Abstract
Microbial eukaryotes hold a key role in aquatic ecosystem functioning. Yet, their diversity in freshwater lakes, particularly in high-mountain lakes, is relatively unknown compared with the marine environment. Low nutrient availability, low water temperature and high ultraviolet radiation make most high-mountain lakes extremely challenging habitats for life and require specific molecular and physiological adaptations. We therefore expected that these ecosystems support a plankton diversity that differs notably from other freshwater lakes. In addition, we hypothesized that the communities under study exhibit geographic structuring. Our rationale was that geographic dispersal of small-sized eukaryotes in high-mountain lakes over continental distances seems difficult. We analysed hypervariable V4 fragments of the SSU rRNA gene to compare the genetic microbial eukaryote diversity in high-mountain lakes located in the European Alps, the Chilean Altiplano and the Ethiopian Bale Mountains. Microbial eukaryotes were not globally distributed corroborating patterns found for bacteria, multicellular animals and plants. Instead, the plankton community composition emerged as a highly specific fingerprint of a geographic region even on higher taxonomic levels. The intraregional heterogeneity of the investigated lakes was mirrored in shifts in microbial eukaryote community structure, which, however, was much less pronounced compared with interregional beta-diversity. Statistical analyses revealed that on a regional scale, environmental factors are strong predictors for plankton community structures in high-mountain lakes. While on long-distance scales (>10 000 km), isolation by distance is the most plausible scenario, on intermediate scales (up to 6000 km), both contemporary environmental factors and historical contingencies interact to shift plankton community structures.
Keywords: alpine lakes, biogeography, diversity, fungi, next-generation sequencing, protistan plankton
Introduction
Protists are single-cell organisms, which in terms of cytological diversity and abundance (if not species diversity) dominate the kingdom Eukarya (Patterson 1999). Inhabiting our planet at least twice as long as multicellular life (Knoll et al. 2004; Berney & Pawlowski 2006), they have conquered nearly any habitat on earth and have evolved multiple modes of life. Protists include mixotrophs, autotrophs, heterotrophs and osmotrophs, and together with bacteria, they account for the majority of the biomass in aquatic environments: for example, the entire pelagic microbial food web, including protozoan microzooplankton, is typically five to ten times the mass of all multicellular aquatic organisms (Pomeroy et al. 2007). This points to the key role protists have in aquatic biogeochemical cycles (Sanders 2009). Traditionally, the description of protists has been microscopy-based and their structural diversity has been recorded since more than two centuries (Weisse 2008), resulting in an invaluable legacy. The recurrent observation of the same morphotypes in freshwater habitats has led some researchers to the conclusion that the global protistan diversity could be relatively low (Finlay & Clarke 1999; Fenchel & Finlay 2004). Furthermore, this assertion led to the concept of a cosmopolitan distribution of most, if not all, protists (Finlay 2002). However, results from more recent molecular-based studies on protistan diversity and distribution patterns (e.g. Bass et al. 2007a; Ryšánek et al. 2015) have prompted most ecologists to abandon the ubiquitous dispersal concept for protists (Gast 2015). Yet, studies with strong evidence for spatial structures in whole protistan communities are still scarce. Likewise, genetic diversity surveys showed that protistan diversity is still far from being described (Massana et al. 2015; de Vargas et al. 2015).
In particular, inland (fresh)water bodies are largely uncharted. Compared to the high number of studies targeting marine protistan communities (e.g. López-García et al. 2001; Lovejoy et al. 2006; Worden et al. 2006; Countway et al. 2007; Massana & Pedrós-Alió 2008; Alexander et al. 2009; Amaral-Zettler et al. 2009; Stoeck et al. 2010; Stock et al. 2012; de Vargas et al. 2015), studies describing genetic protistan diversity from freshwater habitats are rare (e.g. Šlapeta et al. 2005; Lepère et al. 2007; Triadó-Margarit & Casamayor 2012; Taib et al. 2013; Kammerlander et al. 2015; Oikonomou et al. 2015; Simon et al. 2015). Freshwater ecosystems are usually more heterogeneous than the marine realm and thus are expected to hold an even higher genetic protistan diversity than oceans (Logares et al. 2009a; Auguet et al. 2010; Barberán & Casamayor 2010; Barberán et al. 2011). Because marine–freshwater transitions are relatively rare among protists (Logares 2009b; Forster et al. 2012), freshwater communities are distinctively different from marine protistan communities.
Among the inland freshwater bodies, high-mountain lakes hold an exceptional position. Located remotely above the tree line, these ecosystems are traditionally considered as extreme for life (Catalan et al. 2006). Challenges for the biota of high-mountain lakes, such as protists, include in the temperate zone low availability of nutrients, strong changes in incident light and low water temperature (Sommaruga 2001). This requires specific adaptations to osmoregulation and coping with nutrient limitation, which are not necessarily mandatory in the survival toolbox of protists in lowland lakes. The usually low density of organisms in such highly diluted waters (Catalan et al. 2006) entails low encounter rates. Low encounter rates not only are crucial in foraging theory (Jumars 1993), but also are an evolutionary force exerting specific selection pressure and new adaptive pathways (Catalan et al. 2006). Furthermore, high-mountain lake biota is subjected to significantly higher ultraviolet radiation (UVR) compared with lowland lakes, due to a thinner atmosphere and usually high water transparency (Sommaruga 2001). Being harmful to various lake inhabitants, including protists (Sommaruga et al. 1996), intense UVR requires further specific adaptations to prevent DNA damage. Strong UVR exposure alternates at least in the temperate zone with periods of limited light availability and snow/ice-cover time in winter, which may last for many months, or intense short rains in the Altiplano. Not surprisingly, such environmental changes cause pronounced microbial community shifts in those high-mountain lakes (Felip et al. 2002; Dorador et al. 2003; Márquez-García et al. 2009).
In view of these extreme environmental conditions in high-mountain lakes, it is reasonable to assume that these ecosystems raise a protistan genetic diversity that is different from the one found in other inland freshwater lakes. Further, it is highly likely that protistan communities in high-mountain lakes are extremely diverse because a high genomic potential would allow these organisms to buffer shifts in environmental conditions. The very few available genetic protistan diversity studies from high-mountain freshwater lakes support these assumptions, but are not conclusive because of data scarcity and low sample coverage (Triadó-Margarit & Casamayor 2012; Kammerlander et al. 2015). In this study, we therefore sampled 13 lakes from three different mountain regions (Austrian Alps, Chilean Altiplano and Ethiopian Bale Mountains) to analyse and statistically compare microbial eukaryote plankton diversity. Specific aims of the study were (i) to reveal the phylogenetic diversity of eukaryotic microbes to investigate their biogeographic patterns among high-mountain lakes, (ii) to uncover the environmental factors, which might be responsible for the partitioning of diversity in this three distant mountain ranges and (iii) to analyse the degree of genetic novelty within the plankton communities.
Materials and methods
Sample sites
The three selected high-mountain areas strongly differ in important environmental characteristics and temperature regimes. While the lakes in the Alps are ice-covered for several months and are typically dimictic (i.e. water column mix twice a year), those in the Altiplano present a very thin ice cover in winter developing only in the littoral zone during night-time and disappearing usually by midday. Those systems are described as cold polymictic (Mühlhauser et al. 1995). Similarly, the shallow systems in the Bale Mountains are also polymictic and eventually night-time freezing is also observed (Löffler 1978). Whereas lakes in the Chilean Altiplano have a negative water balance (Dorador et al. 2003), those in the Alps and Bale Mountains have a positive one, although in the latter area, rainfall is highly seasonal with a clear rainy season going from March to May and from October to November (Löffler 1978). The lakes in the Chilean Altiplano and in the Bale Mountains are naturally more productive and have high phosphorus concentrations (Löffler 1978; Mühlhauser et al. 1995; Márquez-García et al. 2009), which strongly contrast with the oligotrophic character of the lakes in the Alps. Further, because of the different geology and hydrology, the lakes in the three areas have a strong gradient in salt content as indicated by the concentration of major ions and specific conductivity (Table 1). Thus, the endorheic character of the lakes in the Altiplano is paralleled by the highest conductivity values among the studied lakes (Table 1).
Table 1.
