Abstract
Background
Bile duct carcinoma is a common digestive tract tumor with high morbidity and mortality. As a kind of important non-coding RNA, microRNA (miR) plays an important role in post-transcriptional regulation. MiR-122 is the most abundant miR in the liver. Multiple studies have shown that miR-122 level is reduced in a variety of liver tumors and can be used as a specific marker for liver injury. P53 is a classic tumor suppressor gene that can induce tumor cell apoptosis through various pathways. Whether miR-122 affects p53 in bile duct carcinoma still needs investigation.
Material/Methods
miR inhibitor or mimics was transfected to bile duct carcinoma cells to evaluate its function on proliferation, invasion, apoptosis, and p53 expression.
Results
MiR-122 overexpression reduced cell invasion and migration ability, and inhibited cell apoptosis and p53 expression. Inhibiting miR-122 caused the opposite results.
Conclusions
Upregulating miR-122 can suppress bile duct carcinoma cell proliferation and induce apoptosis. MiR-122 could be used as a target for bile duct carcinoma treatment, which provides a new strategy for cholangiocarcinoma patients.
MeSH Keywords: Apoptosis; Apoptosis Inducing Factor; Cholangiocarcinoma; Genes, p53; Nicardipine
Background
Bile duct carcinoma refers to the malignant tumor derived from the extrahepatic bile duct, including the lower segment of the bile duct. Its etiology may be related to bile duct stones and primary sclerosing cholangitis. Similar to other tumors, it can be treated by surgery, radiotherapy, chemotherapy, and other methods, but the prognosis is poor [1].
In recent years, studies have shown that microRNA (miR) plays an important role in occurrence and development of a variety of diseases. miR is a small non-coding single-stranded RNA with length of 18–24 bp and can bind with the 3′UTR of target mRNA. Under the effect of RNA excision enzyme, it can selectively degrade mRNA to inhibit or activate the downstream genes [2]. MiR regulates downstream genes mainly through inhibiting mRNA transcription. It can also impact cell function and oncogene activation by mediating mRNA degradation and activation [3]. MiR-122 is especially expressed in the liver in large amounts [4], and has been confirmed to participate in liver cell cycle and fat metabolism [6]. MiR-122 is considered to be a marker of liver injury. Recent studies have shown that miR-122 is closely related to hepatocellular carcinoma (HCC) [7].
P53 was the first discovered tumor suppressor gene that can inhibit cancer cells through multiple pathways, such as promoting cancer cell apoptosis and restraining the cell cycle [8]. MiR-122 expression decreased in liver cancer and might be a biomarker of HCC occurrence and development. Its level could be treated as one of the diagnosis criteria of HCC differentiation and malignant degree. Some studies also suggested that miR-122 content increased significantly in the apoptotic cells [9]. Its role in intrahepatic cholangiocarcinoma proliferation and effect on p53 and cell apoptosis is still unclear.
This study aimed to investigate the effect of miR-122 on cholangiocarcinoma proliferation, invasion, apoptosis, and p53 expression by transfection.
Material and Methods
Main reagents
Cholangiocarcinoma cell line QBC939 was bought from the cell bank of the Chinese Academy of Sciences (Shanghai, China). DMEM-F12 medium, penicillin, streptomycin, fetal calf serum, and PBS buffer were from Hyclone (UT, USA). POLD deliver3000 and Opti-MEM medium were purchased from Invitrogen (CA, USA). β-actin antibody was from KangCheng Biotechnology Company (Shanghai, China). P53 antibody was from Abcam (Hong Kong, China). Rabbit Anti-Mouse IgG (H+L) and Rabbit Anti-Mouse IgG (H+L) were from ProteinTech (Wuhan, China). SYBR Green PCR Master Mix was from Takara (Dalian, China). MiR-122 mimics and inhibitor were from Genepharma (Shanghai, China).
MTT
After transfection with miR-122 mimics or inhibitor, the cells in logarithmic phase were seeded in a 96-well plate at 5~10×103/well with 10 replications. The cells were cultured for 24 h and 10 μl MTT was added. We added 100 μl triple fluid to each well after 4-h incubation. The well was tested at 450 nm for calculation.
Cell transfection
The cells were seeded in 6-well plates 1 day before transfection. Ten pmol miR-122 inhibitor, mimics, or negative control were mixed with 5 μl POLDdeliver3000 in 100 μl for 5 min. After instilling the mixture to the plate, the cells were incubated for 48 h for proliferation determination and protein detection.
