Abstract
Inhibition of a main regulator of cell metabolism, the protein kinase mTOR, induces autophagy and inhibits cell proliferation. However, the molecular pathways involved in the cross-talk between these two mTOR-dependent cell processes are largely unknown. Here we show that the scaffold protein AMBRA1, a member of the autophagy signalling network and a downstream target of mTOR, regulates cell proliferation by facilitating the dephosphorylation and degradation of the proto-oncogene C-MYC. We found that AMBRA1 favors the interaction between C-MYC and its phosphatase PP2A and that, when mTOR is inhibited, it enhances PP2A activity on this specific target, thereby reducing the cell division rate. As expected, such a de-regulation of C-MYC correlates with increased tumorigenesis in AMBRA1-defective systems, thus supporting a role for AMBRA1 as a haploinsufficient tumour suppressor gene.
Keywords: cancer, cell cycle, cell metabolism, mTOR, phosphatases
Introduction
As a response to nutrient deprivation and other cell stressors, autophagy is often induced in the context of reduced or arrested cell growth.1 A plethora of signalling molecules and pathways have opposing effects on cell growth and autophagy, supporting the idea that these processes might represent mutually exclusive cell fates.2 The serine/threonine kinase mTOR (mammalian target of Rapamycin) integrates signals affecting both these pathways.3, 4 Indeed, mTOR has a major role in the regulation of protein synthesis, this in turn leading to cell growth inhibition, while also phosphorylating the pro-autophagic complex ULK1 and AMBRA1, resulting in the inhibition of autophagy.4–8 Thus, autophagy inhibition by mTOR is known to occur independently of the mTOR targets related to cell growth.2
A phosphatase known as a cell cycle regulator, the Protein Phosphatase 2A (PP2A), has been recently shown to also exert both a positive and negative effect on autophagy, depending on the particular step on which it acts and on its composition in subunits.9–11 PP2A is usually a complex, containing a catalytic, a scaffold and a regulatory subunit. At least 100 different PP2A heterotrimeric complexes formed through combinatorial association of these subunits have been identified to date, thus mediating different specific physiological functions.12, 13
Recent studies have identified PP2A targets, whose dephosphorylation is critical for this phosphatase’s tumour suppressor activity:14–16 the transcription factor C-MYC is one such target. Inhibition of PP2A activity induces C-MYC Serine 62 (C-MYCS62) phosphorylation and C-MYC protein stabilization, enhancing cell proliferation and impacting on tumorigenesis.17–19 Upon growth-factor withdrawal, a massive dephosphorylation of C-MYC occurs, targeting the protein at the proteasome.19
In this work, we characterize the pro-autophagic protein AMBRA1, a member of the autophagy signalling network in vertebrates,8, 20, 21 as an endogenous PP2A-interacting protein. In particular, upon autophagy induction, AMBRA1 is phosphorylated by the kinase ULK1 and promotes autophagosome formation through the activation of the Class III PI3K complex.20, 21 Moreover, AMBRA1-mediated stabilization of ULK1 through regulative ubiquitination enhances autophagy in a positive feedback loop.8
In vivo inactivation of Ambra1 gives rise to defects in the developing nervous system and results in embryonic death (Ambra1gt/gt mice).20, 22 In addition, an evident hyperproliferative phenotype has been associated with Ambra1 depletion, both in vitro and in vivo even though Ambra1’s role in cell cycle regulation remains completely unexplored.20, 23 Our results show that AMBRA1 enhances PP2A activity in C-MYCS62 dephosphorylation and thereby destabilizes C-MYC. Also, we demonstrate that Ambra1 monoallelic deficiency is responsible for hyperproliferation, mainly dependent on the interaction with PP2A and on the stabilization of C-MYC. Moreover, this AMBRA1- and PP2A-mediated regulation of C-MYC is controlled by mTOR. As expected, such a de-regulation of the oncogene C-MYC correlates with increased tumorigenesis in AMBRA1-defective systems, unravelling AMBRA1 as a haploinsufficient tumour suppressor gene.