Geographic properties (coordinates, altitude) and lake characteristics of the European Alps, Chilean Altiplano and Ethiopian Bale Mountain lakes
| Lake | Lake code | Coordinates | Altitude (m) |
Water temperature (°C) |
pH | Conductivity (μS/cm) |
SO42− (mg/L) |
Cl− (mg/L) |
Na+ (mg/L) |
K+ (mg/L) |
Mg2+ (mg/L) |
Ca2+ (mg/L) |
DOC (μg/L) |
DN (μg/L) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European Alps | N | E | |||||||||||||
| Drachensee | DRA | 47°21′36 | 10°56′0 | 1874 | 7.48 | 8.3 | 146 | 1.06 | 0.23 | 0.09 | 0.12 | 2.59 | 25.84 | 356 | 284 |
| Oberer Plenderlessee | OPL | 47°12′11 | 11°2′17 | 2417 | 11.25 | 7.35 | 22.3 | 3.35 | 0.06 | 0.38 | 0.23 | 0.29 | 3.52 | 290 | 187 |
| Gossenköllesee | GKS | 47°14′0 | 11°01′0 | 2344 | 7.69 | 7.26 | 43 | 11.44 | 0.06 | 0.54 | 0.72 | 0.79 | 5.42 | 178 | 178 |
| Drachensee | DRA II* | 47°21′36 | 10°56′0 | 1874 | 7.45 | 8.33 | 145 | 1.03 | 0.33 | 0.08 | 0.11 | 3.224 | 25.25 | 551 | 253 |
| Oberer Plenderlessee | OPL II* | 47°12′11 | 11°2′17 | 2344 | 5.07 | 7.2 | 59.5 | 17.09 | 0.11 | 0.67 | 0.79 | 1.227 | 7.58 | 237 | 217 |
| Gossenköllesee | GKS II* | 47°14′0 | 11°01′0 | 2417 | 7.21 | 7.36 | 22.1 | 3.32 | 0.2 | 0.39 | 0.24 | 0.282 | 3.65 | 384 | 170 |
| Mutterbergersee | MUT II* | 47°0′58 | 11°7′41 | 2483 | 5.37 | 6.18 | 5.08 | 0.95 | 0.1 | 0.18 | 0.19 | 0.102 | 0.59 | 575 | 68 |
| Schwarzsee ob Sölden | SOS II* | 46°57′53 | 10°56′48 | 2796 | 3.78 | 6.24 | 35.1 | 12.66 | 0.1 | 0.88 | 0.19 | 0.808 | 3.58 | 325 | 50 |
| Chilean Altiplano | S | W | |||||||||||||
| Cotacotani | COT | 18°12′08.6 | 69°13′43.3 | 4550 | 10.5 | 9.8 | 840 | 58.27 | 46.11 | 63.64 | 9.16 | 57.78 | 28.69 | 6920 | 644.9 |
| Piacota | PIA | 18°11′49.8 | 69°15′56.9 | 4400 | 12.6 | 9.3 | 660 | 44.53 | 40 | 60.22 | 13.6 | 30.96 | 22.32 | 8436 | 704 |
| Chungara | CHU | 18.14° | 69.09° | 4600 | 9.9 | 9.22 | 1543 | 94.53 | 76.23 | 142.9 | 28.1 | 104.18 | 40.73 | 10904 | 969 |
| Cotacotani | COT II* | 18°12′08.6 | 69°13′43.3 | 4550 | 5.7 | 8.96 | 849 | 59.15 | 51 | 62.34 | 8.45 | 59.52 | 36.71 | 6725 | 567 |
| Piacota | PIA II* | 18°11′49.8 | 69°15′56.9 | 4400 | 6.4 | 9.9 | 640 | 44.54 | 47 | 55.6 | 9.32 | 42.52 | 20.74 | 8966 | 364.1 |
| Chungara | CHU II* | 18.14° | 69.09° | 4600 | 4.8 | 9.12 | 1400 | 119.39 | 85 | 144.86 | 26.66 | 113.26 | 39.12 | 10486 | 794 |
| Ethiopian Bale Mountains | N | E | |||||||||||||
| Koromi | KOR | 06°53.561′ | 39°54.458′ | 3917 | 8.4 | 7.57 | 21.2 | 1.8 | 0.46 | 0.9 | 0.25 | 0.65 | 3.04 | 3025 | 169 |
| Hara-Lukas | HaLu | 06°51.498′ | 39°53.054′ | 4121 | 13.4 | 8.82 | 50.9 | 2.98 | 1.29 | 1.61 | 0.85 | 1.85 | 6.37 | 7121 | 406 |
| Central | CEN | 06°51.107′ | 39°52.411′ | 4123 | 5.8 | 7.74 | 39 | 3.44 | 1.12 | 1.36 | 0.82 | 1.36 | 5.1 | 7318 | 350 |
| Crane | CRA | 06°51.285′ | 39°53.580′ | 4047 | 10.1 | 8.85 | 49.5 | 2.47 | 0.88 | 1.58 | 0.53 | 1.79 | 6.31 | 5709 | 273 |
| Togona | TOG | 06°53.095′ | 39°53.720′ | 4029 | 10.9 | 9.51 | 21.1 | 1.33 | 0.5 | 0.81 | 0.21 | 0.61 | 2.93 | 3552 | 170 |
‘II*’ indicates sampling during October (Alps) or March (Chile).
Sampling
Samples for three of the five lakes selected in the Austrian Alps were collected in July and October 2013 during the stratification and overturn period, respectively. The lakes Mutterbergersee (MUT II) and Schwarzsee ob Sölden (SOS II) were sampled only in October 2013 as they can only be reached by helicopter. The three lakes in the Chilean Altiplano were sampled in August 2013 (cold, dry season) and in March 2014 (warm after the rainy season) and those in the Bale Mountains, Ethiopia, were collected in October 2013 during the rainy season. In all lakes, a composite sample was collected to obtain representative information for the whole water column. For that, the same volume of lake water collected at different depths (number depended on lake maximum depth) was pooled. Depths close to the sediment were avoided. In all lakes, three replicated composite samples were obtained. Water samples for DNA extraction were filtered on the same day onto 0.65-μm Durapore membranes (Millipore) using a peristaltic pump until clogging was observed. Samples were prescreened to exclude zooplankton through a sieve with a 100-μm mesh size nylon net. Filters were immediately placed into cryovials with RNAlater (Qiagen) and stored as recommended by the manufacturer until further processing.
Measurement of environmental variables and geographic distances
A multiparameter probe (YSI) was used in the European Alps and in Chile to make profiles in the water column for temperature, conductivity, pH and oxygen. Because of the shallowness of the systems in the Bale Mountains, this was not possible and measurements were taken with single probes (Hach). From the same composite samples, several auxiliary parameters were also measured in the dissolved fraction. The dissolved fraction was defined as the filtrate passing through a glass fibre filter (GF/F Whatman), which was previously burned at 450 °C for 2 h. Dissolved organic carbon (DOC) was measured in acidified samples (pH = 2) with a total carbon analyser Shimadzu TOC-VCPH, while dissolved nitrogen (DN) was measured in a total nitrogen measuring unit (Shimadzu TNM–1). All these analyses were performed in Innsbruck. Ion concentrations in samples from the Alps and Bale Mountains were measured by ion chromatography in a Dionex ICS 1000 and 1010 system, whereas those from the Altiplano by atomic absorption spectrophotometry in a Shimadzu 6800 system with a GFA-EX graphite furnace.
Geographic distances were determined based on the longitude/latitude data using the ‘haversine’ formula as implemented in the script provided by Chris Veness (2002–2015) at http://www.movable-type.co.uk/scripts/latlong.html. The ‘haversine’ formula calculates the great-circle distance between two points on a sphere, that is the shortest distance over the earth’s surface. To account for altitude differences, the altitudes of the sample pairs were subtracted and the difference was added to the resulting value of the ‘haversine’ formula for the respective sample pair.
Nucleic acid extraction, PCR and sequencing
As described previously (Oikonomou et al. 2015), each filter was cut with a sterile scalpel into smaller pieces and transferred to a Lysis E Matrix tube (MP Biomedicals, Germany), followed by the addition of 600 μL RLT buffer and 6 μL β-mercaptoethanol. Filters were then shaken at 30 Hz for 45 s using a mixer mill (MM200, Retsch, Germany). The tubes were centrifuged (14 000 g, 3 min), the supernatant was retained, and DNA extraction was carried out using Qiagen’s (Hilden, Germany) All Prep DNA/RNA Mini kit according to the manufacturer’s protocol (after step 4). The concentration of bulk DNA was measured spectrophotometrically (NanoDrop 2000, Thermo Scientific, Wilmington, DE, USA). The hypervariable V4 region of the SSU rDNA was amplified using the primer pair TAReukV4F and TAReukREV (Stoeck et al. 2010), yielding ca. 500-base pair (bp) fragments. The polymerase chain reaction (PCR) mixture, containing 1 U of Phusion High Fidelity DNA Polymerase (New England Biolabs), was heated to 98 °C for 30 s, and the V4 region was amplified with a PCR protocol described in Stoeck et al. (2010). To minimize PCR bias, three individual reactions per filter were prepared. The three individual reactions were pooled during the PCR product purification using Qiagen’s MinElute PCR purification kit. Paired-end sequencing of purified V4 amplicons was conducted on an Illumina MiSeq platform by SeqIT (Kaiserslautern, Germany). The paired-end reads generated from the same amplicon were merged using a custom script.
Sequence data processing and taxonomic assignment
Raw paired-end Illumina reads were quality-filtered using the split_libraries.py script implemented in QIIME v.1.8.0 (Caporaso et al. 2010). Only reads with exact barcodes and primers, unambiguous nucleotides and a minimum length of 300 bp were retained and subjected to a de novo chimera detection using uchime (Edgar et al. 2011). Nonchimeric reads were clustered with SWARM v1.2.6 (Mahé et al. 2014) using d = 1. Other than popular de novo clustering methods, SWARM does not rely on arbitrary global clustering thresholds (as lineages evolve at variable rates, no single sequence similarity value can accommodate the entire tree of life) and input order dependency. Instead, it uses a local clustering threshold to assign amplicons to an operational taxonomic unit (OTU). OTUs grow iteratively by comparing each generation of assigned amplicons to the remaining amplicons. An OTU is closed when no new amplicon can be integrated in the OTU. This strategy produces robust and high-resolution OTUs that allow for accurate, meaningful and interpretable biological results. Using a custom script (Appendix S1, supporting information), an OTU contingency table was created based on the output files of SWARM. Representative sequences (i.e. the most abundant sequence) from each OTU were extracted and analysed with the software package JAGUC (Nebel et al. 2011) and GenBank’s nr nucleotide database release 202 as reference database. The tabular taxonomic information and the OTU contingency table were then merged to the final OTU table. Nontarget OTUs (metazoans, embryophytes) as well as singletons and doubletons [unique/double amplicons that occur exclusively in only one sample are most likely erroneous sequencing products (Bokulich et al. 2013; Nelson et al. 2014)] were excluded. The resulting file was used as a basis for statistical and network analyses.
