ABSTRACT
The development of new phenotypes is key to the commercial development of the main floricultural species and cultivars. Important new phenotypes include features such as multiple-flowers, color variations, increased flower size, new petal shapes, variegation and distinctive petal margin colourations. Although their commercial use is not yet common, the transgenic technologies provide a potentially rapid means of generating interesting new phenotypes. In this report, we construct 5 vectors which we expected to change the color of the flower anthocyanins, from purple to blue, regulating vacuolar pH. When these constructs were transformed into purple torenia, we unexpectedly recovered some genotypes having slightly margined petals. These transgenic lines expressed a chimeric repressor of the petunia PhPH4 gene under the control of Cauliflower mosaic virus 35 S RNA promoter. PhPH4 is an R2R3-type MYB transcription factor. The transgenic lines lacked pigmentation in the petal margin cells both on the adaxial and abaxial surfaces. Expressions of Flavanone 3-hydroxylase (F3H), Flavonoid 3′-hydroxylase (F3′H) and Flavonoid 3′5′-hydroxylase (F3′5′H) genes were reduced in the margins of these transgenic lines, suggesting an inhibitory effect of PhPH4 repressor on anthocyanin synthesis.
KEYWORDS: Anthocyanin, chimeric repressor, DNA extraction, MYB transcription factor, margined flower, PH4, torenia
Abbreviations
- ANS
Anthocyanin synthase
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- DFR
Dihydroflavonol 4-reductase
- F3H
Flavanone 3-hydroxylase
- F3′H
Flavonoid 3′-hydroxylase
- F3′5′H
Flavonoid 3′5′-hydroxylase
- 3GT
UDP glucose flavonoid 3-O-glucosyltransferase
- HPT
Hygromycin B phosphotransferase
- HSP
Heat-shock protein
- MYB
Myeloblastosis
- RD
repression domain
- TCP
Teosinte branched 1, Cycloidea, and PCF
Introduction
Our research institute is using a number of strategies to modify flower phenotype in torenia (Torenia fournieri Lind. ex Fourn. and related species). As well as being a suitable model flower for transformation, torenia is also popular in summertime as a domestic pot plant. The first round of trials with torenia included modification of the expressions of genes associated with anthocyanin synthesis. The purple anthocyanin of the cultivar ‘Crown Violet’ becomes reddish when expression of the Chalcone synthase (CHS) gene is repressed by an antisense construct, whereas the anthocyanin becomes bluish when expression of the Dihydroflavonol 4-reductase (DFR) gene is repressed by an antisense construct.1,2 These transgenic flowers with altered colouration were also subjected to ion beam treatment to induce additional mutations and thus broaden the spectrum of phenotypes.3
As well as altering flower color, a new strategy was tested which modified flower shape and pigmentation pattern. ‘Chimeric repressor’ is a synthetic gene sequence, combining transcription factors and a small peptide called ‘repression domain’ (RD). Attachment of RD changes a transcription activator into a transcription repressor. As the repressor (or ‘chimeric repressor’) inhibits functions of endogenous homologues, chimeric repressor acts as a dominant-negative genetic factor.4 AtTCP3 repressor (TCP: Teosinte branched 1, Cycloidea, and PCF) derived from the TCP3 gene of Arabidopsis (Arabidopsis thaliana (L.) Heynh.) is a typical chimeric repressor which has been used previously to modify flower phenotype in torenia. AtTCP3 repressor reduces petal pigmentation, generates undulated petals with rough edges, and shortens stems. AtTCP3 repressor also changes torenia leaves from dentate to crenate. Leaves are somewhat undulated in transformants.5,6 Many other chimeric repressors were also tested by bulk transformation. In a way similar to AtTCP3 repressor, other repressors also have pleiotropic effects on flower and leaf phenotypes, including the generation of bizarre flowers.7
In this study with torenia, we sought new constructs that might affect vacuolar pH and so change flower color - especially toward blue. Against expectation, anthocyanin color did not change to blue with these constructs. Instead, one of these generated flowers having petals with pale margins. Possible reasons why this construct generated such margins are examined and discussed.
Results
Five genes affecting anthocyanin color
There is a large body of research on the genes affecting anthocyanin color in flowers, including antisense CHS and antisense DFR. Fig. 1A shows photographs of petunia and morning glory flowers. PhAN1, PhAN2 and PhAN11 are genes controlling both pigmentation and vacuolar pH in petunia petals.8,9,10 Petunia also has a series of PhPH genes regulating vacuolar pH in the petals.11,12,13 Because anthocyanins are accumulated in the vacuole and their colors change with pH (reddish at lower pH and bluish at higher pH), the PhPH genes also regulate petal color. PhAN2 and PhPH4 are among these genes and both are MYB (Myeloblastosis) family transcription factors. Vacuolar pH is elevated by any dysfunction in these genes. In morning glory, a more direct method for increasing vacuolar pH (to make anthocyanins bluish) is by expression of Na+/H+ antiporters. A high vacuolar pH is responsible for the blue color of European morning glory,14 and a Na+/H+ antiporter gene InNHX1 has been identified from a mutant of blue Japanese morning glory, being responsible for vacuolar alkalisation.15 Japanese morning glory also has a homolog InNHX2, whereas European morning glory has a counterpart ItNHX1.16,17 Expression of these NHX genes in petals is expected to increase vacuolar pH by pumping H+ ions out of the vacuole. Therefore, we prepared 5 constructs in torenia to express 3 NHX genes and chimeric repressors of PhAN2 and PhPH4 under the control of Cauliflower mosaic virus 35 S RNA promoter (Fig. 1B, C). Repression domains were attached to the 3′ termini of cloned PhAN2 and PhPH4 sequences to generate chimeric repressors.
Figure 1.