Results
Ambra1 dosage affects cell proliferation
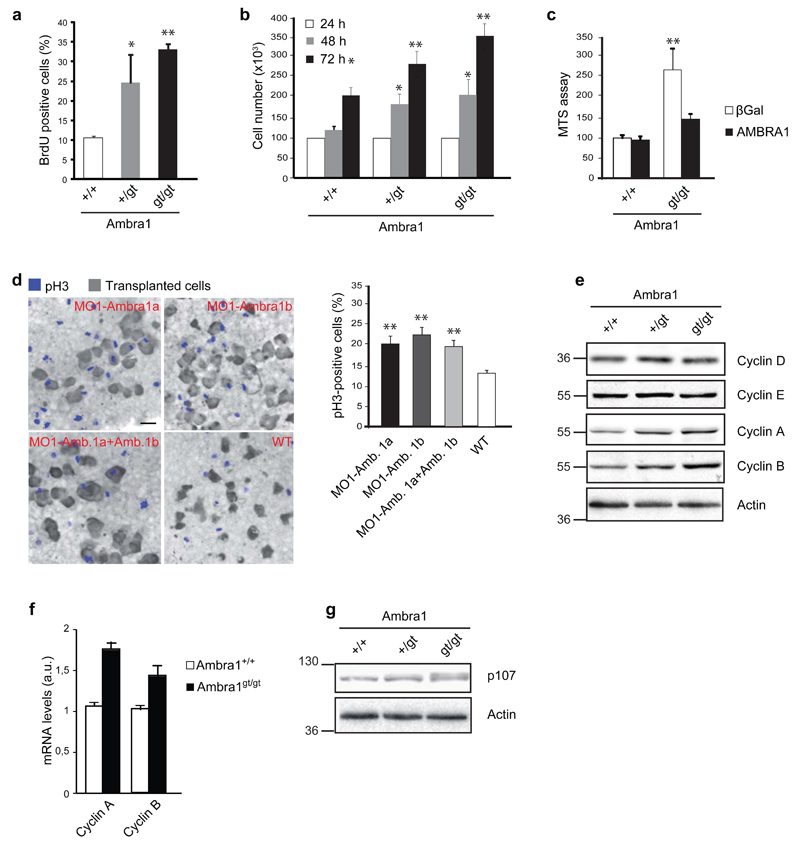
In order to functionally characterize the AMBRA1’s role in cell proliferation, we generated Mouse Embryonic Fibroblast (MEFs) isolated from embryos wild-type (Ambra1+/+), heterozygous (Ambra1+/gt) or homozygous (Ambra1gt/gt) for the gene-trap muation in the Ambra1 locus,20 and we evaluated the cell growth rate by BromodeoxyUridine (BrdU) staining and cell counting (Fig. 1a-b). We confirmed the hyperproliferative phenotype of Ambra1gt/gt cells,20, 23 and highlighted an increased proliferation rate in Ambra1+/gt with respect to wild-type cells. Importantly, the Ambra1 depletion-elicited increase in cell growth is almost completely abolished by reconstitution of AMBRA1 levels in transformed (by RasV12/E1A expression) Ambra1gt/gt MEFs, as demonstrated by an MTS assay (Fig. 1c). Moreover, the cell-autonomous capability of Ambra1-depleted cells to hyperproliferate is supported by the transplantation of zebrafish morphant ambra1a or ambra1b cells23 in a wild-type acceptor embryo (Fig. 1d).
Fig. 1. Ambra1 hemizygousity affects cell proliferation.
a) The proliferation rate of MEFs wild-type (+/+), heterozygous (+/gt) and homozygous (gt/gt) for the gene-trap mutation in the Ambra1 locus was measured by BrdU-incorporation assay. A staining by using anti-BrdU antibody was performed and BrdU-positive cells were counted. Data are presented as means±s.d. and significance is *P<0.05, **P<0.005 (n=3 independent experiments). b) Cell counting of +/+, +/gt and gt/gt MEFs after 24, 48 and 72 hours of growth. Data are presented as means±s.d. and significance is *P<0.05, **P<0.005 (n=3 independent experiments). c) MEFs +/+ and gt/gt were immortalized through infection with RasV12 and E1A oncogenes. Subsequently, gene-trap MEFs were reconstituted for AMBRA1 by lentiviral infection; wild-type cells were infected with lentiviruses encoding for βGal, as a control. Data are presented as means±s.d. and significance is **P<0.005 (n=3 independent experiments). d) Zebrafish-embryo cells injected with Morpholinos (MOs) against Ambra1 mRNA (MO1-Ambra1a, MO1-Ambra1b or both) were transplanted into wild-type embryos. The proliferation of the injected cells, reported in the graph, was calculated by counting pH3-positive cells (blue cells) with respect to the total implanted cells (grey cells). Scale bar, 20μm. Data are presented as means±s.d. and significance is **P<0.005 (n=3 independent experiments). e) Protein extracts of +/+, +/gt and gt/gt MEFs were analysed by western blot, using antibodies against Cyclin D, E, A, B and Actin. f) mRNA levels of Cyclin A and B were analysed by real-time PCR in MEFs +/+ and gt/gt. Data are presented as means±s.e.m. and significance is *P<0.05, **P<0.005 (n=3 independent experiments). g) Electrophoretic mobility of p107 was analysed by western blot analysis in +/+, +/gt and gt/gt MEFs. An anti-p107 and an anti-Actin antibody were used.
Further, the steady-state expression levels of some positive regulators of cell cycle were evaluated in lysates from MEFs of different genotype. All proteins analyzed (Cyclin D, E, A and B) are responsible for the exit from the G0 phase and for the transition from one to the subsequent phase of the cell cycle.24 Unexpectedly, only Cyclin A and B were up-regulated in Ambra1 defective MEFs (Fig. 1e), suggesting an enrichment in cells at S and M phase of the cell cycle.24 Given that such up-regulation could be due to transcriptional or post-translational control of Cyclins, we analyzed Cyclin gene expression in Ambra1+/+ and Ambra1gt/gt MEFs. As shown in Figure 1f, the Cyclin A and B mRNAs are upregulated in Ambra1-deficient cells.
Primed by this result, we analyzed the phosphorylation of a Retinoblastoma family protein, p107, an event related to Cyclins A and B transcription, in Ambra1-defective MEFs. A p107 band with lower electrophoresis mobility appears in Ambra1gt/gt MEFs (Fig. 1g), suggesting that Ambra1 can be important for the dephosphorylation of p107.26 Intriguingly, p107 has been identified as a target of the phosphatase PP2A, involved in autophagy regulation.9, 10, 25
To summarize, these analyses demonstrate that the loss of a single Ambra1 allele is sufficient to increase cell proliferation and that this hyperproliferative phenotype correlates with increased transcription of Cyclin A and B and inactivation of their transcriptional inhibitor p107.
