Abstract
Registration of multiple 3D ultrasound sectors in order to provide an extended field of view is important for the appreciation of larger anatomical structures at high spatial and temporal resolution. In this paper, we present a method for fully automatic spatio-temporal registration between two partially overlapping 3D ultrasound sequences. The temporal alignment is solved by aligning the normalized cross correlation-over-time curves of the sequences. For the spatial alignment, corresponding 3D Scale Invariant Feature Transform (SIFT) features are extracted from all frames of both sequences independently of the temporal alignment. A rigid transform is then calculated by least squares minimization in combination with random sample consensus. The method is applied to 16 echocardiographic sequences of the left and right ventricles and evaluated against manually annotated temporal events and spatial anatomical landmarks. The mean distances between manually identified landmarks in the left and right ventricles after automatic registration were (mean ± SD) 4.3 ± 1.2 mm compared to a reference error of 2.8 ± 0.6 mm with manual registration. For the temporal alignment, the absolute errors in valvular event times were 14.4 ± 11.6 ms for Aortic Valve (AV) opening, 18.6 ± 16.0 ms for AV closing, and 34.6 ± 26.4 ms for mitral valve opening, compared to a mean inter-frame time of 29 ms.
1. INTRODUCTION
Ultrasound is the image modality of choice for assessing the heart in clinical routine, offering high frame rate imaging of the beating heart. As 3D ultrasound is being more widely studied, new applications for imaging, visualization and quantification of the heart are emerging.
Registration of 3D ultrasound images has many potential uses, including motion estimation and extended field of view. Several methods have been proposed using elastic registration to study the motion of the heart walls, which can be used to estimate the strain in the myocardium [1, 2, 3, 4]. As 3D ultrasound images are typically acquired in smaller sectors, to maintain adequate spatial and temporal resolution, registration can be used to fuse multiple 3D sectors together. This extends the field of view, allowing the quantification of larger structures while preserving resolution. Methods for spatial registration of 3D ultrasound include optical flow [5], feature-based registration such as Scale Invariant Feature Transform (SIFT) [6], voxel-wise similarity measures such as Normalized Cross-Correlation (NCC) [7], and similarity measures based on local orientation and phase [8].
Ni et al. used 3D SIFT features to create a panorama image from several 3D ultrasound images by rigid registration [6]. This work was extended by Schneider et al. who used efficient rotation-variant features to provide a transform between consecutive frames in real-time during acquisition [9]. This can be useful for compensating for slight movement of the probe during acquisition, or to cancel movement of anatomy.
Because the heart undergoes a complex contraction that is non-linear with respect to changes in the heart rate, establishing a temporal correspondence between sequences is important for accurate registration. Temporal registration for cardiac images often relies heavily on external recording from ECG. Utilizing image based measurement allows the registration to be independent from this external measurement.
Perperidis et al. used Normalized Cross Correlation (NCC) over time for temporal alignment and normalized mutual information for spatial alignment to provide a free-form spatio-temporal registration between MRI sequences [10]. Expanding on this, Zhang et al. used a similar approach for 3D ultrasound to MRI registration, using NCC over time for temporal alignment and 3D SIFT features for spatial alignment [11].
There are many aspects of ultrasound image processing that present challenges compared to other medical imaging modalities. Ultrasound often contain artifacts such as acoustic shadows, speckles, and reverberations. This means that several of the intensity-based matching methods used for registering images of other modalities are less appropriate for ultrasound processing. Furthermore, acquisitions can have very different gain settings, temporal and spatial resolutions, and can be acquired from different locations depending on the patient, in order to improve the acoustic window.
In this paper we present a method of registering two partially overlapping 3D cardiac ultrasound sequences in space and time. The main contribution of this work is to solve both the spatial and temporal alignment problems for 4D ultrasound, and to solve the spatial alignment with no user interaction and without any a priori assumptions on either the temporal or spatial alignment of the sequences. The temporal alignment is solved by aligning the NCC-over-time functions of the floating and reference sequences. We then use corresponding 3D SIFT features between all frames, without any assumptions on the temporal alignment, to extract a single rigid transform for the whole cardiac cycle by minimizing a least squares problem using Random Sample Consensus (RANSAC). The method was validated by registering 3D sectors of the left and right ventricles in 16 clinical cases.