Diversity statistics
Indices of alpha -diversity and similarities among the samples (beta-diversity) were calculated in R (R-Team 2008) using the program package VEGAN (Oksanen et al. 2013). We used the Shannon entropy as an index, as it provides robust quantification and meaningful comparison of the microbial diversity in molecular data sets (Haegeman et al. 2013). Because the shannon index is biased towards the most abundant taxa (phylotypes), we used log-transformation to down-weigh the most abundant taxa. For an interpretation of a diversity index, it was recommended to convert any given index into ‘true diversity’ as an equivalent, termed as effective number of species (=equally common species) (MacArthur 1965; Jost 2006, 2007). We conducted this conversion according to Jost et al. (Jost 2006).
The analysis of variance (one-way ANOVA) was calculated in R using the STATS package (R-Team 2008).
The Jaccard index was used as a measure of similarity between the samples. Here, we used the binary Jaccard index to avoid bias based on abundance values resulting from different gene copy numbers in protists and fungi (Casamayor et al. 2002; Zhu et al. 2005; Amaral-Zettler et al. 2009; Dunthorn et al. 2014). Prior to beta-diversity estimation, the number of sequences per sample was rarefied to the smallest sample size (n = 36.744). Jaccard similarity values were transformed to distance matrixes for a nonmetric multidimensional scaling (NMDS) analysis. Environmental vectors were fitted to the ordination using the envfit function of the VEGAN package in R. The fit (R2) of each variable to the ordination using the envfit function was assessed with a Monte Carlo analysis of 10000 permutations.
Testing for geographic distance effects
The Mantel test was used to evaluate the relationship between genetic distance and geographic distance (Mantel 1967), which is the most commonly used method to address spatial processes driving population structures (Diniz-Filho et al. 2013). Analyses were conducted with the ibd program package (http://ibdws.sdsu.edu, Jensen et al. 2005) running 5000 randomizations of the rows and columns of the genetic community and geographic distance matrices to generate the distribution of correlations under the null hypothesis. The community distance was calculated as the Sørensen distance between two samples. Positive Mantel correlations and significant P-values are conclusive that nearby communities tend to be more similar than expected by chance and that (genetic) differences in communities increase with geographic distance (Diniz-Filho et al. 2013). Because zero distances are invalid in the Mantel test, the data set(s) used for this analysis included each lake only once. Therefore, Mantel tests were conducted with different data sets, one of them including the data from summer samplings (DRA, OPL, GKS, COT, PIA, CHU and all Ethiopian lakes) and one from winter samplings (DRA II, OPL II, GKS II, MUT II, SOS II, COT II, PIA II, CHU II and all Ethiopian lakes). Details of samples included in a specific analysis are given in the respective figure legends. Furthermore, a variation of the Mantel test (Mantel correlogram) was applied. While the simple Mantel test shows the overall relationship between the geographic and community distance matrices, the Mantel correlogram allows the analyses of relationships between community distances and specific geographic distances across space (Diniz-Filho et al. 2013). Therefore, the geographic distance matrix was divided into three submatrices, each one describing pairs of communities within a bounded interval of geographic distances. This strategy is useful to identify possible variations in the correlation between community distance and geographic distance classes. Our data sets were subdivided into the following distance classes: class 1 – up to 50 km, corresponding to ‘short distances’ (local scale); class 2 – up to 6000 km, corresponding to ‘intermediate distances’; and class 3 – up to 12 000 km, corresponding to ‘large distances’. Three Mantel correlations were then obtained by performing a Mantel test between the community distance matrix and the three geographic distance matrices. The Mantel correlogram was constructed by plotting Mantel correlations between the community distance matrix and each geographic distance class matrix against the mid-point of the respective distance class (Oden & Sokal 1986; Legendre & Legendre 2012).
Identification of novel diversity
The construction of novel diversity networks followed the description of Filker et al. (2015). To infer genetically divergent diversity in the three biogeographic regions, data sets from individual lakes within a specific biogeographic region were pooled. All sequences were truncated to the same length (300 bp) and dereplicated using a custom script (Appendix S1, Supporting information). Unique reads were clustered using SWARM v1.2.6 (Mahé et al. 2014) with d = 1. To build the network, the representative reads from the 200 most abundant OTUs were extracted and subjected to BLASTn analyses (Altschul et al. 1990) against GenBank’s nr nucleotide database (release 202) to identify their taxonomic affiliation. The seed reads were then aligned in Seaview (Galtier et al. 1996), followed by the calculation of sequence similarities between each pair of seed sequences using PAIRALIGN (see Appendix S1, Supporting information). The network was build using the IGRAPH R package (Csárdi & Nepusz 2006) and was based on the sequence similarity values. In the network, two nodes were connected by an edge if they shared a sequence similarity of at least 90%. The resulting network was visualized and modified with GEPHI v.0.8.2-beta (Bastian et al. 2009) according to the OTU taxonomic affiliation and BLAST hit value.
Results
Replicate sequencing of the same sample from Drachensee, Austria (DRAa and DRAb), produced nearly identical results (Fig. S1, Supporting information). For further comparative analyses, we therefore used only one of the two replicate samples (DRAa), for which we obtained nine more phylotypes (n phylotypes DRAa = 284) compared to the replicate sample DRAb.
Microbial eukaryote diversity at local scale (alpha-diversity)
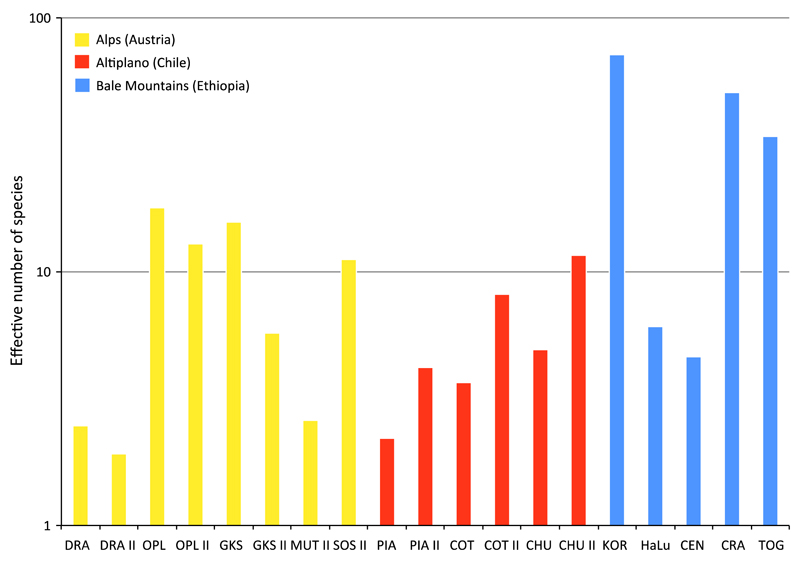
For the 20 samples (including the replicate sequenced sample DRAb) analysed in this study, we obtained between 36 744 and 184 273 high-quality protistan (including fungal) V4 amplicons after Illumina sequencing and data cleaning (Table 2). The number of distinct OTUs for each sample varied from 99 to 1166. Rarefaction analyses predicted that all samples were sampled to near saturation (Fig. S2, Supporting information). Shannon entropy-based effective number of species varied from 1.9 in the Austrian DRA lake to 71.5 in the Ethiopian KOR lake (Fig. 1). One-way ANOVA revealed a significant difference in the effective number of species among the three geographic regions (P < 0.05, for details of ANOVA results, see Fig. S3, Supporting information).
Table 2.
Number of sequences and operational taxonomic units generated after Illumina data processing. Target sequences comprise eukaryotic unicellular organisms and fungi
| Sample ID | Raw sequences | Cleaned sequences | Cleaned OTUs | Target sequences | Target OTUs | Target sequences without singletons/doubletons | Target OTUs without singletons/doubletons |
|---|---|---|---|---|---|---|---|
| DRAa | 70 147 | 65 788 | 4899 | 62 746 | 4723 | 58 156 | 284 |
| DRAb | 93 303 | 71 466 | 5322 | 67 873 | 5119 | 62 817 | 275 |
| OPL | 115 640 | 98 252 | 15 404 | 98 199 | 15 397 | 82 450 | 395 |
| GKS | 103 724 | 89 716 | 12 787 | 89 689 | 12 772 | 76 820 | 532 |
| OPL II | 95 449 | 69 823 | 6990 | 61 216 | 6603 | 54 681 | 348 |
| GKS II | 125 219 | 110 538 | 18 646 | 110 505 | 18 632 | 91 449 | 630 |
| DRA II | 127 949 | 124 754 | 8160 | 124 643 | 8139 | 116 526 | 336 |
| MUT II | 99 639 | 95 503 | 5627 | 89 124 | 5139 | 81 154 | 99 |
| SOS II | 87 461 | 79 774 | 9479 | 78 713 | 9332 | 69 293 | 382 |
| COT | 172 037 | 168 199 | 6518 | 164 832 | 6349 | 158 571 | 210 |
| PIA | 168 593 | 161 554 | 5449 | 160 981 | 5383 | 155 628 | 263 |
| CHU | 213 932 | 200 024 | 9406 | 193 343 | 9045 | 184 273 | 388 |
| COT II | 71 666 | 61 887 | 5896 | 61 732 | 5840 | 56 027 | 420 |
| PIA II | 147 042 | 135 163 | 7697 | 109 392 | 5620 | 103 945 | 353 |
| CHU II | 69 328 | 56 699 | 5378 | 54 399 | 5245 | 49 252 | 340 |
| KOR | 130 088 | 108 526 | 13 069 | 96 414 | 12 206 | 84 439 | 1166 |
| HaLu | 87 025 | 84 689 | 5750 | 82 505 | 5457 | 77 261 | 467 |
| CEN | 54 105 | 52 530 | 2331 | 50 670 | 2177 | 48 624 | 186 |
| CRA | 173 658 | 166 489 | 9843 | 69 801 | 6775 | 63 409 | 934 |
| TOG | 125 227 | 114 330 | 9469 | 42 606 | 6217 | 36 744 | 692 |
Fig.1.