AMBRA1 a interactor of PP2A
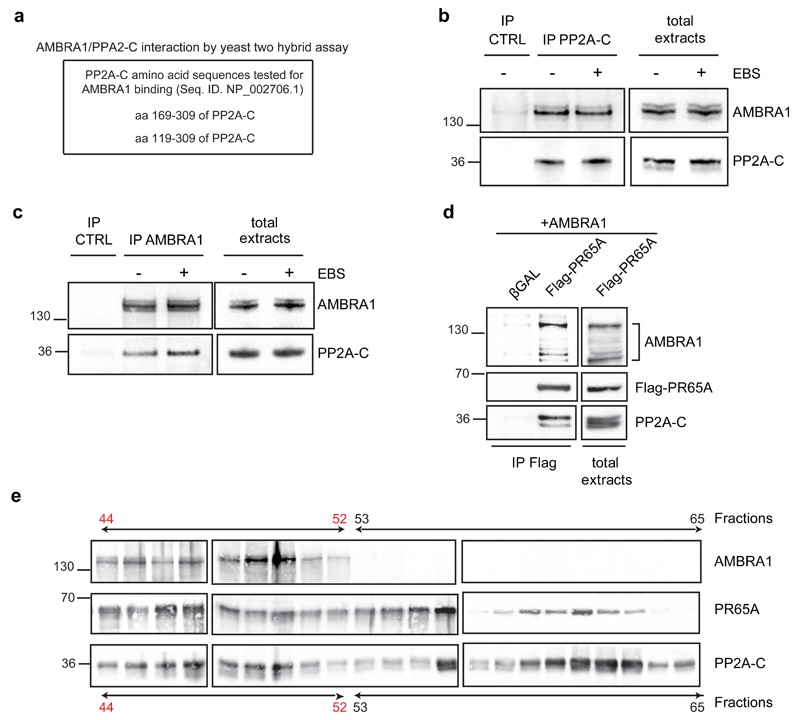
We identified the catalytic subunit of PP2A as an AMBRA1 interactor by both yeast two-hybrid and tandem affinity purification approaches (Fig. 2a, Supplementary Table 1). To verify this interaction, co-immunoprecipitation analyses were performed from endogenous extracts of HeLa cells (Fig. 2b-c). We found that, independently of autophagy modulation, endogenous PP2A-C interacts with AMBRA1. To map the AMBRA1/PP2A-C interaction we tested a number of AMBRA1 constructs for their capacity to co-immunoprecipitate with PP2A-C. Only fragments corresponding to amino acids 181-532 and 946-1269 were able to bind PP2A-C (regions F1WD40 and F3b, Supplementary Fig. 1). Of note, even though the regulative B-subunits of PP2A are usually characterized by WD40 domains, allowing both their association to the PP2A complex and the substrate recognition,12, 13 the AMBRA1 regions here identified contain no WD40 domains (otherwise present at its N-terminus).
Fig. 2. AMBRA1 is an interactor of PP2A.
a) AMBRA1/PP2A-C interaction was identified by yeast two-hybrid assay. Yeast cells were co-transfected with a plasmid coding for AMBRA20 and one coding for the reported regions of PP2A-C protein. The unities of βGal and the number of the clones positive for the screening are 5.7 and 4, respectively. b) Endogenous protein extracts were immunoprecipitated using an anti-PP2A-C antibody (IP PP2A-C) and mouse Immonuglobulins as control (IP CTRL). Purified complexes and corresponding total extracts were analysed by western blot using an anti-AMBRA1 and anti-PP2A-C antibodies. EBS: Earle’s Balanced Salts; an autophagy-inducer medium. c) Endogenous protein extracts were immunoprecipitated using an anti-AMBRA1 antibody (IP AMBRA1) and rabbit Immonuglobulins as control (IP CTRL). Purified complexes and corresponding total extracts were analysed as in a. d) HEK293 cells were co-transfected with vectors encoding AMBRA1 and Flag-PR65A. Protein extracts were immunoprecipitated using an anti-Flag antibody. Purified complexes and corresponding total extracts were analysed by western blot using an anti-AMBRA1, anti-Flag and anti-PP2A-C antibodies. The bracket indicates the bands corresponding to Flag-AMBRA1. e) Protein extracts of HeLa cells was separated in different fractions and the presence of AMBRA1, PR65A and PP2A-C in the different fractions was assessed by western blot, using an anti-AMBRA1, anti-PR65A and anti-PP2A-C antibodies.
As mentioned above, the catalytic subunit of PP2A (PP2A-C), together with the regulatory (B subunit) and the scaffold (PR65A) subunits, constitutes a holoenzyme which also other PP2A-C interactors can join.12, 13 Indeed, AMBRA1 can also interact with PR65A as long with PP2A-C (Fig. 2d, Supplementary Fig. 2a-b), and the three proteins are in a high molecular weight multimeric complex (Fig. 2e, fractions 44-53, in red). As expected, heterodimers of PP2A-C and PR65A can also associate in the absence of AMBRA1 (Fig. 2e, fractions 53-63).