Total RNA extraction
The cells were washed with PBS 3 times at 48 h after transfection. One ml Trizol was added to the cells on ice for 5 min and then the lysate was moved to a 1.5-ml EP tube. We added 200 μl chloroform and the tube was shaken for 15 s. After 3 min at room temperature, the tube was centrifuged at 12 000g and 4°C for 15 min. The aqueous phase was moved to a new EP tube together with 500 μl isopropanol. After 10 min at 4°C, the tube was centrifuged at 12 000g and 4°C for 10 min. After removing the supernatant, the mixture was washed with 1 ml ethanol 3 times. We added 20 μl DEPC water to get the mRNA.
Real-time PCR
MiR-122 primer was designed and synthesized by Sigma, and the sequence was as follows: forward, 5′-TTGAATTCTA ACACCTTCGTGGCTACAGAG-3′; reverse, 5′-TTAGATCTCATTTA TCGAGGGAAGGATTG-3′. U6 was treated as reference, forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAAT TTGCGT-3′ [10]. Real-time PCR was performed at 50 μl system according to the manual. PCR reaction contained at 50 °C for 30 min and 95°C for 5 min, followed by 40 cycles including 95°C for 30 s, 55°C for 30 s, and 72°C for 50 s, and 72°C for 5 min. Amplification curve and melting curve were verified for quality control. Gene expression levels were quantified relative to the expression of reference gene using the optimized comparative Ct (2−ΔCt) value method.
Transwell detection of cell migration
Transwell chamber and related instruments were precooled at 4°C overnight. ECM matrigel were thawed at 4°C. We placed 50 μl diluted ECM matrigel into the transwell chamber and the chamber was at 37°C for 4 h. The cells in logarithmic phase were seeded into the upper chamber, while 10% fetal calf serum was added to the lower chamber. The cells were grouped as normal control, miR-122 mimics group, miR-122 mimics negative control group, miR-122 inhibitor group, and miR-122 inhibitor negative control group, 3 times in each group. After incubation, the chamber was stained with 0.1% crystal violet and observed under an inverted microscope. Five views in each well were counted for calculation.
Total protein extraction
The cells were washed with PBS 3 times and we added 10 μl 100 mM PMSF. After being cracked on ice for 5–10 min, the cells were moved to an EP tube and centrifuged at 12 000 rpm and 4°C for 5 min. The supernatant was whole protein solution.
Western blot
After BCA qualification, the protein solution was standardized to unified concentration. The protein was degenerated at 95°C for 5 min after adding buffer. The protein was then separated by 10% SDS-PAGE electrophoresis and transferred to a PVDF membrane at 300 mA for 1 h. The membrane was incubated with p53 antibody (1:1000) at 4°C overnight and secondary antibody (1:1000) at 37°C for 2 h. The binding was detected by chemiluminescence.
Tunnel method for detecting cell apoptosis
We seeded 2×105 cells in a 24-well plate and transfected them for 24 h. After being fixed by paraformaldehyde, the cells were infiltrated in 3% hydrogen peroxide methanol for 10 min. After being perforated by 0.5% Triton for 5 min, the cells were added with 50 μl Tunnel reaction liquid and incubated at 37°C for 1 h. The cells were observed under a fluorescence microscope and 5 views were selected for positive cell calculation.
Statistical analysis
All statistical analyses were performed using SPSS10.0 software (Chicago, IL). Numerical data are presented as means and standard deviation (χ̄±S). Differences between multiple groups were analyzed by one-way ANOVA or SNK-Q test.
Results
miR-122 expression in cholangiocarcinoma cells
Real-time PCR was used to detect miR-122 expression in each group after miR-122 mimics or inhibitor transfection. As shown in Figure 1, miR-122 level decreased or increased significantly compared with negative control after transfecting miR-122 inhibitor or mimics (p<0.05), suggesting that miR-122 inhibitor or mimics was successfully transfecting into the cells.
Figure 1.
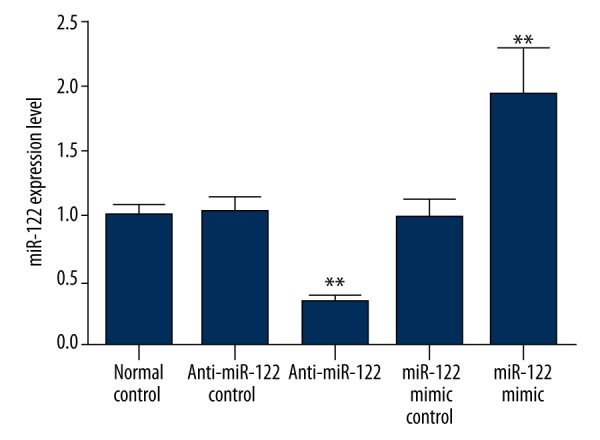
miR-122 expression after transfection. ** p<0.05, compared with normal control.
miR-122 transfection impact on cell proliferation
The cells were counted at 24 h after transfection and seeded in a 96-well plate. MTT method was performed to evaluate the effect of miR-122 inhibitor or mimics on cell proliferation. We found that cell proliferation rate in miR-122 inhibitor or mimics negative control showed no obvious difference from normal control. miR-122 inhibitor transfection significantly increased the cell proliferation rate, while miR-122 mimics transfection markedly reduced it (p<0.05) (Figure 2).