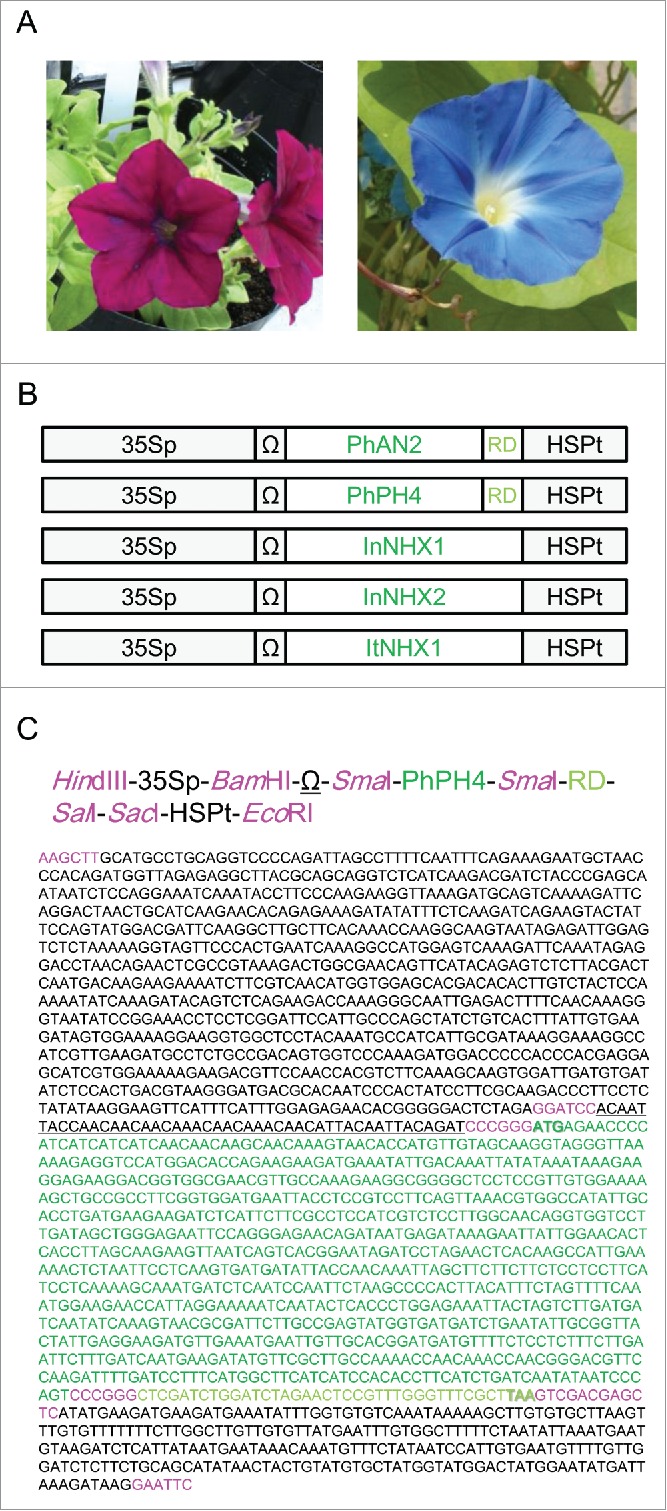
Genetic constructs for transformation. (A) Petunia (left) and morning glory (right). (B) Illustration of the constructs, not to scale. 35Sp, Cauliflower mosaic virus 35 S RNA promoter; Ω, translation enhancer sequence from Tobacco mosaic virus; RD, repression domain (SRDX sequence); HSPt, HSP terminator. (C) Precise sequence information for a representative construct (PhPH4-RD). Sequence profiles are written, up to DNA sequence. The same colors and underlining represent the same parts of the construct. All restriction sites are colored magenta.
Development of a high-throughput method for DNA extraction
During the transformation processes, we developed a high-throughput method for DNA extraction from torenia. In contrast to the more popular model plant, Arabidopsis,18,19 torenia research has previously lacked such a high-throughput method. With this newly developed method, T-DNA insertion and the construct in T-DNA can be determined by PCR immediately. Fig. 2 illustrates the procedure for DNA extraction. Leaf discs (typically 5 × 5 mm) are crushed in 200 µL of extraction buffer (we call this ‘SDSR buffer’: Tris-HCl pH 7.5 (200 mM), EDTA (25 mM), NaCl (250 mM), SDS (sodium dodecyl sulfate, 0.5%), RNase A (5 µg mL−1)) in a 1.5-mL plastic tube with a plastic rod, then heated to 55°C for 5 min. This stock solution can be preserved at −20°C. Stock solutions are diluted 10 times with sterilised, purified water. A volume of 1 µL of the diluted solution is used for one PCR reaction (20–50 µL). PCR reactions were successful with a high-fidelity polymerase (KOD Plus Neo, Toyobo, Osaka, Japan) but were not successful with a standard polymerase (Ex-Taq HS, Takara, Kyoto, Japan). Leaf discs taken from potted plants were also found to be better than those from sterilized plant culture when around 3-kb fragments of T-DNA sequences were amplified. Both types of leaf disc served well when fragments of around 500-bp were amplified.
Figure 2.
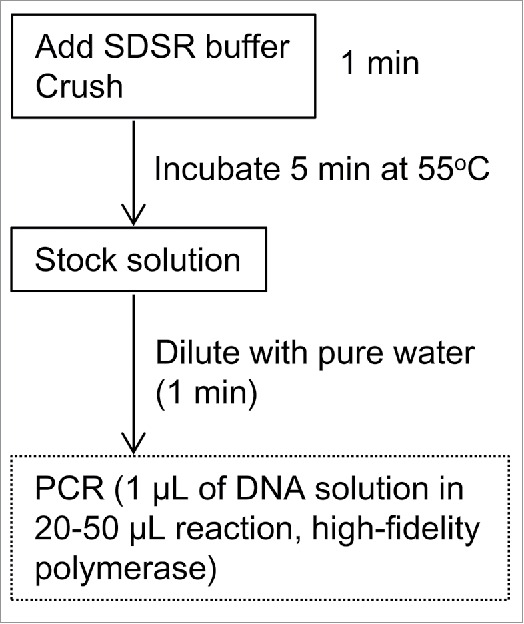
High-throughput DNA extraction from torenia. Illustration of the extraction procedure.
PCR analysis was performed to determine T-DNA insertion in isolated lines. Four primers were combined to amplify 2 target sequences at a time. A 336-bp fragment of Hygromycin B phosphotransferase (HPT) gene was amplified to detect T-DNA inserts. A 573-bp fragment of endogenous DFR gene promoter sequence was amplified as an indicator of the success of the PCR reaction.
