improved on/off-target ratios at all 3 assessed sites when compared to co-delivery with Cas9 plasmid or mRNA (Hendel et al., 2015). This specificity benefit is possibly due to the burst-like kinetics of RNP delivery and associated Cas9 exposure as opposed to more stable plasmid-driven expression (Kim et al., 2014).
Cas9 dosage can also affect the kinetics and modulate specificity, both in vitro (Pattanayak et al., 2013; Fu et al., 2014a) and in cells (Hsu et al., 2013). For example, by plasmid transfection, a 5-fold drop in dose led to a 7-fold increase in the specificity ratio at the cost of a ~2-fold decrease in on-target efficiency, emphasizing the importance of titrating delivery conditions to optimize specificity for any given application (Hsu et al., 2013). Using the weaker H1 promoter for guide RNA expression gives a greater decrease in off-target activity than on-target activity (Ranganathan et al., 2014). Such non-linearity in the on/off-target editing rate relationship with Cas9 or guide RNA concentration reflects the significant, yet incomplete, coupling of the target-finding and nuclease activities of the enzyme. Single molecule imaging studies performed with very low Cas9 concentrations revealed undersampling of heterochromatin regions, suggesting that DNA site accessibility can also play a role in specificity under certain conditions (Knight et al., 2015).
Small molecules can also modulate the kinetics of CRISPR editing. For example, a small molecule-dependent Cas9 was developed by genetically inserting an evolved ligand-dependent intein, which increased specificity upon induction by 2 – 25 fold at 11 assessed off-target sites for 3 guide RNAs, while showing very limited nuclease activity in the absence of the small molecule (Davis et al., 2015). However, the on-target efficiencies were also generally less than that of wild-type Cas9, again demonstrating the cost associated with some efforts to engineer improved specificity. Small molecules can also be used to modulate the cellular response to genome editing. For example, the rate of homology-directed repair over non-homologous end-joining has been improved by using a DNA ligase IV inhibitor (Maruyama et al., 2015) and other molecules with poorly understood mechanisms of action (Yu et al., 2015). Additionally, synchronizing the cell cycle has been suggested to increase the homology-directed repair rate with RNP delivery in human cells (Lin et al., 2014).
Broader Implications of Specificity
Proper consideration of Cas9 genomic specificity should include not only the aggregate number of potential off-target sites for a given guide RNA, but also the physiological impact of individual off-target events (both observed and not). While tumorigenesis and unwanted editing of oncogenic tumor suppressors is a common concern, editing events that negatively impact the cell’s viability or functional capabilities need to also be considered. Moreover, the ability to measure off-target editing differs significantly by tissue. While blood can be sampled relatively easily for genomic DNA sequencing, other tissues cannot be so simply biopsied. As a result, linking observed phenotypes with Cas9-induced genome modifications could be difficult in certain organs. The ‘acceptable’ thresholds for off-target rates could also differ in terminally differentiated cells, as opposed to dividing cells. Certainly, ‘acceptable’ specificity thresholds may not be universal across applications, if they are even determinable, as ultimately empirical demonstration of safety will be of paramount importance in the clinic.
Further, the specificity of Cas9 nucleases is not necessarily confined to genomic specificity, especially for in vivo applications. For example, tissue and cell type specificity are also important considerations when targeting genetic disorders that primarily affect certain organs or cells. However, selective distribution may be mediated by a careful choice of the delivery method. For example, different AAV serotypes present different cell-type tropisms and blood clearance rates when administered systemically (Zincarelli et al., 2008). Alternative routes of administration can also provide localized or tissue-specific expression, as well as immunogenicity benefits (Ge et al., 2001; Limberis and Wilson, 2006). Whereas gene augmentation therapy approaches previously demanded viral serotypes that provided sustained and high transgene expression (Hastie and Samulski, 2015), Cas9 applications may be safer with vectors that deliver short-term expression in a highly specific cell-type.