Figure 2.
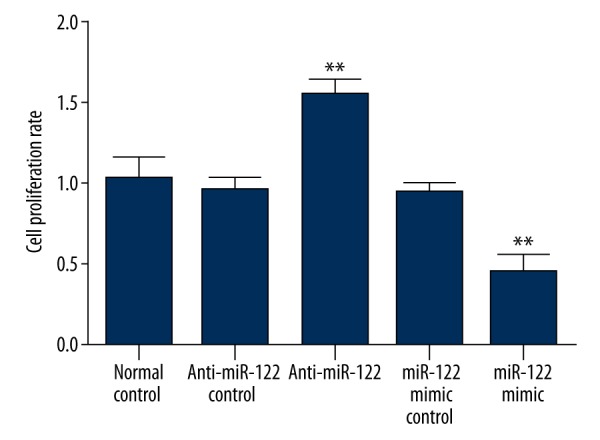
The effect of miR-122 inhibitor or mimics transfection on cell proliferation.
miR-122 transfection impact on cell invasion
As shown in Figure 3, the cell numbers penetrating the membrane gradually increased from 12 h over time and slowed after 36 h. Compared with miR-122 inhibitor negative control and normal control, cell invasive ability was significantly enhanced after transfection with miR-122 inhibitor (p<0.05). In contrast, it clearly weakened in cells transfected with miR-122 mimics compared with miR-122 mimics negative control and normal control (p<0.05).
Figure 3.
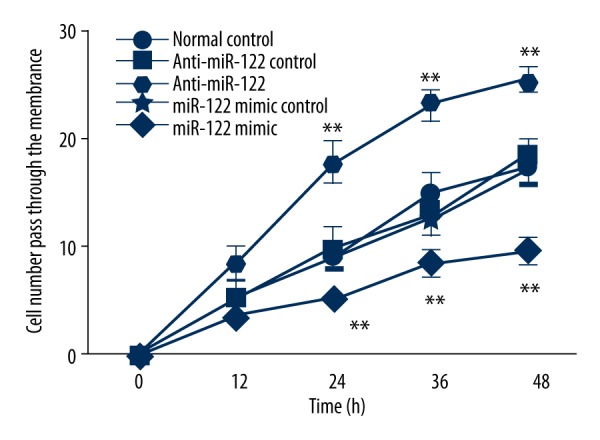
Invasive cell number at 48 h after miR-122 inhibitor or mimics transfection. ** p<0.05, compared with normal control.
Effect of miR-122 transfection on p53 expression
p53 is a common cancer suppressor gene that plays an important role in tumor occurrence and development [11]. As shown in Figure 4, miR-122 inhibitor transfection can significantly decrease p53 expression level compared with normal control and miR-122 mimics negative control (p<0.05), whereas miR-122 mimics transfection can elevate p53 level (p<0.05). The 2 negative controls showed no significant difference from normal control.
Figure 4.
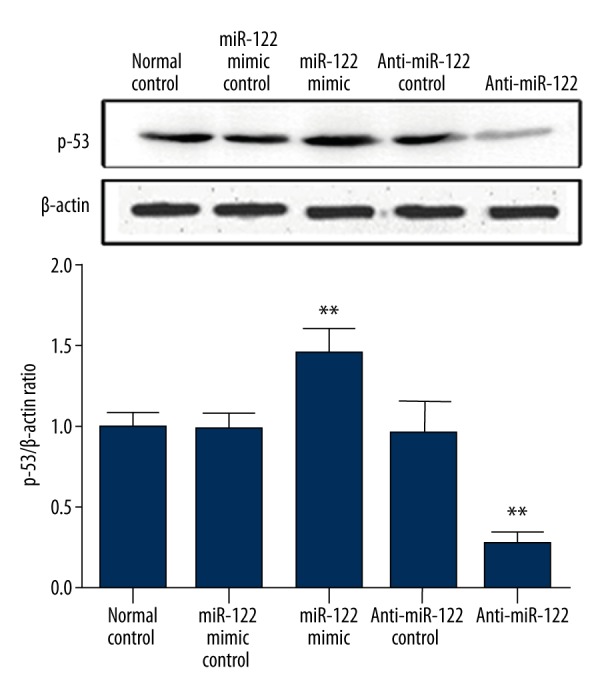
Effect of miR-122 transfection on p53 expression.** p<0.05, compared with normal control.