Generation of transgenic torenia lines
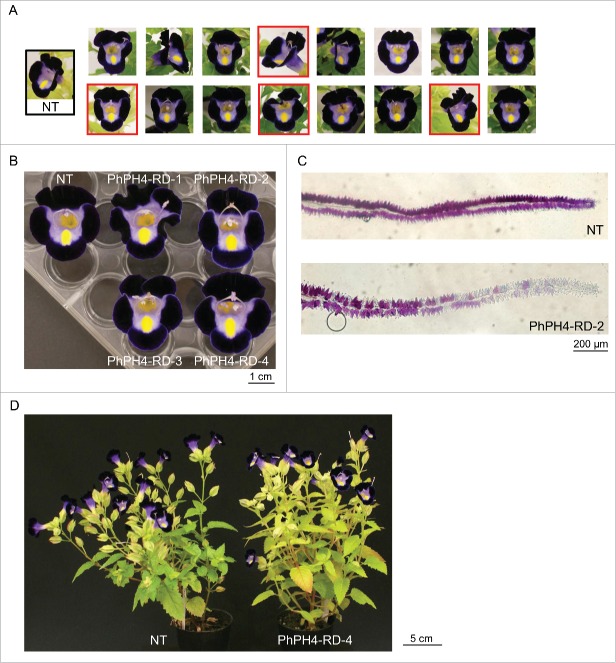
Five vectors were transformed as a bulk (mixture), into the torenia cultivar ‘Crown Violet’. Mixtures of equal concentrations of plasmids were transformed into agrobacterium. After growing transformed colonies of agrobacterium, colonies were combined in sterile water and inoculated onto leaf discs of torenia. After inoculation on co-existence media and incubations on selection/regeneration media, transgenic shoots were grown in plastic containers and later transferred to pots. Flowers where observed after plants had set at least 5 flowers. We observed 16 independent transgenic lines (Fig. 3A). Contrary to expectation, none of the lines set blue flowers. Flower colors were quite similar to those of non-transgenic (NT) plants. Our constructs do not seem to be able to generate blue flowers. In Fig. 3A, 4 lines (marked with red frames) shared the same distinct phenotypic change. This new phenotype is more clearly shown in Fig. 3B. The petals of these lines have narrow, pale margins. Genomic PCR analyses revealed T-DNA insertion of PhPH4 repressor (PhPH4-RD) in all 4 of these lines. We named these lines PhPH4-RD-1, PhPH4-RD-2, PhPH4-RD-3 and PhPH4-RD-4. A new composition of agar was developed, modified for slicing torenia petals. Using this, cross-sections of the ventral limbs were prepared from NT and PhPH4-RD-2, and were observed under a light microscope (Fig. 3C). Anthocyanin accumulation was observed in both adaxial and abaxial epidermal cells in NT. The adaxial epidermal cells (upside in Fig. 3C) were found to accumulate pigment more densely than the abaxial epidermal cells. All epidermal cells of the NT limbs accumulated anthocyanins, including those along the very edge. In contrast, the epidermal cells along the edge of the PhPH4-RD-2 limbs lacked anthocyanins both in the adaxial and the abaxial surfaces. Fig. 3D shows whole plants of NT and PhPH4-RD-4. The chimeric repressor of PhPH4 does not have any further distinguishable effects on plant structure or appearance.
Figure 3.
Phenotypes of transgenic lines. (A) Flowers of independent transformants. NT, non-transgenic line. Lines having pale margins are framed with red squares. (B) PhPH4-RD flowers photographed on a black background. (C) Cross-section of NT and PhPH4-RD-2 limbs. The adaxial petal surfaces are orientated upwards. (D) Whole plants of NT and PhPH4-RD-4.
Expression analysis of genes involved in anthocyanin synthesis
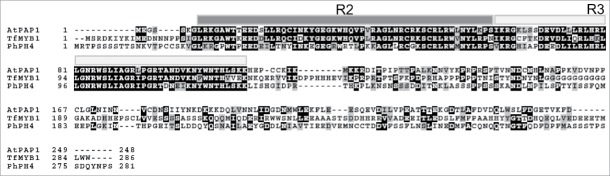
How does the PhPH4 repressor function in torenia? The amino acid sequence of PhPH4 possesses R2R3-type MYB domain,12 harbouring homology to the MYB transcription factors regulating anthocyanin accumulations such as AtPAP1 in Arabidopsis and TfMYB1 in torenia.20 Fig. 4 shows the amino acid alignment in these R2R3-type MYB proteins.
Figure 4.
Sequences of R2R3 MYB transcription factors. R2 and R3 domains26 are indicated on the amino acid sequences with gray bars.
AtPAP1 and TfMYB1 positively regulate the expression of genes for the anthocyanin synthesis pathway which could be considered in 3 parts (Fig. 5A): The first step transforms coumaroyl-CoA and malonyl-CoA into dihydrokaempferol with Chalcone synthase (CHS), Chalcone isomerase (CHI) and Flavanone 3-hydroxylase (F3H). In the second step, dihydrokaempferol is transformed into other dihydroflavonols, dihydroquercetin and dihydromyricetin, with Flavonoid 3′-hydroxylase (F3′H) and Flavonoid 3′5′-hydroxylase (F3′5′H). Dihydroflavonols are then catalyzed into anthocyanins by Dihydroflavonol 4-reductase (DFR) and Anthocyanin synthase (ANS).21 These anthocyanins then undergo further modification with enzymes such as UDP glucose flavonoid 3-O-glucosyltransferase (3GT). Torenia genes corresponding to the genes involved in anthocyanin synthesis above have been cloned and are the subject of an earlier report.22
Figure 5.