Cell-type specificity may also be achieved with transcriptional regulatory elements. RNA polymerase II tissue- and cell-type specific promoters (e.g. TBG, Synapsin, GFAP) capable of driving protein-coding transcripts like Cas9 could be combined with novel guide RNA promoters and expression strategies beyond the U6 and H1 RNA polymerase III promoters long used in the RNAi field for expressing siRNA and shRNA transcripts. A Csy4-dependent method allows RNA polymerase II promoters to express guide RNAs (Nissim et al., 2014), permitting simple multiplexing as well as expression restricted to certain cell types. Additionally, microRNA binding sites could be added to the 3′ end of the Cas9 mRNA to limit expression in certain cells. Reported to be antigen presenting cell (APC)-specific, mir-142-3p has been exploited in this manner to effectively limit antibody and cellular responses to transgenes by repressing expression and peptide presentation in APCs (Majowicz et al., 2013).
Finally, another means of controlling the specificity of Cas9-mediated activity may be through optogenetic control. As previously reported with TALEs (Konermann et al., 2013), harnessing light-inducible heterodimerizing proteins to dCas9 (CIB1-dCas9) and a transcriptional activator (CRY2-VP64) provides for optogenetically controlled transcriptional activation (Polstein and Gersbach, 2015). Although background activity remains to be optimized, this approach could facilitate effective manipulation of the spatial, temporal, and genomic specificity of a dCas9 synthetic transcription factor.
The Need for Standardization
As demonstrated, the field of Cas9 specificity is developing rapidly and has already accomplished dramatic improvements over the first generation methods. In thinking about developing medicines, it is important to keep in mind that all methods for manipulating therapeutic targets – ranging from genome editing reagents and siRNAs to antibodies and small molecules – are prone to unintended off-target effects. The expectation for the specificity of biological engineering and Cas9 in particular should be continuous improvement and appropriate vigilance. Striving for ever-improving enzymes and measurement technologies with improved lower levels of detection while carefully assessing candidate therapies using state of the art functional models and assays will be important to making appropriate pre-clinical decisions for genome editing therapies.
To rigorously benchmark progress, the genome editing community should strive to standardize the best-performing methods for measuring and reporting off-target activity (Joung, 2015). For example, the commonly used Surveyor assay inherently underreports high levels of indels due to the enzymes’ heteroduplex dependency (Vouillot et al., 2015). Moreover, Surveyor quantification by gel image analysis or electrophoresis devices like the Qiagen Qiaxcel or Agilent TapeStation has limited sensitivity compared to targeted deep sequencing. As such, targeted sequencing should become the standard assay for validating off-target events.
Targeted deep sequencing of off-target events is particularly amenable to standardization and reporting of developments in the field, although the method is variably applied across the literature in terms of experimental execution and computational analysis. In our view, it is important to minimize PCR cycle number, use error-correcting DNA polymerases, consider the sequencing coverage per input genome copy, and compare reads to appropriate reference sequences that take into account genetic variation from the standard reference genomes (Box 1). Many groups currently use custom analysis pipelines, so raw data and code should be shared when possible via web repositories such as Github. Ideally, robust analysis tools will be widely adopted as standard pipelines, as exemplified by the Tuxedo suite (Kim et al., 2013; Trapnell et al., 2013) for RNA-seq. CRISPR-GA (Guell et al., 2014) and CRISPResso (http://crispresso.rocks) are two publically available indel analysis packages that are available as online or command line tools.
Box 1. Specific for which genome?
The simplest way to avoid most off-target effects is to design guide RNAs that have few off-target sites with high sequence homology (ie. 0, 1, or 2 mismatches). However, mutations in the genome or the guide RNA can theoretically create off-target sites with greater sequence homology than expected. Mutations in the genome: It can be important to consider the reference genome used with these design tools. The current standard practice is to design guide RNAs against the species of interest’s standard reference genome (ie. hg38), yet variation in the particular genome of the edited tissue, cell line, or strain can be an important determinant of guide specificity. In fact, one group recently observed an off-target event of unexpectedly high activity (36.7%) when targeting human iPSCs, due to a SNP that converted a 3-mismatch site into a 2-mismatch site on one allele (Yang et al., 2014). For CRISPR-Cas9 applications that require exquisite specificity, whole genome sequencing and an understanding of patient or genotypic variation may become an important initial step of guide design. Mutations in the guide RNA: Guide RNAs are transcribed or synthesized in many ways, with varying fidelity. For example, DNA oligos are prone to small deletions, as the coupling reaction that adds each base is 99% efficient. Strikingly, Digenome-seq discovered “RNA-bulge” type off-targets with guides transcribed from an annealed-oligo template, but no such off-target sites with guides transcribed from a clonal plasmid DNA template (Kim et al., 2016b). Effectively, 1-nt deletions in the oligo pool were leading to ‘false positives’ at off-target sites that would not be edited with other delivery methods. When using RNP delivery, note that RNA oligo synthesis is less efficient (~98%) and thus more prone to deletions. As an alternative, one can transcribe RNA in vitro from sequence verified plasmid with T7 polymerase, which has a more acceptable error rate of ~1/20,000 (Huang et al., 2000). The error rate of Pol III transcription is ~1e-7 in yeast and may be similarly low for guide RNAs transcribed from the human U6 promoter (Alic et al., 2007). So, guide RNAs expressed by plasmid and viral delivery likely boast high fidelity, but direct delivery of synthesized oligos may be cause for concern in genomes with 1-nt RNA bulge off-target sites.