AMBRA1 promotes PP2A activity on C-MYC
PP2A-C is known to bind scaffold proteins or different regulatory subunits that mediate specific substrate docking.15 This evidence, together with our findings concerning the roles of AMBRA1 and its interaction with PP2A-C, suggests that AMBRA1 may regulate PP2A activity on some substrates, thus controlling cell proliferation.
To verify this hypothesis, and given the effect of Ambra1 depletion on the PP2A substrate p107,25 we also analysed the phosphorylation level of C-MYC, identified as another crucial target of PP2A.18
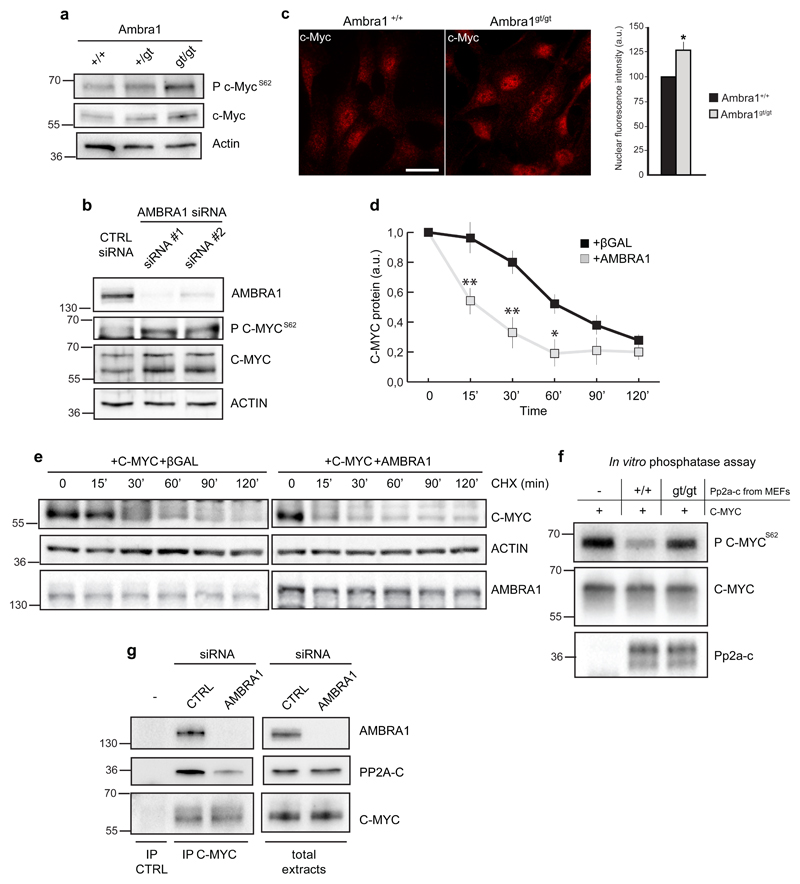
Interestingly, we found that Ambra1 depletion in MEFs or AMBRA1 downregulation in HeLa cells increases C-MYCS62 phosphorylation (P C-MYCS62) relative to total C-MYC (Fig. 3a-b). In addition, a slight increase in the levels of total C-MYC is observed in cells devoid of Ambra1 (Fig. 3a-b and Supplementary Fig. 2c-d), likely as a consequence of C-MYC stabilization through Ser62 phosphorylation; Further, c-Myc nuclear fraction (functional in controlling cell proliferation) is also increased in Ambra1-depleted cells (Fig. 3c). By contrast, no marked differences were observed in the expression or phosphorylation of other known targets of PP2A-C (Supplementary Fig. 3a-b),15 arguing for high specificity of AMBRA1 role in this regulation. Also, we ruled out the possible involvement of the c-Myc protein kinases17 in the observed de-regulation of c-Myc upon Ambra1 depletion, by analysing Erk1/2 phosphorylation levels (Supplementary Fig. 3b). Finally, increased AMBRA1 levels are able to induce a diminished half-life of total C-MYC (Fig. 3d-e). Taken together, these results demonstrate that AMBRA1 promotes C-MYC dephosphorylation and destabilization. Primed by these findings, we set out to establish, by means of an in vitro phosphatase assay, whether AMBRA1 enhanced PP2A-C activity on C-MYC. To this end, C-MYC immunoprecipitates from P C-MYCS62-enriched cells were incubated with independent pools of Pp2a-c isolated from wild-type (Ambra1+/+) or Ambra1-deficient (Ambra1gt/gt) MEFs (Fig. 3f). Interestingly, Ambra1 depletion clearly decreased PP2A-C activity on P C-MYCS62.
Fig. 3. AMBRA1 affects C-MYC de-phosphorylation at Ser62.