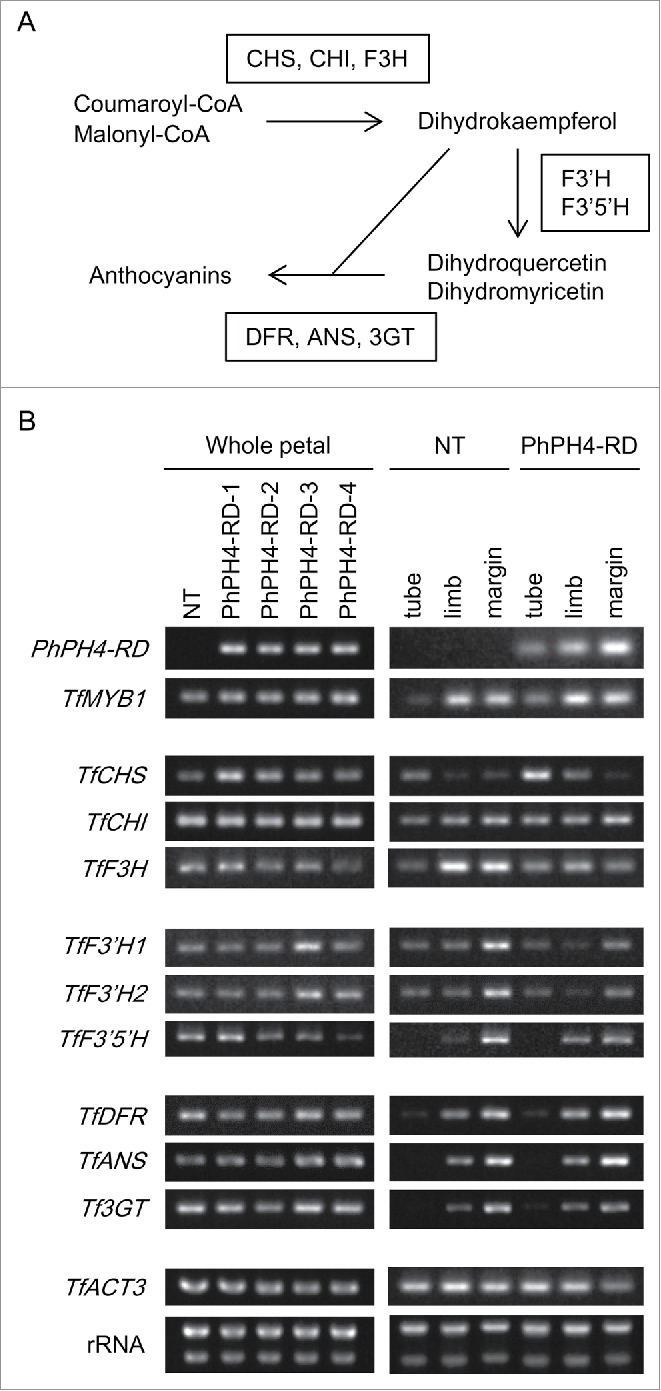
Expression analysis of genes involved in anthocyanin synthesis. (A) Simplified illustration of the pathway of anthocyanin synthesis. (B) Semi-quantitative RT-PCR analysis. RNA samples were examined from whole petals (left) and from parts of petals (tube, limb, petal margin: right). rRNA, rRNA bands observed after electrophoresis of extracted RNA.
Expressions of genes involved in anthocyanin synthesis were compared between NT and PhPH4-RD lines by semi-quantitative RT-PCR analysis (Fig. 5B). Expression levels of PhPH4-RD transgene were similar between all 4 PhPH4-RD lines in the whole petals, consistent with similar phenotypes of these lines. TfCHS expression seems to be up-regulated in all PhPH4-RD lines compared with NT in whole petals. Some other genes are also up-/down-regulated in some specific lines but, in the whole petals, no uniform up-/down-regulation of genes in PhPH4-RD lines was observed except for TfCHS. Clearer differences in gene expression were observed when flower tubes, limbs and petal margins were compared. Expression of PhPH4-RD was stronger in limbs than in tubes, and it was elevated even in the margin in the PhPH4-RD line. TfCHS expression in the tube was higher in the PhPH4-RD line than in NT, whereas TfCHS expression was much lower in the limb and margin both in the NT and PhPH4-RD lines. The expression of no other genes was significantly different in the tube between the NT and PhPH4-RD lines. Interestingly, genes in the ‘second step’ and ‘third step’ of anthocyanin synthesis were more highly expressed in the petal margins than in the whole limbs of NT flowers. The third-step genes maintained high levels of expression, similar to NT, in the petal margins in the PhPH4-RD line, whereas the expressions of the second-step genes were downregulated in PhPH4-RD line compared with NT in the margins. In addition to expression of the second-step genes expression of TfF3H, one of the first-step genes, was also down-regulated in PhPH4-RD in the limb and petal margins.
Discussion
Significance of vacuolar pH modification for petal color modification
The original goal of this report was to generate blue flowers in torenia with a simple genetic modification. Many floricultural species accumulate red, magenta, purple or, less frequently, violet anthocyanin in the vacuoles of the petal cells. Species accumulating completely blue anthocyanins are quite rare (e.g., morning glory, hydrangea, delphinium, blue poppy). Considering the importance of color variation for sales of ornamental flowers, the generation of truly blue flowers is an important goal for many flower breeders. Modifications of anthocyanin structure by genetic transformation have been successful in generating bluer flowers from reddish ones. However, these were obtained at massive experimental effort and cost. Multiple genes had to be introduced by transformation in some cases. In addition, many genes and promoters must usually be tested to find a proper construct which successfully catalyzes anthocyanin modification.23,24 Even after all this effort, anthocyanin color is never truly blue, but is more usually purple or violet.
Simpler strategies for modifying anthocyanin color have already been presented in an earlier study, as mentioned in the introduction. Aida et al.1,2 introduced antisense sequences of CHS or DFR in torenia. As well as reducing the density of pigmentation, antisense CHS changed the color (hue) of torenia from purple to reddish purple, whereas antisense DFR changed the color from purple to violet. These color changes were caused by an altered balance of the concentrations of anthocyanins and flavones (co-pigments). Anthocyanin can be made bluer in this simple manner, but this strategy is not universally effective in other floricultural species. Also, the colors of the transgenic flowers are not really blue.
Some cultivars of morning glory set completely blue flowers (Fig. 1A) and the InNHX1 gene is necessary for this color. With a mutation in this gene, flower color of blue morning glory can change to purple.15 The InNHX1 gene is believed to transport H+ ions from the vacuolar lumen to the cytosol, and this increases vacuolar pH during flower opening. The anthocyanins of purple flowers of other species also undergo the same bluish color shift under neutral and weakly-alkaline pHs. A truly blue pigment can be generated by crushing purple torenia petals and adding the resulting paste to water. This indicates that the pigments in purple torenia flowers can be modified to blue, merely by raising the pH to about 7. In other words, anthocyanin color is a pH indicator. Unfortunately, flower color change was not observed in this study, suggesting that the 5 constructs did not raise vacuolar pH significantly. The pH values of petal exudates from the 3 transgenic lines examined (2 of them possessing the PhPH4-RD construct and the other the InNHX2 construct) were substantially the same as in NT (i.e. pH 5.0 to 5.4). Further effort is required to render these genes functional in torenia petals.