In addition, standard guide RNAs for specificity experiments already help researchers compare methods, and should continue to be used as benchmarks. For example, each of the BLESS, HTGTS, GUIDE-seq, IDLV capture, and Digenome-seq papers assess at least one shared guide RNA (targeting either EMX1 or VEGFA). The different methods identified intersecting but unequal sets of off-targets for the standard guide RNAs, confirming that none of the unbiased methods are yet perfectly sensitive and reliable. However, the off-target sites detected across multiple studies are also generally the most frequently detected and edited while the others are very rare – an encouraging result for the field at large (Gabriel et al., 2015).
We propose that a set of guidelines for reporting CRISPR-Cas9 specificity research should be established, much like the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines for qPCR (Bustin et al., 2009), the MIAME for microarrays(Brazma et al., 2001), the MIAPE for proteomics (Taylor, 2006; Taylor et al., 2007; Martinez-Bartolome et al., 2014), the MISFISHIE for FISH experiments (Deutsch et al., 2006; Deutsch et al., 2008), as well as others (Chervitz et al., 2011). Such guidelines are directed toward ensuring unambiguous communication of experimental designs and results, such that independent scientists can fairly interpret their conclusions in the context of the broader field while reproducing those results without requiring additional information. These ‘minimum information’ requirements should include the number of off-target sites assessed, the off-target prediction constraints, cell type, delivery method, time point, dosage, analysis method, computational tools, sample size, and other key factors known to impact specificity.
The research community could begin to report guide RNA on- and off-target activity to shared databases, like EENdb (Xiao et al., 2013) or CrisprGE (Kaur et al., 2015). Such standardized information, in combination with continued progress in off-target detection methods and Cas9 optimization, could greatly improve guide selection, enable fair comparison and benchmarking across publications while ultimately enabling precise and safe genome-editing. Importantly, such standards must be developed with a focus on maximizing end-user adoption rates, minimizing usability hurdles, and incorporating user feedback, because “a standard that is not widely used is not really a standard” (Chervitz et al., 2011). Historically, working group scientific consortiums have led the development of ‘Omics’ standards while scientific journals and funding agencies have been critical in enforcing their broad compliance. Aligning genome-editing stakeholders towards these goals would represent a discrete step forward in the maturation of this field.
Conclusion
The scientific landscape around Cas9 specificity is rapidly evolving with marked improvements recently achieved in guide selection, protein and guide engineering, off-target detection methods, and more. Notably, two recent protein engineering approaches (eSpCas9 and SpCas9-HF) have elegantly increased SpCas9 specificity, most likely by reducing tolerance for mismatched DNA binding, although their overall on-target efficiency relative to the wild-type enzyme remain to be studied by more groups in different applications and organisms. These highly specific variants should encourage development of refined off-target detection methods with limits of detection well below 0.1%. Cas9 orthologs and non-Cas9 CRISPR enzymes are also emerging as genome editing tools, but will ultimately face the same challenges (Shmakov et al., 2015; Zetsche et al., 2015). In order to propagate these advances throughout the broader research community, the genome-editing field will greatly benefit from discussion and consensus about best practices for future research as well as data standardization. Beyond specificity, key challenges and opportunities for the field lie in improving delivery and increasing efficiency, especially for precise edits.
Box 2. Recommended practices for minimizing, identifying, and measuring Cas9 off-target cleavage.