Shannon-based effective number of species (alpha-diversity) between the sampled lakes in the European Alps, the Chilean Altiplano and the Ethiopian Bale Mountains. Results are based on normalized data sets (n = 36 744).
Taxonomy of phylotypes
Austrian Alps
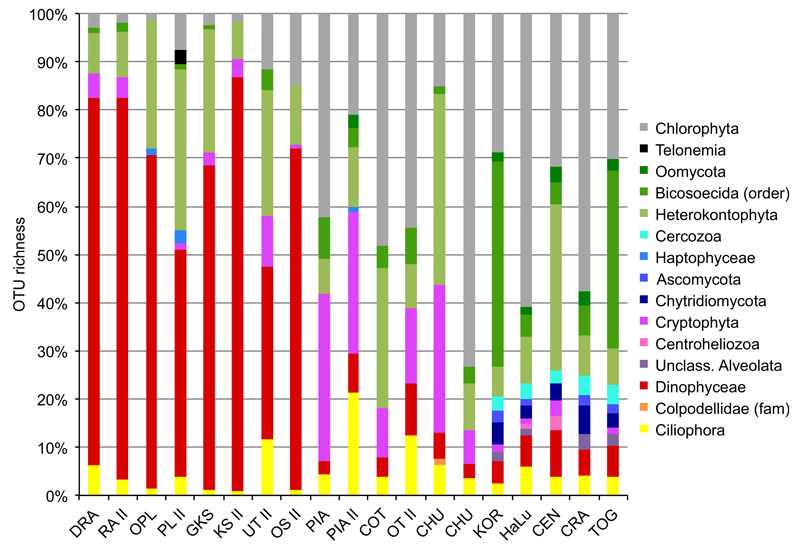
In terms of phylotype diversity (measured as OTU richness), there was little variation among the different lakes when considering high taxonomic levels (phylum level). All lakes in this region were strongly dominated by dinoflagellates (Fig. 2). This group accounted for up to 86% (531 phylotypes (OTUs), in Gossenköllesee, GKS II) of the total phylotype diversity in these lakes. The second most diverse taxon group was Heterokontophyta (mostly Chrysophyceae and Synurophyceae) accounting for up to 33% (112 phylotypes, in Oberer Plenderlessee, OPL II). Fungi and nonpigmented protist phylotypes such as ciliates and heterokont flagellates were underrepresented in terms of diversity. Seasonal variations in higher taxon diversity patterns were observed, but not pronounced. For example, in OPL, the number of ciliate phylotypes increased from 5 to 13 from OPL (July) to OPL II (October). Such minor discrepancies could, however, be a result of patchy distribution during sampling or of undersampling.
Fig. 2.
Distribution of the protistan and fungal communities among the analysed lakes in the European Alps, the Chilean Altiplano and the Ethiopian Bale Mountains. The bars display the relative number of distinct operational taxonomic units (OTUs) per taxonomic group (i.e. OTU richness). Only those taxonomic groups (phylum level) were considered that contain at least 1% of the total number of OTUs per sample.
Chilean Altiplano
In this region, phylotype diversity was more variable among the three lakes and seasons than in the Alps. In general, lakes from this region were characterized by a high diversity of chlorophytes, cryptophytes and Heterokontophyta. In individual lakes (PIA II), also ciliates accounted for up to 21% of the observed phylotype diversity. Dinoflagellates accounted for a maximum of 11% in COT II.
Ethiopian Bale Mountains
Similarly to the lakes in the Altiplano, they showed unique diversity signatures at the phylum level. Even though the higher-rank taxon diversity in these lakes was also largely coined by pigmented lineages (with the exception of lake KOR), also nonpigmented taxon groups were remarkably diverse. These include bicosoecid flagellates, oomycetes, centroheliozoans, cercozoans and fungi (ascomycetes and chytridiomycetes). Variation among the lakes in the relative proportions of different higher-rank taxon orders was pronounced. For example, while KOR and TOG harboured a relatively high proportion of nonpigmented flagellates (bicosoecids accounting for 42% of the observed phylotypes in KOR and 37% in TOG), chlorophytes dominated the lakes HaLu (59%) and CRA (55%).
Partitioning of diversity among biogeographic regions
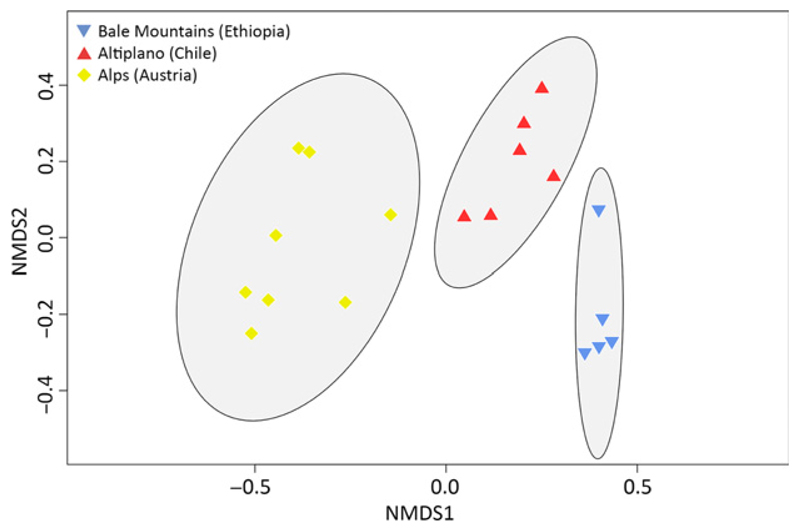
The pronounced differences in microbial eukaryote community composition in the distinct geographic regions on higher taxonomic ranks were statistically confirmed at the phylotype level. In a nonmetric multidimensional scaling (distance measure: Jaccard), geographic plankton community clusters with their 95% confidence intervals did not overlap (Fig. 3). The biogeographic clusters followed a gradient along axis 1. Within each of the geographic regions, the individual lakes were arranged along a gradient of axis 2. The 2D stress (Kruskal) of the NMDS analysis was 0.11.
Fig. 3.
Nonmetric multidimensional scaling analysis of the European, Chilean and Ethiopian high-mountain lake samples under study based on Jaccard similarities. For each community cluster, the ellipses represent 95% confidence intervals.
Environmental effects on diversity partitioning
Because the lakes in the different geographic regions were significantly different in a number of environmental parameters, we tested whether the observed partitioning of diversity was due to biogeography or to environmental selection, or a combination of both. Although ANOVA also revealed significant differences in altitude, pH, all ions measured, DOC and DN (P < 0.01 in all cases) among the lakes in the three geographic regions, none of these parameters correlated significantly with the effective number of species (Pearson R for all comparisons >−0.3 or <0.3). Fitting environmental variables into the NMDS analyses revealed that all but water temperature (P > 0.5) have a significant effect on the observed diversity partitioning patterns (Table 3). Interestingly, the vast majority of environmental parameters showed a much stronger affinity to NMDS axis 2. Only altitude correlated strongly with NMDS axis 1. Thus, altitude was the only factor, which was meaningful for the observed gradient along NMDS axis 1 (separation of geographic regions). The remaining environmental variables contributed to explaining the detected differences in microbial eukaryote community structures within each of the individual biogeographic regions.
Table 3. Environmental variables fitted into nonmetric multidimensional scaling analysis. P-values <0.05 are considered as significant effect.