Genetic factors inducing margined flowers
To our knowledge there has been only one report on a genetic factor inducing margined flowers. Morita et al.25 examined a margined (picotee) flower of petunia. siRNA was generated from a tandem repeat of 2 CHS genes in this flower. For reasons unknown, gene silencing seems to occur due to siRNA accumulation only in the margins of petals. Although the PhPH4-RD lines generated in this study also induced color change in the petal margins, they modified gene expression by a chimeric repressor (artificial transcription repressor), not by gene silencing. The target genes (downregulated genes) of the PhPH4 repressor were 3 hydroxylases, F3H, F3′H and F3′5′H, and not CHS. Our study thus revealed a new type of genetic factor inducing margined flowers. This is also the first report of a transgenic modification being successful in inducing margined flowers.
PhPH4 is an R2R3-type MYB transcription factor. R2R3 is a major family of MYB in plants. The number of MYB genes in the torenia genome is not clear at this stage, but there are certainly more than 100 MYB genes in the Arabidopsis genome.26 The original function of PhPH4 in petunia is vacuolar pH modification, whereas PhPH4-RD down-regulates anthocyanin synthesis in torenia, and does not seem to affect the pH of the whole petal. This result is consistent with its homology to AtPAP1 and TfMYB1, which regulate anthocyanin accumulation in Arabidopsis and torenia.27,20 The AtPAP1 repressor also cancels sucrose-responsive accumulation of anthocyanins in Arabidopsis.4 The reasons for the differential functions of PhPH4 in petunia and torenia are not clear. For example, the torenia genome may lack some unknown endogenous target gene for PhPH4, involved in regulation of vacuolar pH. Also, PhPH4 may partially regulate other genes involved in anthocyanin synthesis in place of the actual target genes. Regulation of the expression of genes involved in anthocyanin accumulation is not restricted to R2R3-type of MYB. The chimeric repressor of AtRL2, a single-type MYB gene from Arabidopsis, seems to strongly down-regulate anthocyanin accumulation in torenia petals in an earlier report.7 A contrastingly restricted activity of PhPH4-RD in downregulation of genes involved in anthocyanin synthesis, could lead to margined flowers.
If the function of PhPH4-RD is restricted, it is not yet clear why anthocyanin synthesis is stopped only in the marginal cells. Because PhPH4-RD expression was stronger in the petal margins, than in the tubes and whole limbs, a possible explanation may be that PhPH4-RD inhibits the transcription of target genes only when it is strongly expressed - that is, only in the marginal cells. Another plausible idea is that the factor leading to the induction of pale margins in the PhPH4-RD lines has to do with the nature of the torenia flower. Petal margin cells seem to be more ‘susceptible’ to genetic change (and so tend to lose pigmentation) than the more central cells. Pale margins appear in transgenic torenia flowers harbouring T-DNA insertions of the chimeric repressors of AtAGL6 and AtGNL genes, together with other phenotypic alterations.7 Although transgene expressions and reproducibility of the phenotype were not examined, these chimeric repressors could be said to induce pale margins. Margined flowers also appear in the bulk of ion-beam induced torenia mutants.3 These observations would suggest that marginal cells of torenia petals have a more fragile anthocyanin synthesis machinery and so more easily lose their ability to synthesize anthocyanin after genetic perturbation – the central region of the limb being not so susceptible to genetic perturbation.
Materials and methods
Plant materials
Petunia (Petunia x hybrida, an unnamed cultivar setting red-purple flowers, Fig. 1A) and morning glory (Ipomoea tricolor Cav. ‘Heavenly Blue’, Fig. 1A) were used for cDNA isolation. A sterilised culture of the torenia cultivar Crown Violet was incubated in transparent plastic containers (7×7×10 cm) at 25°C under fluorescent light (PPFD = 150 µmol m−2 s−1). Media contained half-strength Murashige-Skoog salt and vitamin, 3% sucrose, 50 mg L−1 shiomarin and was solidified with 0.2% gellam gum. After incubation for at least 2 months, plants were transferred to soil, in pots, in a naturally lit greenhouse.
Transformation
The transformation procedure followed a previous report.28 Briefly, leaf discs prepared from the sterilised culture were inoculated with agrobacterium, incubated in the dark on co-existence media for one week, and transferred to selection/regeneration media containing 150 mg L−1 hygromycin and 300 mg L−1 carbenicillin (in this study, the hygromycin concentration was higher than that usually used for torenia viz. 20 mg L−1).
cDNA isolation
RNA was extracted with the RNeasy Plant Mini-Kit (Qiagen K. K., Tokyo, Japan), from the petals of petunia and morning glory. After reverse transcription with oligo-(dT)20 primer using ReverTra Ace α (Toyobo, Osaka, Japan), partial cDNA sequences including whole coding sequences of PhAN2 (petunia), PhPH4 (petunia) and ItNHX1 (morning glory) were amplified by PCR with KOD Plus Neo (Toyobo, Osaka, Japan). Amplified DNA sequences were TA cloned and sequenced. The InNHX1 and InNHX2 clones (derived from Japanese morning glory, Ipomoea nil) were provided by the National Institute for Basic Biology. Primer sequences are listed in Table 1. Compared with the original database sequences (AF146702.1, AY973324.1, AB054979.1), the cloned PhAN2 sequence had 8 nucleotide substitutions within the coding sequence, leading to 8 amino acid substitutions in the encoded protein. Similarly, PhPH4 had 3 nucleotide substitutions within the coding sequences, and ItNHX1 had 11, but these mutations did not result in amino acid substitutions.
Table 1.