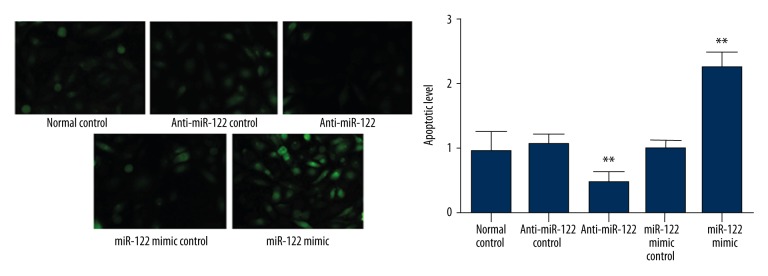
Effect of miR-122 mimics or inhibitor transfection on cell apoptosis
Promoting cancer cell apoptosis is an important strategy in cancer therapy [12]. We used Tunnel method to evaluate the effect of miR-122 inhibitor or mimics on cell apoptosis. Compared with normal control and negative control, miR-122 inhibitor or mimics transfection can significantly decrease or increase cell apoptotic rate (p<0.05), indicating that miR-122 overexpression can promote intrahepatic cholangiocarcinoma cell apoptosis (Figure 5).
Figure 5.
Effect of miR-122 mimics or inhibitor transfection on cell apoptosis. ** p<0.05, compared with normal control.
Discussion
In this study we transfected miR-122 inhibitor or mimics to cholangiocarcinoma cells by liposome to determine the effect of miR-122 on p53 and cell apoptosis. We aimed to evaluate the effect of miR-122 on cholangiocarcinoma by detecting cell proliferation rate, invasive ability, p53 protein expression, and cell apoptosis.
We discovered that miR-122 overexpression can suppress cholangiocarcinoma cell invasion and migration, while elevating p53 level and promoting cell apoptosis, suggesting that miR-122 plays an important role in cholangiocarcinoma occurrence and development.
Cholangiocarcinoma is a type of malignant tumor in the bile duct, with high morbidity and mortality [13]. Complications of cholangiocarcinoma include obstructive jaundice, cholangitis, and biliary sepsis [14]. It is difficult to treat because of the specific pathogenic site. MiR is a type of non-coding RNA at the length of 20–24 bp that newly discovered in recent years. It can bind with 3′ UTR of the mRNA to post-transcriptionally regulate mRNA expression level. MiR is quite important in regulating protein synthesis under the physiological condition. Its dysregulation is associated with multiple cancers, including thymic carcinoma [15], hepatic carcinoma, and bile duct carcinoma [16]. Different miRs play important roles in different malignant tumors. MiR-122 is rich and specific in the liver, and it accounts for 72% of the total miRs [17]. It has an important regulating effect in physiological and pathological conditions and is also considered as the marker of liver injury [18]. Previous research showed that miR-122 is significantly decreased in liver cancer tissue, suggesting it could be a biomarker reflecting HCC development [19].
p53 is an important tumor suppressor gene in the cell. Its upregulation facilitates cell survival by promoting cancer cells apoptosis directly and by changing the AKT phosphorylation level. Therefore, p53 expression level can directly accelerate tumor cell apoptosis [19]. MiR-122 has been reported to regulate p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma [20]. Through modulating its target gene, cyclin G1, miR-122 can also influence p53 protein stability and transcriptional activity and reduces invasion capability of HCC-derived cell lines [21]. In addition, overexpression of miR-122 triggers doxorubicin-induced apoptosis of HCC-derived cell lines. However, the exact mechanism by which miR-122 overexpression induces p53-dependent apoptosis in cholangiocarcinoma cells (possibly through cyclin G1) was not investigated in the present study and remains unclear. Several target genes of miR-122 have been identified to be involved in hepatocarcinogenesis, epithelial mesenchymal transition, and angiogenesis, including cyclin G1, Bcl-w, Wnt1, transcription factor CUTL1, a disintegrin and metalloprotease (ADAM)10, ADAM17, serum response factor, insulin-like growth factor-1 receptor, the embryonic isoform of pyruvate kinase (Pkm2), pituitary tumor-transforming gene 1 binding factor, Cut-like homeobox 1, and c-myc [22]. Among these targets, Bcl-w, an anti-apoptotic Bcl-2 family member, has also been identified as the miR-122 target gene involved in apoptosis of human hepatocellular carcinoma cell lines, apart from cyclin G1 [23]. Several other molecules are also involved in cell apoptosis, such as p21 [24] and Dock180 [25]. Further research is needed to determine which gene is involved in the miR-122-induced apoptosis in cholangiocarcinoma cells and which signaling molecules connect miR-122 with p53. Experimental results suggested that miR-122 participates in cell proliferation, invasion, and migration in renal cell carcinoma cells through PI3K/AKT signaling pathways [10,26]. Thus, in this study, we examined miR-122 regulation function on p53. We found that the effect of miR-122 on apoptosis is dependent on p53, and we also found that miR-122 upregulation or downregulation can decrease or increase cholangiocarcinoma cell invasive ability.