| NMDS1 | NMDS2 | R 2 | Pr (>r) | |
|---|---|---|---|---|
| Altitude | 0.9399 | 0.3412 | 0.76 | 0.0002 |
| Water temperature | 0.3384 | −0.9410 | 0.28 | 0.0748 |
| pH | 0.7812 | 0.6243 | 0.43 | 0.0134 |
| Conductivity | 0.2489 | 0.9685 | 0.57 | 0.0020 |
| SO42− | 0.2401 | 0.9708 | 0.51 | 0.0038 |
| Cl− | 0.2885 | 0.9575 | 0.58 | 0.0019 |
| Na+ | 0.2795 | 0.9602 | 0.53 | 0.0025 |
| K+ | 0.2852 | 0.9585 | 0.48 | 0.0037 |
| Mg2+ | 0.2647 | 0.9643 | 0.56 | 0.0019 |
| Ca2+ | 0.1122 | 0.9937 | 0.71 | 0.0002 |
| DOC | 0.7299 | 0.6836 | 0.73 | 0.0003 |
| DN | 0.3844 | 0.9232 | 0.49 | 0.0041 |
Geographic distance effects on diversity partitioning
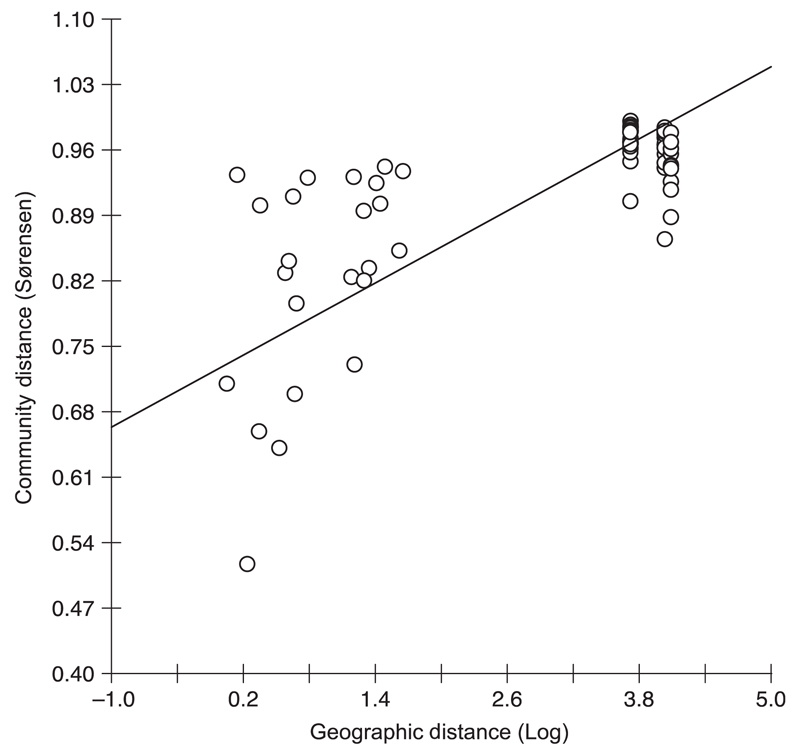
To evaluate the effect of geographic distance on (genetic) community structure, we used variations of the Mantel test. The correlation between community distance and geographic distance (Fig. 4) showed a spatial pattern. The Mantel correlation between the two matrices was equal to 0.739 when using log geographic distances. This showed an increase in community distance with geographic distance. None of the values from 5000 randomizations was larger than the observed value of 0.739, so that the chance of obtaining a value as large as the observed is smaller than 1/5000. This resulted in a highly significant P-value of < 0.0002 for this Mantel test, suggesting a significant increase in community distance with geographic distance. Further, when using the natural distances (nonlog) in the Mantel test, the P-value was < 0.0002 at a Mantel correlation r of 0.536.
Fig. 4.
Relationship between the pairwise community (Sorensen distance) and geographic distances (log-transformed) for the analysed high-mountain lakes (including winter samples).
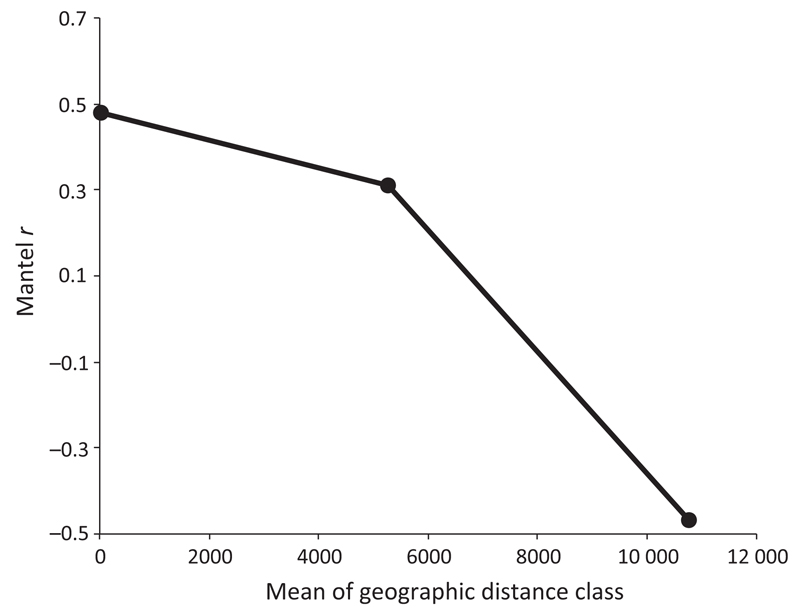
To test whether distance effects are more or less pronounced at different distance scales (as suggested by a nonlinear relationship between geographic distance and community distance, Fig. 4 and Fig. S4, Supporting information), we used a Mantel correlogram, which considers nonoverlapping distance classes. In the first distance class (0–50 km), microbial eukaryote communities tend to be more similar (rm = 0.478; P < 0.01 with 5000 permutations) than in the second distance class (5235–5278 km; rm = 0.315; P = 0.05 with 5000 permutations) (Fig. 5). In the third distance class (10 741–12 270 km), Mantel correlation decreased to −0.469 (P = 0.016). This negative correlation value indicates that communities that are located at such high distances apart tended to be more dissimilar than communities of the first (up to 50 km) and second (up to 5278 km) distance classes.
Fig. 5.
Mantel correlogram displaying spatial structuring of the high-mountain lake communities within the following nonoverlapping distance classes: 0–50 km, 5235–5278 km and 10 741–12 270 km.
Degree of novel diversity
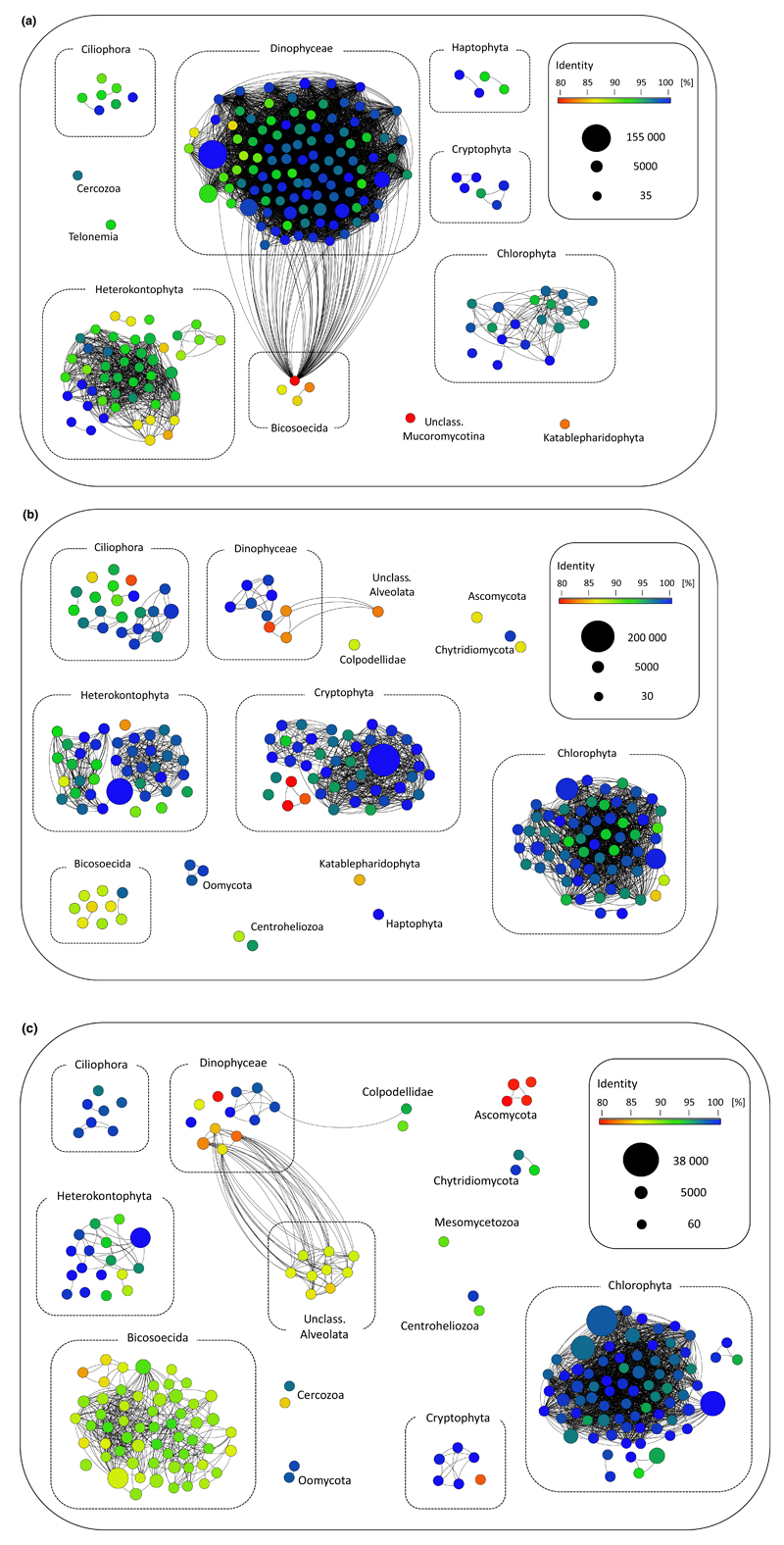
Austrian Alps: The 200 most abundant phylotypes detected in the alpine lakes were relatively well represented in NCBI’s nucleotide database (Fig. 6a). The mean sequence similarity of all 200 phylotypes to previously deposited sequences was 96.5%. Only 16 phylotypes had a sequence similarity of <90% to previously deposited sequences. Specifically, several pigmented heterokonts (stramenopiles) (mean sequence divergence to previously described sequences: 94.8%) as well as all bicosoecid flagellates (mean sequence divergence to previously described sequences: 88%), one katablepharid and one mucoromycote phylotype among the most abundant 200 phylotypes were highly divergent to previously deposited sequences. Among the dinoflagellates, which accounted for the largest proportion of diversity in the 200 most abundant phylotypes, only few phylotypes were highly divergent to previously deposited sequences (mean sequence divergence to previously described sequences: 97.4%).
Fig. 6.