Primers for genomic PCR, cloning and sub-cloning.
| Primer name* | Sequence | Target gene |
|---|---|---|
| [Genomic PCR] | ||
| HPT-F1 | 5′-ATTCCGGAAGTGCTTGACAT-3′ | HPT (T-DNA) |
| HPT-R3 | 5′-CAGTCCTCGGAAAGCATCAG-3′ | |
| PDFR-F1 | 5′-TGGCGTTCCTTAAACAGCTT-3′ | TfDFR promoter |
| PDFR-R2 | 5′-TGAACACCGTTGCTTTTTGA-3′ | |
| 35S-F1 | 5′-CGCACAATCCCACTATCCTT-3′ | 35 S promoter |
| PhAN2-R1 | 5′-GGTCCATGCACCTTTCCTTA-3′ | PhAN2 |
| PhPH4-R1 | 5′-TGATGATGATGGGGTTCTCA-3′ | PhPH4 |
| InItNHX1-R1 | 5′-CCGAATTTTGGAGCAAAGAA-3′ | InNHX1, ItNHX1 |
| InNHX2-R1 | 5′-AGTGTCCCAACGTCAACTCC-3′ | InNHX2 |
| [Cloning] | ||
| AN2-UP-F3 | 5′-CTTTCTTTGTCCTTTAGTTGTCAGTTGCAGTGAG-3′ | PhAN2 |
| AN2-UP-R1 | 5′-AATTTCAAATGTCCAACGATTTCAACTGTAGTGTTT-3′ | |
| PH4-UP-F1 | 5′-CTCCCCTCCCAAATTTAGTACTATGACTTTATC-3′ | PhPH4 |
| PH4-UP-R3 | 5′-AACCCTCACATACATATATATATAGCTACAACATCA-3′ | |
| InNHX1-UP-F1 | 5′-CCCCACATCTCACCTTTCAAGTGATTTGTATGTTTT-3′ | ItNHX1 |
| InNHX1-UP-R1 | 5′-TGAGAAGACTTTCACCCAGCCCATTTCATAAGAG-3′ | |
| [Subcloning] | ||
| AN2-F1-SmaI | 5′-AATAATCCCGGGATGAGTACTTCTAATGCATCAACATCAGGA-3′ | PhAN2 |
| AN2-R1-SmaI | 5′-AATAATCCCGGGACTAACTAAATCCCATATGTCATCAATATCAACTG-3′ | |
| PhPH4-F1-SmaI | 5′-AATAATCCCGGGATGAGAACCCCATCATCATCATCAACAA-3′ | PhPH4 |
| PhPH4-R1-SmaI | 5′-AATAATCCCGGGACTGGGATTATATTGATCAGATGAAGGTGTGGA-3′ | |
| InNHX1-F1-SmaI | 5′-AATAATCCCGGGATGGCGTTCGGGTTGTCTTCTT-3′ | InNHX1 |
| InNHX1-R1-SalI | 5′-AATAATGTCGACTCATCTAGGGCTCTGCTCAACTGG-3′ | |
| InNHX2-F1-DraI | 5′-AATAATTTTAAAATGGGAGTTGACGTTGGGACAC-3′ | InNHX2 |
| InNHX2-R1-SalI | 5′-AATAATGTCGACTCACTGCCATTGTGGTTCATTCTG-3′ | |
| ItNHX1-F1-SmaI | 5′-AATAATCCCGGGATGGCGTTCGGATTGTCTTCTTT-3′ | ItNHX1 |
| ItNHX1-R1-SalI | 5′-AATAATGTCGACTCATCTAGGGCTCTGCTCAGCTG-3′ | |
| HSPt-F-SacI | 5′-AAAGAGCTCATATGAAGATGAAGATGAAA-3′ | HSP terminator |
| HSPt-R-EcoRI | 5′-AAAGAATTCCTTATCTTTAATCATATTCC-3′ |
‘F’ represents forward primer and ‘R’ represents reverse primer.
Construction of plasmids
For construction of the PhAN2-RD and PhPH4-RD vectors, coding sequences excluding stop codons were amplified from plasmids with restriction sites (SmaI sites) attached on both DNA ends by primers. Amplified fragments were TA cloned and sequenced. A new vector containing the terminator sequence of a heat shock protein (HSP) gene was prepared as follows. A DNA fragment containing the HSP terminator (HSPt)29 was amplified by PCR, with SacI and EcoRI sites attached to either end of the DNA. The amplified HSPt fragment was digested with SacI and EcoRI. The SacI-EcoRI portion of the p35SSRDXG vector30 was replaced with this fragment, to produce a pG35S-SRDX-HSPt vector. After digestion with SmaI, the coding sequences were introduced into pG35S-SRDX-HSPt. SRDX, a typical repression domain encoding 12 amino acid residues,4 was attached to the 3′ end of the coding sequences in the frame of these constructs. To construct the 3 NHX vectors, coding sequences including stop codons, were amplified with restriction sites (SmaI (or DraI for InNHX2) and SalI sites) attached by primers, TA cloned, digested with SmaI (or DraI) and SalI, and then introduced into pG35S-SRDX-HSPt. These five vectors were LR cloned into a binary vector pBCKH.31 Binary vectors were transformed into agrobacterium strain EHA105 by electroporation. The primer sequences used for sub-cloning are listed in Table 1.
Genomic PCR
Crude DNA was extracted with ‘SDSR buffer’ as described in the text. Constructs were separately amplified with a forward primer binding to the 35 S promoter sequence and specific primers binding to each coding sequence. The InNHX1 and ItNHX1 constructs were simultaneously amplified with the same primer. PCR conditions were the same both for the detection of genomic DNA and for the detection of the constructs: PCR was performed with KOD Plus Neo in 3-step cycles. The annealing temperature was 55°C, extension was for 1 min and the number of thermal cycles was 40. The other conditions followed the manufacturer's instructions. The primer sequences used for genomic PCR are listed in Table 1.
Cross-section of petals
The ventral flower limbs (limb, flat parts on flower tubes; ventral, the lower part) were excised. A newly-developed ‘petal slicing agar’ (28.4 g trehalose, 110 mg CaCl2-2H2O, 3 g agar and 300 µL Tween-20 per 150 mL of water) was melted in a microwave oven and cooled to approx. 60°C. After pouring the melted agar into small plastic dishes, the excised ventral limbs were embedded. The agar blocks were later sectioned (55 µm) parallel to the petal's longitudinal axes with an electric micro-slicer (DTK-1000, Dosaka EM Co., Ltd., Kyoto, Japan). Slices were observed with a light microscope (Provis AX70, Olympus, Tokyo, Japan).
Protein alignment
Amino acid sequences were aligned with ClustalW32 and shades supplemented with BoxShade.