a) Protein extracts of MEFs wild-type (+/+), heterozygous (+/gt) and homozygous (gt/gt) for the gene-trap mutation in the Ambra1 locus were analysed by western blot, using antibodies against c-Myc phosphorylated at the Ser62 (P c-MycS62), total c-Myc and Actin. b) Protein extracts of HeLa cells knocked-down for AMBRA1 (siRNA #1 and #2) or treated with aspecific oligonucleotides (CTRL siRNA), as a control, were analysed by western blot, using antibodies against P C-MYCS62, total C-MYC and ACTIN. c) Immunostaining of c-Myc in +/+ and gt/gt MEFs. Scale bar, 20 μm. Fluorescence intensity per cell was quantified by imagej software, as previously described.53 Data are presented as means±s.e.m. and significance is *P<0.05 (n=100 cells pooled from 3 independent experiments in which 6-8 fields were assessed per experiment). d-e) Cells co-transfected with C-MYC and AMBRA1 or βGal, as a control, were treated with cycloheximide and harvested at the indicated time points. Protein extracts of cells were analysed by western blot, using antibodies against C-MYC, AMBRA1 and ACTIN. Quantification is shown in d. Data are presented as mean±s.d. and significance is *P<0.5 and **P<0.05 (n=3 independent experiments). f) Different phosphatase reactions were performed in vitro, using Pp2a-c extracted from +/+ or gt/gt cells, incubated with the same amount of C-MYC. C-MYC immunoprecipitation plus mouse immunoglobulin mix, instead of anti-Pp2a-c, was used as a negative control. The products of the reactions were analysed by western blot, using antibodies against P C-MYCS62, total C-MYC and Pp2a-c. g) HeLa cells were knocked-down for AMBRA1 (AMBRA1 siRNA) or treated with aspecific oligonucleotides (CTRL siRNA), as a control. Endogenous C-MYC was immunoprecipitated using an anti-C-MYC antibody (IP C-MYC) and mouse immunoglobulins were used as control (IP CTRL). The immunoprecipitated complexes were analysed by western blot with an anti-AMBRA1, anti-C-MYC and anti-PP2A-C antibodies.
Next, we found that endogenous C-MYC is in the same complex with AMBRA1 and PP2A-C and that AMBRA1 depletion by transient knock-down affects the interaction between C-MYC and PP2A-C (Fig. 3g).
Taken together, these experiments provide biochemical evidence that AMBRA1 favours the recruitment of PP2A to C-MYC, enhances C-MYC-associated PP2A activity, so decreasing P C-MYCS62 and resulting in the de-stabilization of the protein.
mTOR is upstream of AMBRA1 in controlling C-MYC
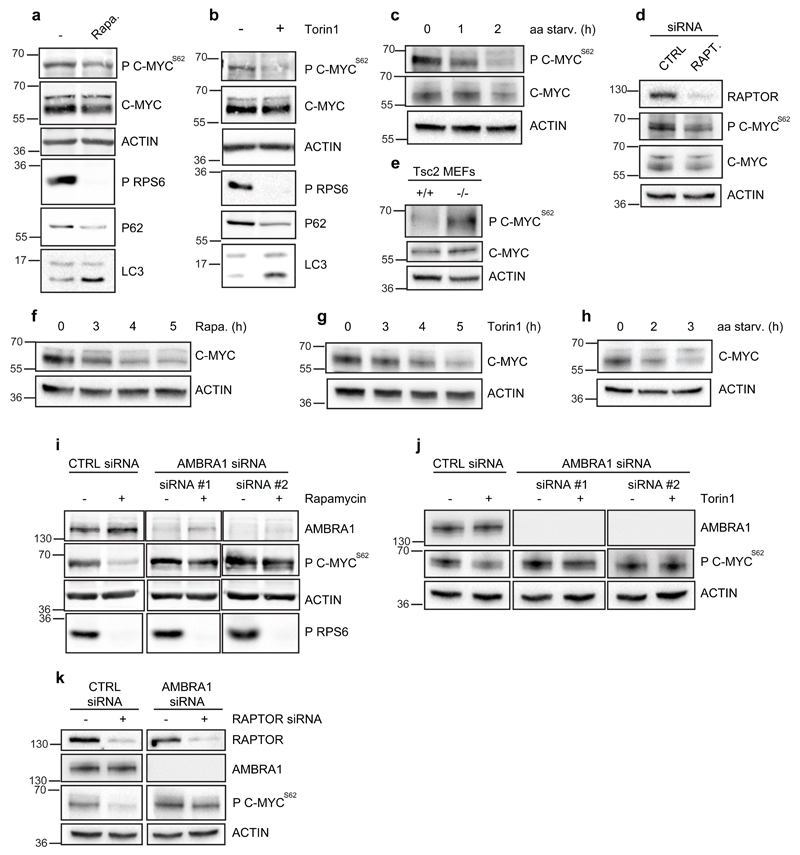
Although mTOR is considered a key regulator of both cell growth and autophagy,2, 3 the mTOR target molecules in this co-regulation have not yet been identified. We have shown that mTOR-mediated phosphorylation on AMBRA1 results in the negative regulation of autophagy.8 Thus, we verified whether mTOR could also affect AMBRA1/PP2A-C-mediated dephosphorylation of C-MYC. We found C-MYC to be dephosphorylated at Ser62 upon mTOR inhibition by both Rapamycin and Torin1 treatments and by amino acid starvation (Fig. 4a-c); We also verified the involvement of mTOR in the regulation of C-MYC by targeting RAPTOR, a component of mTORC1 complex, whose depletion results in mTOR inhibition.26 As expected, also in this case, C-MYC dephosphorylation on Ser62 was observed (Fig. 4d). By contrast, cells lacking the mTORC1 inhibitor Tsc227 are characterized by high levels of P c-MycS62 (Fig. 4e). In line with the role of C-MYCS62 phosphorylation in this protein stability, a decrease of total C-MYC is also observed upon prolonged mTOR inhibition (Fig. 4f-h). Further, by depleting AMBRA1 upon mTOR negative regulation, we also found that AMBRA1 is required to maintain mTOR’s inhibitory effect on C-MYC (Fig. 4i-k).