Conclusions
Our results demonstrated that upregulating miR-122 can suppress bile duct carcinoma cell proliferation and induce apoptosis, suggesting that MiR-122 could be a new target for the treatment of bile duct carcinoma.
Footnotes
Source of support: Departmental sources
References
- 1.Chiang NJ, Shan YS, Hung WC, Chen LT. Epigenetic regulation in the carcinogenesis of cholangiocarcinoma. Int J Biochem Cell Biol. 2015;67:110–14. doi: 10.1016/j.biocel.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–27. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–38. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Chang J, Nicolas E, Marks D, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya SN, Habermacher R, Martine U, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Zhu X, Wu L, et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/beta-catenin pathway. Liver Int. 2012;32:752–60. doi: 10.1111/j.1478-3231.2011.02750.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Chen J, Li Z, et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469:325–46. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 9.Yuan Y, Zhang X, Zeng X, et al. Glutathione-mediated release of functional miR-122 from gold nanoparticles for targeted induction of apoptosis in cancer treatment. J Nanosci Nanotechnol. 2014;14:5620–27. doi: 10.1166/jnn.2014.8735. [DOI] [PubMed] [Google Scholar]
- 10.Lian JH, Wang WH, Wang JQ, et al. MicroRNA-122 promotes proliferation, invasion and migration of renal cell carcinoma cells through the PI3K/Akt signaling pathway. Asian Pac J Cancer Prev. 2013;14:5017–21. doi: 10.7314/apjcp.2013.14.9.5017. [DOI] [PubMed] [Google Scholar]
- 11.Dickson PV, Behrman SW. Distal cholangiocarcinoma. Surg Clin North Am. 2014;94:325–42. doi: 10.1016/j.suc.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Soares ND, Teodoro AJ, Lotsch PF, et al. Anticancer properties of carotenoids in prostate cancer: A review. Histol Histopathol. 2015;30(10):1143–54. doi: 10.14670/HH-11-635. [DOI] [PubMed] [Google Scholar]
- 13.Brandi G, Farioli A, Astolfi A, et al. Genetic heterogeneity in cholangiocarcinoma: A major challenge for targeted therapies. Oncotarget. 2015;6:14744–53. doi: 10.18632/oncotarget.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uppal DS, Wang AY. Advances in endoscopic retrograde cholangiopancreatography for the treatment of cholangiocarcinoma. World J Gastrointest Endosc. 2015;7:675–87. doi: 10.4253/wjge.v7.i7.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–43. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho HJ, Kim JK, Nam JS, et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem. 2015;48:1073–78. doi: 10.1016/j.clinbiochem.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Laterza OF, Lim L, Garrett-Engele PW, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–83. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manfe V, Biskup E, Rosbjerg A, et al. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PLoS One. 2012;7:e29541. doi: 10.1371/journal.pone.0029541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fornari F, Gramantieri L, Giovannini C, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–67. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- 22.Nakao K, Miyaaki H, Ichikawa T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J Gastroenterol. 2014;49:589–93. doi: 10.1007/s00535-014-0932-4. [DOI] [PubMed] [Google Scholar]
- 23.Lin CJ, Gong HY, Tseng HC, et al. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–20. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 24.Chen A, Huang X, Xue Z, et al. The Role of p21 in apoptosis, proliferation, cell cycle arrest, and antioxidant activity in UVB-irradiated human HaCaT keratinocytes. Med Sci Monit Basic Res. 2015;21:86–95. doi: 10.12659/MSMBR.893608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan A, Li G, Zhang X, et al. Pro-survival effect of Dock180 overexpression on rat-derived H9C2 cardiomyocytes. Med Sci Monit Basic Res. 2013;19:12–19. doi: 10.12659/MSMBR.883738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Wang H, Yang Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS One. 2012;7:e47053. doi: 10.1371/journal.pone.0047053. [DOI] [PMC free article] [PubMed] [Google Scholar]