Analyses of the novel diversity within the protistan (and fungal) communities in the European Alps lakes (a), the Chilean Altiplano lakes (b) and the Ethiopian Bale Mountain lakes (c). Each dot represents one operational taxonomic unit (OTU), the different colours indicate the level of sequence similarity to a deposited reference sequence, and the size of on OTU indicates the number of included V4 sequences. An edge weight (sequence similarity) of 90% was chosen to discriminate between the different OTUs.
Chilean Altiplano: The mean sequence similarity of the 200 most abundant phylotypes to previously deposited sequences was 96.5% (Fig. 6b). Of these phylotypes, 22 have a sequence similarity of <90% to previously deposited sequences. Ciliates have a mean sequence divergence of 95.8% to previously deposited sequences, and for dinoflagellates, this value is even lower (94.7%). Sequences assigned to heterokontophyts are on average 96.9% divergent to described sequences and bicosoecids even 90.4%. Further, highly divergent fungi (Ascomycota, Chytridiomycota), Cryptophyta and a katablepharid were detected. Within the most diverse group (Chlorophyta), most of the detected phylotypes were closely related to previously deposited sequences.
Ethiopian Bale Mountains: These lakes harboured the largest proportion of hitherto undescribed diversity (Fig. 6c). The mean sequence similarity of the 200 most abundant phylotypes to previously deposited sequences was only 94.2%. Of these phylotypes, 52 have a sequence similarity of <90% to previously deposited sequences. All of the numerous bicosoecid phylotypes were <92.6% similar to previously described sequences. The mean sequence divergence in the detected ascomycote sequences was as low as 81.9%, and even within the dinoflagellates, half of the detected phylotypes have a remarkably high divergence to described sequences. Our analyses even identified a whole group of unclassified alveolates (with loose affiliation to dinoflagellates), which were very divergent (mean: 87.7%, n = 9) to previously deposited sequences. All detected ciliate phylotypes (n = 7) and the vast majority of the detected chlorophyte phylotypes were relatively closely related to previously deposited sequences (mean divergence ciliates: 98.1%; mean divergence chlorophytes: 98.3%).
Discussion
The protistan and fungal diversity of inland water bodies have received relatively little attention compared with coastal and open ocean ecosystems. Specifically, freshwater lakes located above the tree line or at high altitude are largely uncharted in this respect (Triadó-Margarit & Casamayor 2012; Kammerlander et al. 2015). Therefore, understanding the planktonic diversity of high-mountain lakes and its geographic distribution patterns is fundamental. This is particularly relevant because different global changes strongly affect high-mountain lakes. Knowledge about the diversity of high-mountain lake biota (and subsequently their functions) will help to model the effect of such contemporary environmental change on these sensitive ecosystems and to develop conservation strategies.
Triadó-Margarit & Casamayor (2012) analysed the 18S rDNA gene of protistan plankton obtained from 11 high-mountain lakes in the central Pyrenees. They observed that high-mountain lakes even within the same lake district can be so heterogeneous that highly distinct protistan plankton communities evolve in adjacent lakes. Their analyses included only 953 18S rDNA sequences. Even though these data are very valuable, they left the majority of the protistan diversity undetected because of a low sampling (sequencing) depth. Kammerlander et al. (2015) analysed protistan plankton (including fungi) in two adjacent high-mountain lakes in the Austrian Alps and two more in the Himalaya region. The authors used ultra-deep Illumina sequencing of the V4 fragment of the 18S rDNA as taxonomic marker and revealed a much higher diversity than reported earlier from high-mountain lakes. In agreement with Triadó-Margarit & Casamayor (2012), also Kammerlander et al. (2015) reported a high genetic divergence between microbial eukaryote plankton communities in adjacent lakes. However, these authors showed that interregional genetic differences in plankton communities are much more pronounced than intraregional differences. This tentatively suggested a biogeographic pattern of microbial eukaryote plankton communities in high-mountain lakes. However, the four lakes analysed by Kammerlander et al. (2015) were insufficient to draw more solid conclusions regarding biogeographic distribution patterns.
The higher-rank taxon groups (phylum level) detected in the lakes under the present study were largely the same and also corroborated well previous molecular diversity studies of high-mountain lakes in the central Pyrenees (Triadó-Margarit & Casamayor 2012). Yet, the diversity patterns of these higher-rank taxon groups were vastly different in the three mountain regions (interregional differences). In fact, the protistan and fungal communities in high-mountain lakes are fingerprints, which are specific for each of the three mountain regions. Thus, our study provides strong support for biogeographic patterns in high-mountain lakes. Even though intraregional microbial eukaryote communities were distinct from each other (as observed in Triadó-Margarit & Casamayor 2012 and in Kammerlander et al. 2015), the differences were more pronounced on an interregional scale (Fig. 2). Interestingly, high-mountain lakes in the central Pyrenees have yet again a different community fingerprint than the three mountain regions analysed in our study. In the Pyrenean lakes, stramenopiles, in particular chrysophytes, account for the most diverse phylotypes. To the best of our knowledge, no study has been published that analysed whole microbial eukaryote community structures in high-mountain lakes using microscopy. Most, if not all, microbial ecologists meanwhile agree that at least some single-celled organisms have limited distribution patterns at least on a high-resolution molecular scale (Gast 2015) and literature within (but see Finlay 2002 and Finlay & Fenchel 2004). Mostly, the ‘moderate endemicity hypothesis’ (Foissner 2006) finds support in (both genetic and morphological) studies on individual taxa across the eukaryotic tree. These include, just to mention a few examples, the planktonic testate amoeba Difflugia tuberspinifer (Gomaa et al. 2015), some chrysophyte species (Stoeck et al. 2008), the green algae Klebsormidium (Ryšánek et al. 2015), diverse ciliates (Foissner et al. 2003; Katz et al. 2005; Dolan et al. 2012; Dunthorn et al. 2012), the cercomonads Eocercomonas and Paracercomonas (Bass et al. 2007b), diatoms (Van de Vijver et al. 2005) and the synurophyte Mallomonas (Rezácová & Neustupa 2007). At the whole-community level, relatively few studies have analysed the effect of (large-scale) geographic distance on protistan and fungal community genetic distance. Bates et al. (2012) analysed a pyro-sequenced 18S rDNA fragment (including the V4 region) from 40 soil samples collected along a north–south gradient (North and South America and Antarctica). The analyses of these samples rejected the global dispersal hypothesis because none of the observed phylotypes was found in all samples across the latitudinal gradient. Instead, 84% of the observed phylotypes occurred in five samples or less. However, statistical analyses of the obtained community profiles revealed that distance is not a very strong predictor of community dissimilarity. Instead, shifts in soil protistan community structure are significantly correlated to annual soil moisture availability. Despite the high value of this study by Bates et al. (2012), the obtained data has to be interpreted with caution, because of severe undersampling. For statistical analyses of all samples, the authors had only 150 sequences available for each of the 40 samples. Thus, the full extent of protistan diversity was far from being uncovered.
In our study of high-mountain lake microbial eukaryote communities, we found predictable beta-diversity patterns: geographic distance had a significant effect on shifts in plankton community structures. On a local scale, environmental factors could be identified as structuring components for plankton communities. Heterogeneous community structures on a regional scale resulting from differences in environmental factors were reported in previous studies on protistan communities in high-mountain lakes in Tibet (Wu et al. 2009) and the Pyrenees (Triadó-Margarit & Casamayor 2012). Specifically, the concentration of ions, pH and also nutrients (Wu et al. 2009; Triadó-Margarit & Casamayor 2012) was identified as structuring factors on local scales. Studies in lakes of the Pyrenees showed that cryptophytes are associated with lakes with higher phosphorus concentrations, whereas chrysophytes are characteristic for oligotrophic and ultra-oligotrophic lakes (Catalan et al. 2006). Other algal groups did not show any correlation with nutrients. Furthermore, altitude seems to be a decisive factor for the structuring of high-mountain lake protistan communities (Wu et al. 2009). A critical factor changing with increasing altitude is the increase in harmful UV-B radiation (Sommaruga 2001), which requires specific adaptive mechanisms for inhabitants of high-mountain lakes.
Our analyses showed, however, that the effects of environmental variables as structuring factors for plankton communities at a local scale were much less pronounced than the distance effect on an interregional scale (Fig. 3, Table 3). This corresponds well with the hypothesis of Martiny et al. (2006) on the biogeography of bacteria that at local scales, spatial variation in community structures is largely due to contemporary environmental factors, while at large distance scales, variations in community structures are largely due to historical contingencies. At intermediate scales, a mixture model of both parameters applies. This is supported by the Mantel correlogram, which did not show a linear decrease of community similarity with increasing geographic distance (Fig. 5). Our study is the first to confirm this hypothesis for microbial eukaryote communities. For soil ciliates, Foissner et al. (2008) showed tentative evidence that the split of Pangaea into Laurasia and Gondwana may have been an important historical contingency to relate contemporary ciliate distribution patterns.
Although the dispersal of freshwater protists over very long (intercontinental) geographic distances seems difficult (Ryšánek et al. 2015), one potential vector of dispersal is migratory land- and waterbirds (e.g. Maguire 1963; Figuerola & Green 2002). The Bale Mountains in particular has a large number of migratory birds, and lakes in the Altiplano have important populations of waterbirds. Both, however, are less important in the case of high-altitude lakes in the Alps (Boere et al. 2006). Further, although an African/west Eurasian migratory waterbird flyway is known, it is uncertain how strong this dispersal mechanism can be between the Alps and Bale Mountains. In addition, apparently no bird migratory flyways are known that will connect the three study areas (Boere et al. 2006). Another dispersal mechanism for protists may be transportation by wind. This vector is known to transport microorganisms even between continents (Smith et al. 2013), and for example, dust from areas such as the Sahara has been identified as an important source of fungi, bacteria and even algae (Favet et al. 2013). In particular, this vector could be relevant for lakes near the tropics (Bale Mts. and Altiplano) where an ice cover occurs only on a daily basis. However, airborne dispersal seems more common among terrestrial protists, which can be picked up with dust particles from dry soil, rather than for freshwater protists (Atkinson 1971; Foissner 2006, 2008). Thus, although we cannot totally exclude the importance of these vectors, we argue that isolation by distance seems a more plausible explanation for the observed long-distance patterns in high-mountain lake microbial eukaryote community structures (Fig. 4).