Fig. 4. Inhibition of mTOR affects C-MYC phosphorylation at Ser62 in an AMBRA1-dependent manner.
a) HeLa cells were treated with Rapamycin for 2h and protein extracts were analysed by western blot using antibodies against P C-MYCS62, total C-MYC, ACTIN, phospho-RPS6, p62 and LC3. b) HeLa cells were treated with Torin1 for 2h and protein extracts were analysed by western blot using the same antibody as in a. The bracket indicates bands corresponding to AMBRA1. c) The levels of P C-MYCS62 were measured in HeLa cells upon amino acid starvation (aa starv.); protein extracts were analysed by western blot using antibodies against P C-MYCS62, total C-MYC and ACTIN. d) Protein extracts of HeLa cells knocked-down for RAPTOR (RAPT.) or treated with aspecific oligonucleotides (CTRL), as a control, were analysed by western blot, using the anti-RAPTOR antibody and the same antibodies as in c. e) Protein extracts of immortalized MEFs, wild-type (+/+) and knock-out (-/-) for Tsc2, were analysed by western blot, using the same antibodies as in c. f) HeLa cells were treated with Rapamycin for 3h, 4h and 5h. Protein extracts were analysed by western blot using the antibodies against total C-MYC and ACTIN. g) HeLa cells were treated with Torin1 3h, 4h and 5h. Protein extracts were analysed by western blot using the same antibodies as in f. h) HeLa cells were amino acid starved and protein extracts were analysed by western blot using the same antibodies as in f. i) HeLa cells treated with oligo-interference against AMBRA1 (siRNA #1 and #2) or with aspecific oligonucleotides (CTRL siRNA) were incubated with Rapamycin for 2h. Protein extracts were analysed by western blot using anti-AMBRA1, anti-P C-MYCS62, ACTIN, and phospho-RPS6 antibodies. j) HeLa cells treated as in i were incubated with Torin1 for 2h. Protein extracts were analysed by western blot using antibodies against AMBRA1, P C-MYCS62 and ACTIN. k) HeLa cells were first treated with oligo-interference against AMBRA1 (AMBRA1 siRNA) and, after 24h, with oligos against RAPTOR (+RAPTOR siRNA). For each transfection, suitable aspecific oligonucleotides (CTRL siRNA; -RAPTOR siRNA) were used. Protein extracts were analysed by western blot using antibodies against RAPTOR, AMBRA1, P C-MYCS62 and ACTIN.
AMBRA1 controls cell proliferation by C-MYC regulation
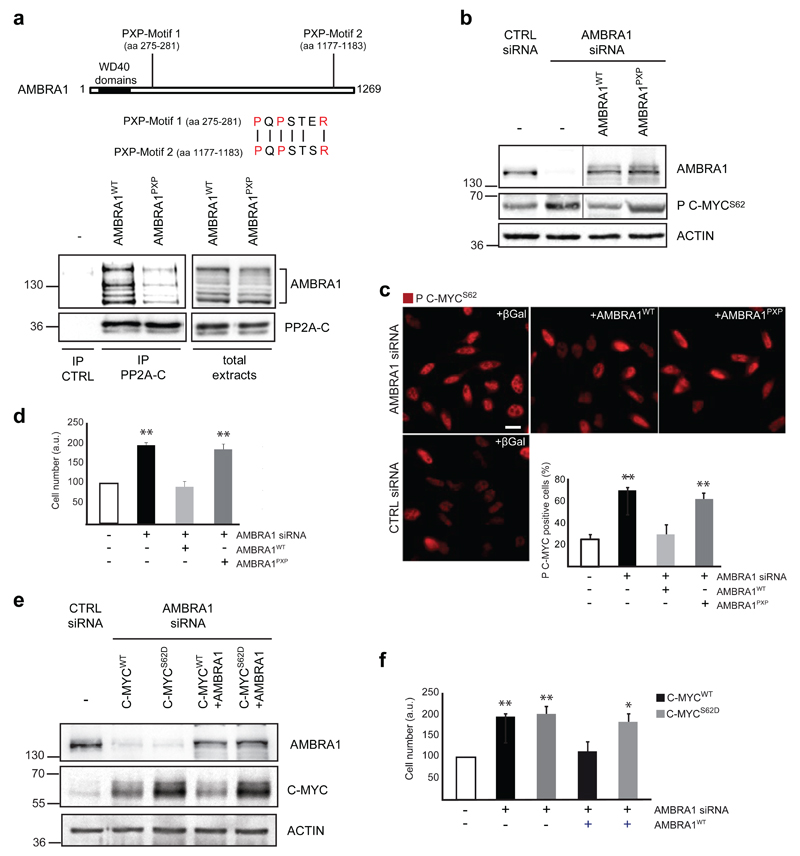
To verify the impact of the AMBRA1/PP2A/C-MYC interaction on cell cycle regulation, we generated a mutant AMBRA1 incapable of binding PP2A-C. In order not to affect AMBRA1 interaction with its additional binding partners, we used a site-specific mutation approach. AMBRA1 is an intrinsically disordered protein,28 having no domains but the WD40s. For this reason, we hypothesized the presence of a linear motif in the AMBRA1 sequence to be responsible for binding PP2A-C. By analysing the amino acid sequences of both regions previously identified as responsible for PP2A-C interaction (F1ΔWD40 and F3b, respectively; Supplementary Fig. 1), and as independently capable of binding PP2A-C, we identified a single conserved peptide with the sequence PQPSTxR. The motif PxPxxxR is reminiscent of SH3 binding motifs, which consist of a PxxP core with a positively charged residue either preceding or following this consensus29 and already shown as relevant for PP2A binding.30 We thus mutated both potential binding sites on AMBRA1 (AMBRA1PXP) (Fig. 5a and Supplementary Fig. 2a-b), and so disrupted AMBRA/PP2A-C interaction, whilst mutagenesis of each single motif (AMBRA1NT-PXP, AMBRA1CT-PXP) did not (Supplementary Fig. 2a-b and S4a-b).