2. METHODS
Given a reference image sequence and a floating sequence , the objective of the registration method is to find a transform such that any voxel in the transformed sequence coincides with the corresponding voxel in .
Following the approach used by others [10, 11], we decouple the registration problem into spatial and temporal domains and solve these separately. We first solve the temporal alignment using the NCC over the cardiac cycle, before extracting a spatial transform from 3D SIFT feature correspondences between all frames.
2.1 Temporal Registration
The heart undergoes a complex contraction during the cardiac cycle. Because this contraction is slightly different for each beat and non-linear with changes of heart frequency, different acquisitions will generally not be synchronized, even if performed within a relatively short period of time without external influences.
The NCC over time has been shown to be a characteristic function describing the events of the cardiac cycle in a consistent manner, and has been used for temporal alignment of MRI sequences [10] and between ultrasound and MRI [11]. In both of these studies, the temporal alignment was achieved by first detecting key cardiac events in the NCC-over-time function, which involves calculating its second order derivative.
However, our experiments indicate that the second order derivation is unstable in ultrasound images of high frame rate, because of the inherently low signal-to-noise ratio and the presence of artifacts such as acoustic shadows, speckles and reverberations. This is specially apparent for images of the right ventricle which generally has lower image quality than the left ventricle, due to the challenging acquisition [12]. We therefore propose to solve the temporal registration by aligning the NCC-over -time functions with an optimization problem.
The temporal transform is modeled as a global affine transform used to scale the sequences to the same length and compensate for global phase shift, and a local transform used to adjust for non-linear difference in the contraction pattern of the sequences. The local transform is modeled as a 1D B-spline with Nt knots,
| (1) |
where T is the length of the sequence, and bi and τi are the B-spline basis functions and control time displacements respectively. The combined transform is given by
| (2) |
The control time displacements τi of the local temporal deformation are calculated by solving the minimization problem
| (3) |
where fr and ff are the NCC functions of the reference and floating sequences. The optimization problem is solved numerically by sequential quadratic programming. By constraining to be monotonically increasing, we guarantee that the frame order remains unchanged.
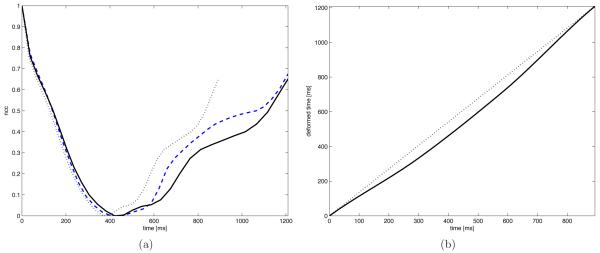
Because 3D cardiac ultrasound is gated by electrocardiography in almost all clinical cases, all image sequences were ordered such that the first frame corresponded to the QRS complex. We therefore assumed a zero phase shift, β = 0 in . Figure 1 shows the NCC functions before and after temporal alignment in an example case.
Figure 1.
The normalized cross-correlation over time for a reference (dashed) and floating sequence before (dotted) and after (solid) temporal alignment (a), resulting temporal registration (b).
2.2 Time-Independent Spatial Registration
When registering sequences of the same patient during the same exam without external influences, it is fair to assume that the true spatial transform between the sectors of two ultrasound sequences is rigid and constant over the cycle. The spatial transform between frames of the reference and floating sequences and at similar points in the cardiac cycle can thus be assumed to be approximately rigid. This assumption leads a novel time-independent feature-based alignment (FBA) method [13], involving 3D SIFT feature extraction and matching.