At least on regional scales, it seems likely that the most abundant organisms (large population sizes) have the highest chances of regional dispersal (Fenchel & Finlay 2004; Finlay & Fenchel 2004) (Fig. 5), corroborating with Hubbells unified neutral theory of biodiversity and biogeography (Hubbell 2001). This may explain the low degree of novel diversity detected in the two hundred most abundant phylotypes of the high-mountain lake microbial eukaryote consortia of the Alps Fig (Fig. 6a). Compared with the Chilean Altiplano lakes and the African Bale Mountain lakes, protistan morphotypes in European mountain lakes are much better studied (e.g. Stoeck et al. 2014, Kammerlander et al. 2015). Therefore, abundant species in European mountain lakes have better chances of being picked up and sequenced previously compared with abundant protists and fungi in the Altiplano and Bale Mountain regions. Protistan diversity in the latter two regions is largely uncharted resulting in a lack of gene data from these regions in public databases. This may be the most plausible explanation for a notably higher degree of novel diversity among the most abundant phylotypes of the Altiplano and Bale Mountain lakes (Fig. 6b,c) compared with the alpine lakes. This result further supports the finding that different geographic regions hold different sets of protistan and fungal diversity. Further diversity surveys in previously largely unexplored biogeographic regions would show how far we are indeed from uncovering earth’s diversity of microbial eukaryote life.
Supporting information
Additional supporting information may be found in the online version of this article.
Fig. S1 Taxonomic distribution (phylum-based) of all protistan OTUs among the replicate sequenced samples from the alpine lake Drachensee, Austria (DRA).
Fig. S2 Rarefaction curves for the different lakes (European Alps, Chilean Altiplano, Ethiopian Bale Mountains) based on Illumina target sequences.
Fig. S3 One-way ANOVA analysis of the Shannon-based effective number of species for each of the 19 high-mountain lake communities under study.
Fig. S4 Relationship between the pairwise community (Sorensen distance) and geographic distances (log-transformed) for the analysed high-mountain lakes (including summer samples).
Appendix S1 Scripts for the dereplication of reads and construction of an OTU contingency table.
Acknowledgements
We thank Josef Franzoi, Gry Larsen and Salvador Morales-Gomez (Austria) and Vilma Barrera (Chile) for water chemical analyses and Anna Waibel, Birgit Stenzel, Hannes Peter, Artur Hepberger (Austria), Eyasu Shuba (Ethiopia), Sergio Scott, Jose Carvajal, Juan Pablo Oyanedel and Ignacio Tobar (Chile) for helping during sampling. Furthermore, we thank four anonymous reviewers, who were extremely helpful and constructive with their comments and notably improved a previous version of this manuscript. Sampling in Chile was supported by a grant from CONICYT 1140543 to I.V., in Ethiopia by the APPEAR program to R.S. and in Austria by the Austrian Science Fund (FWF, Project BACK-ALP P24442-B25) to R.S.
Footnotes
T.S. and R.S. conceived and guided the study. R.S. and I.V. conducted sampling. S.F. performed laboratory work. S.F. and T.S. analysed data. T.S. and S.F. wrote the first version of the manuscript, and R.S. and I.V. added to subsequent versions. All authors read and approved the final manuscript.
Data accessibility
Sequence reads have been deposited to National Center for Biotechnology Information (NCBI)’s Sequence Read Archive and can be found under the Accession No. SRP065150. Custom scripts are provided in Appendix S1 (Supporting information).
References
- Alexander E, Stock A, Breiner HW, et al. Microbial eukaryotes in the hypersaline anoxic L’Atalante deep-sea basin. Environmental Microbiology. 2009;11:360–381. doi: 10.1111/j.1462-2920.2008.01777.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson KM. Further experiments in dispersal of phytoplankton by birds. Wildfowl. 1971;22:98–99. [Google Scholar]
- Auguet JC, Barberán A, Casamayor EO. Global ecological patterns in uncultured Archaea. The ISME Journal. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Barberán A, Casamayor EO. Global phylogenetic community structure and beta-diversity patterns of surface bacte-rioplankton metacommunities. Aquatic Microbial Ecology. 2010;59:1–10. [Google Scholar]
- Barberán A, Fernández-Guerra A, Auguet JC, Galand PE, Casamayor EO. Phylogenetic ecology of widespread uncultured clades of the Kingdom Euryarchaeota. Molecular Ecology. 2011;20:1988–1996. doi: 10.1111/j.1365-294X.2011.05057.x. [DOI] [PubMed] [Google Scholar]
- Bass D, Howe A, Brown N, et al. Yeast forms dominate fungal diversity in the deep oceans. Proceedings of the Royal Society B: Biological Sciences. 2007a;274:3069–3077. doi: 10.1098/rspb.2007.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evolutionary Biology. 2007b;7:1. doi: 10.1186/1471-2148-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. ICWSM. 2009;8:361–362. [Google Scholar]
- Bates ST, Clemente JC, Flores EF, et al. Global biogeography of highly diverse protistan communities in soil. The ISME Journal. 2012;7:652–659. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney C, Pawlowski J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proceedings of the Royal Society B: Biological Sciences. 2006;273:1867–1872. doi: 10.1098/rspb.2006.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boere GC, Galbraith CA, Stroud DA, editors. Waterbids around the World. The Stationery Office; Edinburgh, UK: 2006. p. 960. [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor EO, Massana R, Benlloch S, et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environmental Microbiology. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. [DOI] [PubMed] [Google Scholar]
- Catalan J, Camarero L, Felip M, et al. High mountain lakes: extreme habitats and witnesses of environmental changes. Limnetica. 2006;25:551–584. [Google Scholar]
- Countway PD, Gast RJ, Dennett MR, et al. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream) Environmental Microbiology. 2007;9:1219–1232. doi: 10.1111/j.1462-2920.2007.01243.x. [DOI] [PubMed] [Google Scholar]
- Csárdi G, Nepusz T. The igraph software package for complex network research. International Journal of Complex Systems. 2006;1695:1–9. [Google Scholar]
- Diniz-Filho JAF, Soares TN, Lima JS, et al. Mantel test in population genetics. Genetics and Molecular Biology. 2013;36:475–485. doi: 10.1590/S1415-47572013000400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan JR, Pierce RW, Yang EJ, Kim SY. Southern Ocean biogeography of tintinnid ciliates of the marine plankton. Journal of Eukaryotic Microbiology. 2012;59:511–519. doi: 10.1111/j.1550-7408.2012.00646.x. [DOI] [PubMed] [Google Scholar]
- Dorador C, Pardo R, Vila I. Temporal variations of physical, chemical and biological parameters of a high altitude lake: the case of Chungara lake. Revista Chilena de Historia Natural. 2003;76:15–22. [Google Scholar]
- Dunthorn M, Stoeck T, Wolf K, Breiner HW, Foissner W. Diversity and endemism of ciliates inhabiting Neotropical phytotelmata. Systematics and Biodiversity. 2012;10:195–205. [Google Scholar]
- Dunthorn M, Otto J, Berger SA, et al. Placing environmental next-generation sequencing amplicons from microbial eukaryotes into a phylogenetic context. Molecular Biology and Evolution. 2014;31:993–1009. doi: 10.1093/molbev/msu055. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favet J, Lapanje A, Giongo A, et al. Microbial hitchhikers on intercontinental dust: catching a lift in Chad. The ISME Journal. 2013;7:850–867. doi: 10.1038/ismej.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felip M, Wille A, Sattler B, Psenner R. Microbial communities in the ice and snow cover of an alpine lake: evidence for general patterns, and lake-cover relationship. Aquatic Microbial Ecology. 2002;29:123–134. [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: patterns of local and global diversity. BioScience. 2004;54:777–784. [Google Scholar]
- Figuerola J, Green A. Dispersal of aquatic organisms by waterbirds: a review of past research and priorities for future studies. Freshwater Biology. 2002;47:483–494. [Google Scholar]
- Filker S, Gimmler A, Dunthorn M, Mahé F, Stoeck T. Deep sequencing uncovers protistan plankton diversity in the Portuguese Ria Formosa solar saltern ponds. Extremophiles. 2015;19:283–295. doi: 10.1007/s00792-014-0713-2. [DOI] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Clarke KJ. Apparent global ubiquity of species in the protist genus Paraphysomonas. Protist. 1999;150:419–430. doi: 10.1016/S1434-4610(99)70042-8. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Fenchel T. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist. 2004;155:237–244. doi: 10.1078/143446104774199619. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozoologica. 2006;45:111–136. [Google Scholar]
- Foissner W. Protist diversity and distribution: some basic considerations. Biodiversity and Conservation. 2008;17:235–242. [Google Scholar]
- Foissner W, Strueder-Kypke M, van der Staay G, Moon-van der Staay SY, Hackstein JH. Endemic ciliates (Protozoa, Ciliophora) from tank bromeliads (Bromeliaceae): a combined morphological, molecular and ecological study. European Journal of Protistology. 2003;39:365–372. [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodiversity and Conservation. 2008;17:345–363. [Google Scholar]
- Forster D, Behnke A, Stoeck T. Meta-analyses of environmental sequence data identify anoxia and salinity as parameters shaping ciliate communities. Systematics and Biodiversity. 2012;10:277–288. [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Computational and Applied Biosciences. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gast CJ. Microbial biogeography: the end of the ubiquitous dispersal hypothesis? Environmental Microbiology. 2015;17:544–546. doi: 10.1111/1462-2920.12635. [DOI] [PubMed] [Google Scholar]