Fig. 5. AMBRA1 affects C-MYC phosphorylation and cell proliferation through its direct binding to PP2A-C.
a) HEK293 cells were transfected with AMBRA1 or AMBRA1PXP. Protein extracts have been used for PP2A-C immunoprecipitation and the immunocomplexes have been analysed by western blot, with anti-AMBRA1 and anti-PP2A-C antibodies. A schematic representation of PXP motifs on AMBRA1 and ClustalW alignment of the two motifs present in AMBRA1 (corresponding to aa 275-281 and 1177-1183) are reported. b) HeLa cells knocked-down for AMBRA1 (or with unrelated control siRNA, -) and reconstituted with AMBRA1WT, with AMBRA1PXP or with βGal, as a control, have been analysed for the amount of P C-MYCS62 by western blot. In addition, an antibody against AMBRA1 has been used in order to check for successful AMBRA1 knock–down and reconstitution. Anti-ACTIN antibody has been used as a loading control. Vertical line represents a splice mark. The samples were obtained and processed in the same experiment, and the gels/blots were processed in parallel. c) HeLa cells, transfected as in b, have been analysed for the localization and amount of P C-MYCS62. P C-MYCS62 positive cells have been counted. Data are presented as means±s.d. and significance is **P<0.005 (n=100 cells pooled from 3 independent experiments in which 6-8 fields were assessed per experiment). Scale bar, 10μm. d) HeLa cells knocked-down for endogenous AMBRA1 and reconstituted with AMBRA1WT or with AMBRA1PXP have been analysed by MTS assay. Data are presented as means±s.d. and significance is **P<0.005 (n=3 independent experiments). e) HeLa cells were knocked-down for AMBRA1 (or with unrelated control siRNA, CTRL) and transfected with wild-type C-MYC (C-MYCWT) or phosphomimicking C-MYC (C-MYCS62D). Subsequently, cells were reconstituted with AMBRA1WT or transfected with βGal, as a control. In these cells, we analysed the amount of C-MYC and AMBRA1 by western blot, in order to check for AMBRA1 knock-down/transfection and for C-MYC transfection. Anti-ACTIN antibody has been used as a loading control. f) Cells treated as in (e) have been analysed by MTS assay [see (d)]. Data are presented as means±s.d. and significance is **P<0.005 (n=3 independent experiments).
Next, we used AMBRA1PXP to assess its effect on AMBRA1-mediated regulation of C-MYC and on cell proliferation. As expected, the expression of AMBRA1PXP in AMBRA1 deficient cells does not restore normal P C-MYCS62 levels (Fig. 5b-c), and has no effect on cell numbers (Fig. 5d), at a variance with AMBRA1WT.
By contrast, no difference can be observed in the pro-autophagic capability of the mutant AMBRA1PXP when compared with the wild-type (AMBRA1WT) (Supplementary Fig. 2a-b and 4c) and, indeed, AMBRA1PXP can still bind both BECLIN 1 and ULK1, two AMBRA1 interactors in the autophagy pathway8, 20, 21 (Supplementary Fig. 2a-b and 4d-e).
Last, we verified the significance of C-MYC dephosphorylation in this AMBRA1-dependent regulation. Upon C-MYC constitutive phosphorylation (by means of the phosphomimicking C-MYCS62D mutant construct) and by co-transfecting AMBRA1 in AMBRA1-knocked-down cells, we could not compensate for the AMBRA1-deficient phenotype, in contrast to what we observe with wild-type C-MYC (Fig. 5e-f). This finding implies that C-MYC dephosphorylation is, indeed, absolutely required for this pathway of regulation. In line with AMBRA1’s role in the regulation of C-MYC stability, in this experiment we also observed a decrease of exogenous total C-MYCWT upon AMBRA1 over-expression (Fig. 5e).