- Important: Design guides for uniqueness in the appropriate reference genome
- No off-by-1 or off-by-2 mismatch sites
- Highly ranked by specificity score (Hsu et al., 2013) or CFD (Doench et al., 2016)
- Preferred: Detect off-target sites with one or more unbiased methods
- BLESS, GUIDE-seq, Digenome-seq, etc.
- Important: Validate all detected off-target sites with targeted deep sequencing
- Prepare library from >10,000 cells
- Minimize PCR cycle number and use high-fidelity polymerase
- Determine limit of detection based on ‘indel’ rate in negative control, aim for ~0.1% with current technology
- Iterate steps 2 and 3 to optimize on-target: off-target ratio
- Titrate dose
- Employ Cas9 variants (eCas9, Cas9-HF)
- Optimize guide RNA structure (tru-guides, GGX20)
Acknowledgments
We gratefully acknowledge Alexandra Glucksmann, Hari Jayaram, and Andrew Hack for expert comments on the article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
J.T. and V.M. are employees and P.D.H. is a former employee of Editas Medicine, which is developing therapeutics based on CRISPR-Cas9 genome editing technology.
Optimizing the specificity of the CRISPR-Cas9 technology is a major challenge in genome engineering. Tycko et al. present a comprehensive practical guide to the key tools and strategies for off-target reduction and analysis, while highlighting recent breakthroughs and anticipated future developments in the field.
References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016 doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N, Ayoub N, Landrieux E, Favry E, Baudouin-Cornu P, Riva M, Carles C. Selectivity and proofreading both contribute significantly to the fidelity of RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2007;104:10400–10405. doi: 10.1073/pnas.0704116104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienhoff SE, Allen GK, Berg JN. Release of tumor necrosis factor-alpha from bovine alveolar macrophages stimulated with bovine respiratory viruses and bacterial endotoxins. Vet Immunol Immunopathol. 1992;30:341–357. doi: 10.1016/0165-2427(92)90104-x. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bolukbasi MF, Gupta A, Oikemus S, Derr AG, Garber M, Brodsky MH, Zhu LJ, Wolfe SA. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods. 2015;12:1150–1156. doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolukbasi MF, Gupta A, Wolfe SA. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Methods. 2016;13:41–50. doi: 10.1038/nmeth.3684. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Hu J, Almeida R, Liu H, Balakrishnan S, Covill-Cooke C, Lim WA, Huang B. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016;44:e75. doi: 10.1093/nar/gkv1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Deutsch EW, Field D, Parkinson H, Quackenbush J, Rocca-Serra P, Sansone SA, Stoeckert CJ, Jr, Taylor CF, Taylor R, et al. Data standards for Omics data: the basis of data sharing and reuse. Methods Mol Biol. 2011;719:31–69. doi: 10.1007/978-1-61779-027-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013a;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Lee J, Carroll D, Kim JS, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013b;195:1177–1180. doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Jia G, Choi J, Ma H, Anaya E, Ye C, Shankar P, Wu H. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Ball CA, Berman JJ, Bova GS, Brazma A, Bumgarner RE, Campbell D, Causton HC, Christiansen JH, Daian F, et al. Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE) Nat Biotechnol. 2008;26:305–312. doi: 10.1038/nbt1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Ball CA, Bova GS, Brazma A, Bumgarner RE, Campbell D, Causton HC, Christiansen J, Davidson D, Eichner LJ, et al. Development of the Minimum Information Specification for In Situ Hybridization and Immunohistochemistry Experiments (MISFISHIE) OMICS. 2006;10:205–208. doi: 10.1089/omi.2006.10.205. [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu BX, Hansen LL, Artiles KL, Nonet ML, Fire AZ. Landscape of target:guide homology effects on Cas9-mediated cleavage. Nucleic Acids Res. 2014a;42:13778–13787. doi: 10.1093/nar/gku1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014b;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, von Kalle C, Schmidt M. Mapping the precision of genome editing. Nat Biotechnol. 2015;33:150–152. doi: 10.1038/nbt.3142. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Powell S, Van Roey M, McArthur JG. Factors influencing the development of an anti-factor IX (FIX) immune response following administration of adeno-associated virus-FIX. Blood. 2001;97:3733–3737. doi: 10.1182/blood.v97.12.3733. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D. Delivery and Specificity of CRISPR-Cas9 Genome Editing Technologies for Human Gene Therapy. Hum Gene Ther. 2015;26:443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- Guell M, Yang L, Church GM. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA) Bioinformatics. 2014;30:2968–2970. doi: 10.1093/bioinformatics/btu427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Hastie E, Samulski RJ. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success–a personal perspective. Hum Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brieba LG, Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK. Unwanted mutations: Standards needed for gene-editing errors. Nature. 2015;523:158. doi: 10.1038/523158a. [DOI] [PubMed] [Google Scholar]