First, a set of position-scale pairs {} are extracted in all 3D image frames of each sequence by identifying local maxima and minima of the difference-of-Gaussian function,
| (4) |
where is the convolution of the image with a Gaussian kernel of variance σ2 and κ the scale sampling rate. Following detection, an orientation is assigned to each feature using local gradient orientation information, and finally an appearance descriptor is generated from the patch of voxels within the image region defined by . Appearance descriptors are normalized according to image intensity and local geometry, and can thus be used to compute image-to-image correspondences in a manner invariant to monotonic intensity shifts and global similarity transforms.
For time-independent matching, each sequence is modeled as an unordered bag-of-features with spatial but no temporal information. The FBA method [13] is then used to estimate a set of highly probable sequence-to-sequence feature correspondences. Briefly, a set of nearest-neighbor feature correspondences are identified based on the distance between appearance descriptors, resulting in a set of corresponding feature positions . We then apply RANSAC to solve the minimization problem
| (5) |
while simultaneously rejecting outlier correspondences.
3. VALIDATION
The registration method was evaluated on 3D transthoracic echocardiographic studies of 16 clinical cases of patients with aortic insufficiency. Each case had two acquisitions showing the left and right ventricles in different sectors with varying degree of overlap, both acquired from an apical position. All images were recorded on a Vivid E9 scanner using a 4V-D transducer (GE Vingmed Ultrasound AS, Horten, Norway). Each sequence contained a single heart cycle acquired using multi-beat during breath hold, containing on average 38 frames.
The Mitral Valve (MV) and Aortic Valve (AV) opening and closing events were manually tagged for each recording by an experienced cardiologist. Anatomical landmarks were identified in the MV opening and MV closing frames. These included the AV, MV and Tricuspid Valve (TV) center points, and the AV-MV junction.
4. RESULTS
4.1 Temporal alignment
The signed temporal alignment error between the key cardiac events were (mean ± SD) 4.9 ± 18.2 ms for AV opening, −2.0 ± 24.9 ms for AV closing and −5.9 ± 44.0 ms for MV opening. The corresponding absolute errors were 14.4 ± 11.6 ms, 18.6 ± 16.0 ms, and 34.6 ± 26.4 ms. For reference, the inter-frame time was 29 ± 5.1 ms.
4.2 Spatio-temporal alignment
The average euclidean distance between corresponding manually identified anatomical landmarks under the automatic registration was 4.3 ± 1.2 mm, compared to an average distance of 2.9 ± 0.7 mm with a Procrustes alignment between all manual landmarks.
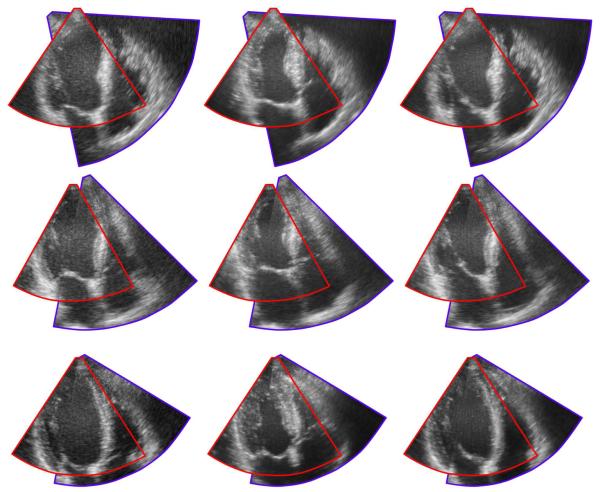
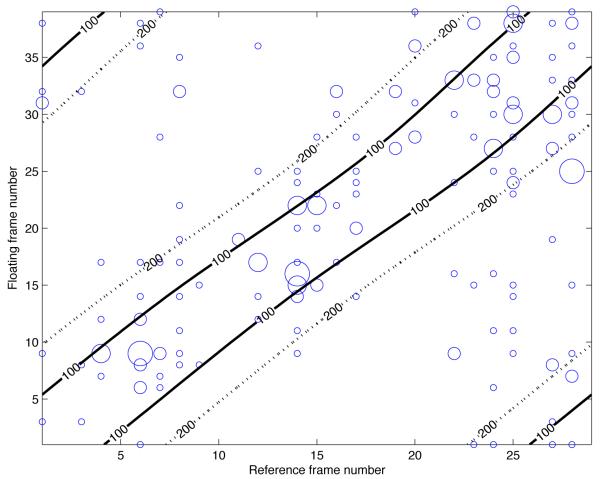
Figure 1(a) shows an example of the NCC curves before and after the temporal alignment, as well as the resulting temporal transform. Figure 1(b) illustrates the densities of 3D SIFT correspondences across the cardiac cycle. It is clear that, although no assumption is made on the temporal alignment, correspondences are more frequent in temporally aligned frames. Finally, examples of the resulting spatio-temporal alignment are shown in figure 3.