- Gomaa F, Yang J, Mitchell EA, et al. Morphological and molecular diversification of Asian endemic Difflugia tuberspinifera (Amoebozoa, Arcellinida): a case of fast morphological evolution in protists? Protist. 2015;166:122–130. doi: 10.1016/j.protis.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Haegeman B, Hamelin J, Moriarty J, et al. Robust estimation of microbial diversity in theory and in practice. The ISME Journal. 2013;7:1092–1101. doi: 10.1038/ismej.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32) Vol. 32 Princeton University Press; Princeton: 2001. [Google Scholar]
- Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genetics. 2005;6 doi: 10.1186/1471-2156-6-13. v.3.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- Jost L. Partitioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- Jumars PA. Concepts in Biological Oceanography: An Interdisciplinary Primer. Oxford University Press; New York: 1993. [Google Scholar]
- Kammerlander B, Breiner HW, Filker S, et al. High diversity of protistan plankton communities in remote high mountain lakes in the European Alps and the Himalayan mountains. FEMS Microbiology Ecology. 2015;91:fiv010. doi: 10.1093/femsec/fiv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, McManus GB, Snoeyenbos-West OLO, et al. Reframing the Microbial ‘Everything is Everywhere’ Debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquatic Microbial Ecology. 2005;41:55–65. [Google Scholar]
- Knoll AH, Walter MR, Narbonne GM, Christi-Blick N. A new period for the geologic time scale. Science. 2004;305:621–622. doi: 10.1126/science.1098803. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology. 3rd edn. Elsevier; Amsterdam: 2012. [Google Scholar]
- Lepère C, Domaizon I, Debroas D. Community composition of lacustrine small eukaryotes in hyper-eutrophic conditions in relation to top-down and bottom-up factors. FEMS Microbiology Ecology. 2007;61:483–495. doi: 10.1111/j.1574-6941.2007.00359.x. [DOI] [PubMed] [Google Scholar]
- Löffler H. Limnological and paleolimnological data on the Bale Mountain Lakes (Ethiopia) Verhandlungen der Internationalen Vereinigung für Theoretische und Angewandte Limnologie. 1978;20:1131–1138. [Google Scholar]
- Logares R, Boltovskoy A, Bensch S, Laybourn-Parry J, Rengefors K. Genetic diversity patterns in five protist species occurring in lakes. Protist. 2009a;160:301–317. doi: 10.1016/j.protis.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Logares R, Bråte J, Bertilsson S, et al. Infrequent marine–freshwater transitions in the microbial world. Trends in Microbiology. 2009b;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- López-García P, Rodriguez-Valera F, Pedrós-Alió C, et al. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Lovejoy C, Massana R, Pedrós-Alió C. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Applied and Environmental Microbiology. 2006;72:3085–3095. doi: 10.1128/AEM.72.5.3085-3095.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur R. Patterns of species diversity. Biological Reviews. 1965;40:510–533. [Google Scholar]
- Maguire BJ. The passive dispersal of small aquatic organisms and their colonisation of isolated bodies of water. Ecological Monographs. 1963;33:161–185. [Google Scholar]
- Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Reserach. 1967;27:209–220. [PubMed] [Google Scholar]
- Márquez-García M, Vila I, Hinojosa LF, et al. Distribution and seasonal fluctuations in the aquatic biodiversity of the southern Altiplano. Limnologica-Ecology and Management of Inland Waters. 2009;39:314–318. [Google Scholar]
- Martiny JB, Bohannan BJ, Brown JH, et al. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Massana R, Pedrós-Alió C. Unveiling new microbial eukaryotes in the surface ocean. Current Opinion in Microbiology. 2008;11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Massana R, Gobet A, Audic S, et al. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environmental Microbiology. 2015;17:4035–4049. doi: 10.1111/1462-2920.12955. [DOI] [PubMed] [Google Scholar]
- Mühlhauser H, Hrepic H, Mladinic P, Montecino V, Cabrera S. Water quality and limnological features of a high altitude Andean lake, Chungara, in northern Chile. Revista Chilena de Historia Natural. 1995;68:341–349. [Google Scholar]
- Nebel ME, Wild S, Holzhauser M, et al. JAGUC: a software package for environmental diversity analyses. Journal of Bioinformatics and Computational Biology. 2011;9:749–773. doi: 10.1142/s0219720011005781. [DOI] [PubMed] [Google Scholar]
- Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS ONE. 2014;9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oden N, Sokal RR. Directional autocorrelation: an extension of spatial correlograms to two dimensions. Systematic Zoology. 1986;35:608–617. [Google Scholar]
- Oikonomou A, Filker S, Breiner HW, Stoeck T. Protistan diversity in a permanently stratified meromictic lake (Lake Alatsee, SW Germany) Environmental Microbiology. 2015;17:2144–2157. doi: 10.1111/1462-2920.12666. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. vegan: Community Ecology Package. 2013 R package version 2.0-10. [Google Scholar]
- Patterson DJ. The diversity of eukaryotes. American Naturalist. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Pomeroy LA, Williams PJL, Azam F, Hobbie JE. The microbial loop. Oceanography. 2007;20:28–33. [Google Scholar]
- Rezácová M, Neustupa J. Distribution of the genus Mallomonas (Synurophyceae) – Ubiquitous dispersal in microorganisms evaluated. Protist. 2007;158:29–37. doi: 10.1016/j.protis.2006.06.004. [DOI] [PubMed] [Google Scholar]
- R-Team. R. A language and environment for statistical computing. Vienna, Austria; 2008. [Google Scholar]
- Ryšánek D, Hrčková K, Škaloud P. Global ubiquity and local endemism of free-living terrestrial protists: phylogeo-graphic assessment of the streptophyte alga Klebsormidium. Environmental Microbiology. 2015;17:689–698. doi: 10.1111/1462-2920.12501. [DOI] [PubMed] [Google Scholar]
- Sanders RW. Protists. In: Likens GE, editor. Encyclopedia of Inland Waters. Elsevier; Oxford, UK: 2009. pp. 252–260. [Google Scholar]
- Simon M, López-García P, Deschamps P, et al. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME Journal. 2015 doi: 10.1038/ismej.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šlapeta JM, Moreira D, López-García P. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proceedings of the Royal Society B: Biological Sciences. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Timonen HJ, Jaffe DA, et al. Intercontinental dispersal of bacteria and archaea by transpacific winds. Applied and Environmental Microbiology. 2013;79:1134–1139. doi: 10.1128/AEM.03029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommaruga R. The role of solar UV radiation in the ecology of alpine lakes. Journal of Photochemistry and Photobiology B: Biology. 2001;62:35–42. doi: 10.1016/s1011-1344(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Sommaruga R, Oberleiter A, Psenner R. Effect of UV radiation on the bacterivory of a Heterotrophic nanoflagellate. Applied and Environmental Microbiology. 1996;62:4395–4400. doi: 10.1128/aem.62.12.4395-4400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A, Breiner HW, Pachiadaki M, et al. Microbial eukaryote life in the new hypersaline deep-sea basin Thetis. Extremophiles: Life under Extreme Conditions. 2012;16:21–34. doi: 10.1007/s00792-011-0401-4. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Jost S, Boenigk J. Multigene phylogenies of clonal Spumella-like strains, a cryptic heterotrophic nanoflagellate, isolated from different geographical regions. International Journal of Systematics and Evolutionary Microbiology. 2008;58:716–724. doi: 10.1099/ijs.0.65310-0. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Molecular Ecology. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Breiner HW, Filker S, et al. A morphogenetic survey on ciliate plankton from a mountain lake pinpoints the necessity of lineage-specific barcode markers in microbial ecology. Environmental Microbiology. 2014;16:430–444. doi: 10.1111/1462-2920.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taib N, Mangot JF, Domaizon I, et al. Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshwater protist diversity. PLoS ONE. 2013;8:e58950. doi: 10.1371/journal.pone.0058950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triadó-Margarit X, Casamayor EO. Genetic diversity of planktonic eukaryotes in high mountain lakes (Central Pyrenees, Spain) Environmental Microbiology. 2012;14:2445–2456. doi: 10.1111/j.1462-2920.2012.02797.x. [DOI] [PubMed] [Google Scholar]
- Van de Vijver B, Gremmen NJM, Bayens L. The genus Stauroneis (Bacillariophyceae) in the Antarctic region. Journal of Biogeography. 2005;32:1791–1798. [Google Scholar]
- de Vargas C, Audic S, Henry N, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348 doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- Weisse T. Distribution and diversity of aquatic protists: an evolutionary and ecological perspective. Biodiversity and Conservation. 2008;17:243–259. [Google Scholar]
- Worden AZ, Cuvelier ML, Bartlett DH. In-depth analyses of marine microbial community genomics. Trends in Microbiology. 2006;14:331–336. doi: 10.1016/j.tim.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wu QL, Chatzinotas A, Wang J, Boenigk J. Genetic diversity of eukaryotic plankton assemblages in Eastern Tibetan Lakes differing by their salinity and altitude. Microbial Ecology. 2009;58:569–581. doi: 10.1007/s00248-009-9526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiology Ecology. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.