Gene accessions
PhAN2 (AB982128), PhPH4 (AB982129), InNHX1 (AB033989.1), InNHX2 (AB194065.1), ItNHX1 (AB982130), TfMYB1 (AB719455.1), AtRL2 (NM_127736). The sequence of the TfDFR promoter will be reported elsewhere (Sasaki K et al.). Accessions of genes involved in anthocyanin synthesis and TfACT3 have been described previously.22
Gene expression analysis
RNA was extracted from whole petals using the RNeasy Plant Mini-Kit. Flower tubes, limbs and margins of limbs (approx. 1 mm of margin) were also gathered and their RNA extracted. The width of the margin samples (1 mm) was the least that could be prepared manually from the fragile petals. The ‘PhPH4-RD’ sample (Fig. 5B, right) derives from the PhPH4-RD-2 line for tube and limb, and this derives from a mixture of PhPH4-RD lines for margin. RNA samples were reverse-transcribed with ReverTra Ace α by oligo-(dT)20 primer. The resulting cDNA samples were diluted to 10% with water and used for PCR amplification. Volumes of 1 µL of the cDNA samples were used in total volumes of 20 µL for the PCR reactions with Ex-Taq HS (Takara, Kyoto, Japan). Primer sequences and PCR cycles in semi-quantitative RT-PCR analysis are presented in Table 2. The RT-PCR primers for TfANS, TfCHI, TfF3′H1, TfF3′H2, TfF3′5′H, Tf3GT and TfACT3 are from Sasaki et al.22 The other RT-PCR primers were designed in this study. TfCHS1 and TfCHS2 (cloned in Sasaki et al.22) shared nearly the same nucleotide sequences and seemed to be alleles. New primers were designed to amplify TfCHS1 and TfCHS2 simultaneously (this gene is here referred to as TfCHS).
Table 2.
Primers for semi-quantitative RT-PCR.
| Primer name* | Sequence | Target gene | PCR Cycles |
|---|---|---|---|
| PhPH4-RD-F2 | 5′-TTGCCGAGTATGGTGATGAT-3′ | PhPH4-RD | 30 |
| PhPH4-RD-R3 | 5′-CGAAACCCAAACGGAGTTCT-3′ | ||
| TfMYB1-F1 | 5′-ATCGGTGCAGAAAGAGTTGC-3′ | TfMYB1 | 35 |
| TfMYB1-R2 | 5′-CCACCACATGAGTGTTCCAA-3′ | ||
| TfANS-F | 5′-CGCCCTCACGTTCATCCTCCAC-3′ | TfANS | 30 |
| TfANS-R | 5′-CTCCACCCATCACTCAACACTC-3′ | ||
| TfCHI-F | 5′-TTCCTCGAAGTGTTCAAGAACG-3′ | TfCHI | 30 |
| TfCHI-R | 5′-TACAAATCCACGATCATTGACC-3′ | ||
| TfCHS-F1 | 5′-CTGAGCGAGTACGGGAACAT-3′ | TfCHS1, TfCHS2 | 30 |
| TfCHS-R3 | 5′-CACACACACACACACCACAAA-3′ | ||
| TfDFR-F1 | 5′-TGATGGGAATGGGATTTACC-3′ | TfDFR | 35 |
| TfDFR-R1 | 5′-CACGACATTTCACCATTTTCC-3′ | ||
| TfF3H-F3 | 5′-AGGCCTGTCTGAACATGGAC-3′ | TfF3H | 30 |
| TfF3H-R4 | 5′-GGGTACACGATAGCCTCTGG-3′ | ||
| TfF3′H1-F | 5′-GCGGGTCGTAGGATCTGCGCC-3′ | TfF3′H1 | 30 |
| TfF3′H-R | 5′-CAGCAGTACAAACTAGATTTATC-3′ | ||
| TfF3′H2-F | 5′-GTCAAGGGGAACGATTTCGAAG-3′ | TfF3′H2 | 30 |
| TfF3′H-R | 5′-CAGCAGTACAAACTAGATTTATC-3′ | ||
| TfF3′5′H-F | 5′-GAGAATGTTGGAGCAGCATGCG-3′ | TfF3′5′H | 30 |
| TfF3′5′H-R | 5′-GAGATAATTTATTTGATAGATTGG-3′ | ||
| Tf3GT-F | 5′-TCGAATGGTGCAAGATGTTTGG-3′ | Tf3GT | 35 |
| Tf3GT-R | 5′-GAAGACGTAGACATTTGACTCG-3′ | ||
| TfACT3-F | 5′-AAATACAGTGTTTGGATCGGAGGTTC-3′ | TfACT3 | 30 |
| TfACT3-R | 5′-GAATAGCACACAGAGAATAGCAAACC-3′ |
‘F’ represents forward primer and ‘R’ represents reverse primer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge valuable discussions with Takaaki Nishijima, Tomoya Niki and Atsushi Hoshino and the technical assistance of Yoshiko Kashiwagi. Norihiro Ohtsubo supported this study in a number of ways. The InNHX1 and InNHX2 clones are kind gifts from the National Institute for Basic Biology.
Funding
This work was supported by the Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (Japan).
Author contributions
Experiments were designed by IK and KS, and carried out by IK. The pG35S-SRDX-HSPt vector was constructed by KS. The manuscript was written by IK and checked by KS.