Figure 3.
Resulting rigid registration for three sequence pairs across the cardiac cycle. Each row shows three frames from a single case throughout the cardiac cycle.
5. DISCUSSION
In this paper, a spatio-temporal registration method for 3D cardiac image sequences has been presented and evaluated on ultrasound images. The method performed close to manual registration in both time and space, while requiring no manual user input.
The temporally aligned valve events were all close to the ground truth, and within clinically acceptable values. The temporal errors were noticeably larger for the MV opening event compared to AV opening and closing, which is expected as MV opening is the last valvular event in the cardiac cycle and thus furthest from the ECG-gated first frame. For the spatial alignment, the aligned anatomical landmarks were close to the ground truth, and comparable to values reported by others for similar feature-based ultrasound to MRI registration [11].
Our experiments indicate that even with near perfect temporal alignment, a rigid transform between 3D SIFT feature correspondences of temporally aligned frame pairs was not robust in all cases. However, by combining all correspondences across the cardiac cycle in a time-independent manner and utilizing the assumption that inter-frame deformations are negligible, the registration was highly robust and accurate in all cases, despite the deformable contraction of the heart. This usage of correspondences can be justified by the following considerations: Firstly, the extracted features inherently contain some temporal information, as feature scale and appearances tend to fade in and out of existence at certain parts of the cardiac cycle. This can be seen in fig. 2, as the majority of the feature correspondences are found on temporally aligned frames. Secondly, although some spurious matches between unrelated points in the cardiac cycle will arise, they will bear no spatial consistency. Furthermore, as the heart’s motion is cyclical, feature correspondences between deformed points during contraction will tend to be canceled out when the heart relaxes. Finally, by employing RANSAC and least squares optimization, we are estimating a rigid transform in a very robust manner.
Figure 2.
Correspondence density of feature matching between all reference and floating frames in one case. The size of each circle is proportional to the number of correspondences for a single frame pair. The black lines are contour lines of the time displacement, in milliseconds, after temporal alignment, illustrating the deformable temporal alignment.
One of the strengths of the presented method is that it requires no prior information on the spatial relationship between the acquisitions. This means that the method could be used for registering different views, such as apical, parasternal or subcostal, allowing the sonographer maximum freedom to find the best acoustic window.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the Research Council of Norway and the Norwegian Research School in Medical Imaging for their research grants, as well as the support of the Center for Cardiological Innovation. Research reported in this publication was also supported by the National Institutes of Health under award numbers P41EB015898 and R01CA138419.