- Kaur K, Tandon H, Gupta AK, Kumar M. CrisprGE: a central hub of CRISPR/Cas-based genome editing. Database (Oxford) 2015;2015:bav055. doi: 10.1093/database/bav055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016a doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim S, Kim S, Park J, Kim JS. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016b;26:406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016a;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015a;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015b;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016b doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson JB, Dahan M, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo T, Lee J, Kim JS. Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Mol Cells. 2015;38:475–481. doi: 10.14348/molcells.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 System Enables Specific Genome Editing in Mammalian Cells. Mol Ther. 2016;24:645–654. doi: 10.1038/mt.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberis MP, Wilson JM. Adeno-associated virus serotype 9 vectors transduce murine alveolar and nasal epithelia and can be readministered. Proc Natl Acad Sci U S A. 2006;103:12993–12998. doi: 10.1073/pnas.0601433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Harrington LB, O’Connell MR, Zhou K, Doudna JA. Single-Stranded DNA Cleavage by Divergent CRISPR-Cas9 Enzymes. Mol Cell. 2015;60:398–407. doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson C. The CRISPR Software Matchmaker: A New Tool for Choosing the Best CRISPR Software for Your Needs. 2015 blog.addgene.org, Addgene.
- Majowicz A, Maczuga P, Kwikkers KL, van der Marel S, van Logtenstein R, Petry H, van Deventer SJ, Konstantinova P, Ferreira V. Mir-142–3p target sequences reduce transgene-directed immunogenicity following intramuscular adeno-associated virus 1 vector-mediated gene delivery. J Gene Med. 2013;15:219–232. doi: 10.1002/jgm.2712. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Annotation and Classification of CRISPR-Cas Systems. Methods Mol Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013b;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013c;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Bartolome S, Binz PA, Albar JP. The Minimal Information about a Proteomics Experiment (MIAPE) from the Proteomics Standards Initiative. Methods Mol Biol. 2014;1072:765–780. doi: 10.1007/978-1-62703-631-3_53. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, Safi A, Crawford GE, Reddy TE, Gersbach CA. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25:1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M. Genome Editing: A New Approach to Human Therapeutics. Annu Rev Pharmacol Toxicol. 2016;56:163–190. doi: 10.1146/annurev-pharmtox-010814-124454. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Rahdar M, McMahon MA, Prakash TP, Swayze EE, Bennett CF, Cleveland DW. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc Natl Acad Sci U S A. 2015;112:E7110–7117. doi: 10.1073/pnas.1520883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013a;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013b;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan V, Wahlin K, Maruotti J, Zack DJ. Expansion of the CRISPR-Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat Commun. 2014;5:4516. doi: 10.1038/ncomms5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kuscu C, Quinlan A, Qi Y, Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43:e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24:125–131. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF. Minimum reporting requirements for proteomics: a MIAPE primer. Proteomics. 2006;6(Suppl 2):39–44. doi: 10.1002/pmic.200600549. [DOI] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, et al. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Vouillot L, Thelie A, Pollet N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 2015;5:407–415. doi: 10.1534/g3.114.015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, Chang T, Huang H, Lin RJ, Yee JK. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wu Y, Yang Z, Hu Y, Wang W, Zhang Y, Kong L, Gao G, Zhu Z, Lin S, et al. EENdb: a database and knowledge base of ZFNs and TALENs for endonuclease engineering. Nucleic Acids Res. 2013;41:D415–422. doi: 10.1093/nar/gks1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Grishin D, Wang G, Aach J, Zhang CZ, Chari R, Homsy J, Cai X, Zhao Y, Fan JB, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One. 2014;9:e108424. doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]