References
- 1.Aida R, Kishimoto S, Tanaka Y, Shibata M. Modification of flower color in torenia (Torenia fournieri Lind.) by genetic transformation. Plant Sci 2000; 153:33-42; PMID:11164576; http://dx.doi.org/ 10.1016/S0168-9452(99)00239-311164576 [DOI] [Google Scholar]
- 2.Aida R, Yoshida K, Kondo T, Kishimoto S, Shibata M. Copigmentation gives bluer flowers on transgenic torenia plants with the antisense dihydroflavonol-4-reductase gene. Plant Sci 2000; 160:49-56; PMID:11164576; http://dx.doi.org/ 10.1016/S0168-9452(00)00364-2 [DOI] [PubMed] [Google Scholar]
- 3.Sasaki K, Aida R, Niki T, Yamaguchi H, Narumi T, Nishijima T, Hayashi Y, Ryuto H, Fukunishi N, Abe T, Ohtsubo N. High-efficiency improvement of transgenic torenia flowers by ion beam irradiation. Plant Biotechnol 2008; 25:81-9; http://dx.doi.org/ 10.5511/plantbiotechnology.25.81 [DOI] [Google Scholar]
- 4.Hiratsu K, Matsui K, Koyama T, Ohme-takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 2003; 34:733-9; PMID:12787253; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01759.x [DOI] [PubMed] [Google Scholar]
- 5.Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007; 19:473-84; PMID:17307931; http://dx.doi.org/ 10.1105/tpc.106.044792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narumi T, Aida R, Koyama T, Yamaguchi H, Sasaki K, Shikata M, Nakayama M, Ohme-Takagi M, Ohtsubo N. Arabidopsis chimeric TCP3 repressor produces novel floral traits in Torenia fournieri and Chrysanthemum morifolium. Plant Biotechnol 2011; 28:131-40; http://dx.doi.org/ 10.5511/plantbiotechnology.11.0121a [DOI] [Google Scholar]
- 7.Shikata M, Narumi T, Yamaguchi H, Sasaki K, Aida R, Oshima Y, Takiguchi Y, Ohme-Takagi M, Mitsuda N, Ohtsubo N. Efficient production of novel floral traits in torenia by collective transformation with chimeric repressors of Arabidopsis transcription factors. Plant Biotechnol 2011; 28:189-99; http://dx.doi.org/ 10.5511/plantbiotechnology.10.1216a [DOI] [Google Scholar]
- 8.Spelt C, Quattrocchio F, Mol J, Koes R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 2002; 14:2121-35; PMID:12215510; http://dx.doi.org/ 10.1105/tpc.003772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 1999; 11:1433-44; PMID:10449578; http://dx.doi.org/ 10.1105/tpc.11.8.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 1997; 11:1422-34; PMID:9192870; http://dx.doi.org/ 10.1101/gad.11.11.1422 [DOI] [PubMed] [Google Scholar]
- 11.Faraco M, Spelt C, Bliek M, Verweij W, Hoshino A, Espen L, Prinsi B, Jaarsma R, Tarhan E, de Boer AH et al.Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep 2014; 6:32-43; PMID:24388746; http://dx.doi.org/ 10.1016/j.celrep.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 12.Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 2006; 18:1274-91; PMID:16603655; http://dx.doi.org/ 10.1105/tpc.105.034041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verweij W, Spelt C, Sansebastiano GPD, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat Cell Biol 2008; 10:1456-62; PMID:18997787; http://dx.doi.org/ 10.1038/ncb1805 [DOI] [PubMed] [Google Scholar]
- 14.Yoshida K, Kondo T, Okazaki Y, Katou K. Cause of blue petal colour. Nature 1995; 373:291; PMID:19521056; http://dx.doi.org/ 10.1038/373291a019521056 [DOI] [Google Scholar]
- 15.Fukada-Tanaka S, Inagaki Y, Yamaguchi T, Saito N, Iida S. Colour-enhancing protein in blue petals. Nature 2000; 407:581; PMID:11034195; http://dx.doi.org/ 10.1038/35036683 [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi T, Fukada-Tanaka S, Inagaki Y, Saito N, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Iida S. Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol 2001; 42:451-61; PMID:11382810; http://dx.doi.org/ 10.1093/pcp/pce080 [DOI] [PubMed] [Google Scholar]
- 17.Ohnishi M, Fukada-Tanaka S, Hoshino A, Takada J, Inagaki Y, Iida S. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol 2005; 46:259-267; PMID:15695437; http://dx.doi.org/ 10.1093/pcp/pci028 [DOI] [PubMed] [Google Scholar]
- 18.Berendzen K, Searle I, Ravenscroft D, Koncz C, Batschauer A, Coupland G, Somssich IE, ⇐lker B. A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods 2005; 1:4; PMID:16270938; http://dx.doi.org/ 10.1186/1746-4811-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasajima I, Ide Y, Ohkama-Ohtsu N, Hayashi H, Yoneyama T, Fujiwara T. A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol Biol Rep 2004; 22:49-52; http://dx.doi.org/ 10.1007/BF02773348 [DOI] [Google Scholar]
- 20.Nishijima T, Morita Y, Sasaki K, Nakayama M, Yamaguchi H, Ohtsubo N, Niki T, Niki T. A torenia (Torenia fournieri Lind. ex Fourn.) novel mutant ‘flecked’ produces variegated flowers by insertion of a DNA transposon into an R2R3-MYB gene. J Japan Soc Hort Sci 2013; 82:39-50; http://dx.doi.org/ 10.2503/jjshs1.82.39 [DOI] [Google Scholar]
- 21.Grotewold . The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 2006; 57:761-80; PMID:16669781; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105248 [DOI] [PubMed] [Google Scholar]
- 22.Sasaki K, Aida R, Yamaguchi H, Shikata M, Niki T, Nishijima T, Ohtsubo N. Functional divergence within class B MADS-box genes TfGLO and TfDEF in Torenia flurnieri Lind. Mol Genet Genomics 2010; 284:399-414; PMID:20872230; http://dx.doi.org/ 10.1007/s00438-010-0574-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A et al.. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 2007; 48:1589-600; PMID:17925311; http://dx.doi.org/ 10.1093/pcp/pcm131 [DOI] [PubMed] [Google Scholar]
- 24.Noda N, Aida R, Kishimoto S, Ishiguro K, Fukuchi-Mizutani M, Tanaka Y, Ohmiya A. Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol 2013; 54:1684-95; PMID:23926063; http://dx.doi.org/ 10.1093/pcp/pct111 [DOI] [PubMed] [Google Scholar]
- 25.Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J 2012; 70:739-49; PMID:22288551; http://dx.doi.org/ 10.1111/j.1365-313X.2012.04908.x [DOI] [PubMed] [Google Scholar]
- 26.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 2001; 4:447-56; PMID:11597504; http://dx.doi.org/ 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- 27.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000; 12:2383-93; PMID:11148285; http://dx.doi.org/ 10.1105/tpc.12.12.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aida R, Shibata M. Agrobacterium-mediated transformation of torenia (Torenia fournieri). Breeding Sci 1995; 45:71-4; http://doi.org/ 10.1270/jsbbs1951.45.71 [DOI] [Google Scholar]
- 29.Nagaya S, Kawamura K, Shinmyo A, Kato K. The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol 2010; 51:328-32; PMID:20040586; http://dx.doi.org/ 10.1093/pcp/pcp188 [DOI] [PubMed] [Google Scholar]
- 30.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005; 17:2993-3006; PMID:16214898; http://dx.doi.org/ 10.1105/tpc.105.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshima Y, Mitsuda N, Nakata M, Nakagawa T, Nagaya S, Kato K, Ohme-Takagi M. Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol 2011; 28:201-10; http://dx.doi.org/ 10.5511/plantbiotechnology.11.0124a [DOI] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673-80; PMID:7984417; http://dx.doi.org/ 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]