References
- [1].Ledesma-Carbayo M, Kybic J, Desco M, Santos A, Suhling M, Hunziker P, Unser M. Spatio-temporal nonrigid registration for ultrasound cardiac motion estimation. Medical Imaging, IEEE Transactions on. 2005 Sep;24:1113–1126. doi: 10.1109/TMI.2005.852050. [DOI] [PubMed] [Google Scholar]
- [2].Elen A, Choi HF, Loeckx D, Gao H, Claus P, Suetens P, Maes F, D’hooge J. Three-dimensional cardiac strain estimation using spatio-temporal elastic registration of ultrasound images: A feasibility study. Medical Imaging, IEEE Transactions on. 2008 Nov;27:1580–1591. doi: 10.1109/TMI.2008.2004420. [DOI] [PubMed] [Google Scholar]
- [3].Kiss G, Barbosa D, Hristova K, Crosby J, Orderud F, Claus P, Amundsen B, Loeckx D, D’hooge J, Torp H. [Ultrasonics Symposium (IUS), 2009 IEEE International] Sep, 2009. Assessment of regional myocardial function using 3d cardiac strain estimation: comparison against conventional echocardiographic assessment; pp. 507–510. [Google Scholar]
- [4].De Craene M, Piella G, Camara O, Duchateau N, Silva E, Doltra A, D’hooge J, Brugada J, Sitges M, Frangi A. Temporal diffeomorphic free-form deformation: application to motion and strain estimation from 3D echocardiography. Medical image analysis. 2012 Feb.16:427–50. doi: 10.1016/j.media.2011.10.006. [DOI] [PubMed] [Google Scholar]
- [5].Danudibroto A, Gerard O, Alessandrini M, Mirea O, D’hooge J, Samset E. [Functional Imaging and Modeling of the Heart], Lecture Notes in Computer Science. Vol. 9126. Springer International Publishing; 2015. 3d farnebäck optic flow for extended field of view of echocardiography; pp. 129–136. [Google Scholar]
- [6].Ni D, Qu Y, Yang X, Chui Y, Wong T-T, Ho S, Heng P. Volumetric ultrasound panorama based on 3d sift. In: Metaxas D, Axel L, Fichtinger G, Székely G, editors. [Medical Image Computing and Computer-Assisted Intervention - MICCAI 2008] Lecture Notes in Computer Science. Vol. 5242. Springer Berlin Heidelberg; 2008. pp. 52–60. [DOI] [PubMed] [Google Scholar]
- [7].Mulder HW, van Stralen M, van der Zwaan HB, Leung KYE, Bosch JG, Pluim JPW. Multiframe registration of real-time three-dimensional echocardiography time series. Journal of Medical Imaging. 2014;1(1):014004. doi: 10.1117/1.JMI.1.1.014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grau V, Becher H, Noble J. Registration of multiview real-time 3-d echocardiographic sequences. Medical Imaging, IEEE Transactions on. 2007 Sep;26:1154–1165. doi: 10.1109/TMI.2007.903568. [DOI] [PubMed] [Google Scholar]
- [9].Schneider RJ, Perrin DP, Vasilyev NV, Marx GR, Del Nido PJ, Howe RD. Real-time image-based rigid registration of three-dimensional ultrasound. Medical image analysis. 2012 Feb.16:402–14. doi: 10.1016/j.media.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Perperidis D, Mohiaddin RH, Rueckert D. Spatio-temporal free-form registration of cardiac mr image sequences. Medical Image Analysis. 2005;9(5):441–456. doi: 10.1016/j.media.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [11].Zhang W, Noble J, Brady J. Spatio-temporal registration of real time 3d ultrasound to cardiovascular mr sequences. In: Ayache N, Ourselin S, Maeder A, editors. [Medical Image Computing and Computer-Assisted Intervention - MICCAI 2007] Lecture Notes in Computer Science. Vol. 4791. Springer Berlin Heidelberg; 2007. pp. 343–350. [DOI] [PubMed] [Google Scholar]
- [12].Valsangiacomo Buechel ER, Mertens LL. Imaging the right heart: the use of integrated multimodality imaging. European Heart Journal. 2012;33(8):949–960. doi: 10.1093/eurheartj/ehr490. [DOI] [PubMed] [Google Scholar]
- [13].Toews M, Wells WM. Efficient and robust model-to-image alignment using 3d scale-invariant features. Medical image analysis. 2013;17(3):271–282. doi: 10.1016/j.media.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]