Ambra1 is a tumour suppressor gene
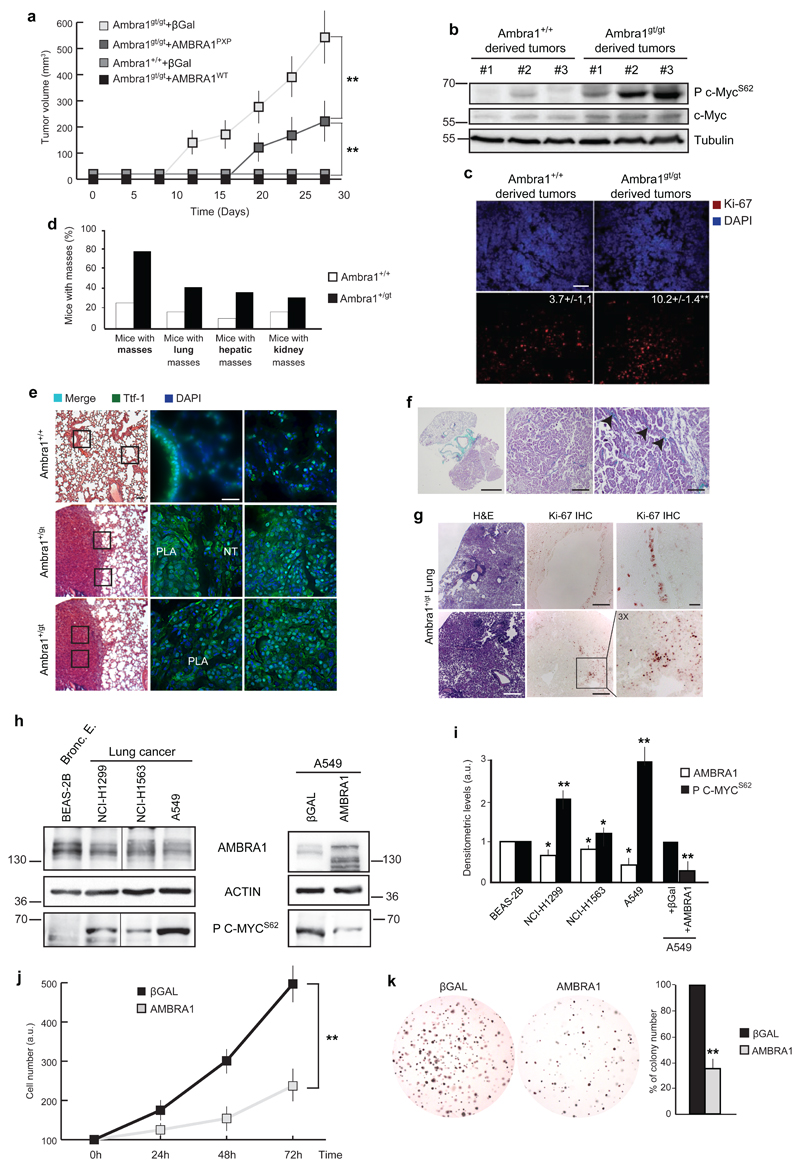
Given Ambra1’s effect on cell proliferation and on C-MYC stability, we set out to assess whether Ambra1 could be considered as a tumour suppressor gene in vivo. We addressed this issue by subcutaneously injecting into athymic mice primary MEFs, wild-type (Ambra1+/+) and null (Ambra1gt/gt) for the Ambra1 locus, transformed by a lentivirus encoding two oncogenes, a constitutive active form of Ras (RasV12) and E1A. Mice xenografted with MEFs devoided of Ambra1 (Ambra1gt/gt) or expressing AMBRA1 mutated in the PP2A binding sites (Ambra1gt/gt+AMBRA1PXP) developed tumours earlier compared to the animals injected with the Ambra1+/+ cells or the Ambra1gt/gt reconstituted with wild-type AMBRA1 (Ambra1gt/gt+AMBRA1WT) (Fig. 6a).
Fig. 6. Ambra1 affects tumorigenesis.
a) MEFs Ambra1+/+ and Ambra1gt/gt have been immortalized through RasV12/E1A oncogene infection. Subsequently, cells were subcutaneously injected into athymic mice and the insurgence and the size of masses were analysed. Data are presented as means±s.d. and significance is *P<0.05, **P<0.005 (n=3 independent experiments). b) Protein extracts have been obtained from +/+ and gt/gt derived tumours (in a). The extracts have been analysed by western blot, with anti-P c-MycS62, total c-Myc and Tubulin. c) Ki-67 staining on tumours derived from +/+ and gt/gt MEFs (shown in a). Scale bar, 50μm. d) Analysis of the frequence of spontaneous tumours in Ambra1+/gt mice between 12 and 17 months of age. e) H&E and TTF-1 (scale bars, 100μm and 50μm, respectively) immunofluorescence of close sections from the same lung mouse (Ambra1+/+ or Ambra1+/gt) are shown. PLA: Papillary Lung Adenocarcinoma; NT: Normal Tissue. f) Trichromic staining on Ambra1+/gt lung tumours. Scale bars: 2mm, 50μm, 100μm. g) Ki-67 staining on lung of Ambra1+/gt mice. H&E and IHC, second row: tumour tissue (scale bars, 100μm and 50μm). IHC, first row: healthy tissue (Scale bars, 50μm and 5μm). h) Western blot analysis of AMBRA1 and P C-MYCS62 in human adenocarcinoma cell lines. On the right panel, A549 cells have been reconstituted for AMBRA1. The levels of P C-MYCS62 have been analysed by western blot. Bronc. E.: Bronchiolar epithelium. The vertical line represents a splice mark of samples from the same gel. i) Graph showing the correlation between AMBRA1 and P C-MYCS62 protein levels in lung cell lines. The densitometric analysis of the western blots reported in (h) is reported in the graph. Data are presented as means±s.d. and significance is *P<0.05; **P<0.005 (n=4 independent experiments). j) A549 cells, overexpressing AMBRA1 or βGal as control, by retroviral infection, were analysed by MTS assay at different time points. Data are presented as means±s.d. and significance is **P<0.005 (n=3 independent experiments). k) The tumorigenicity of the cells used in j was assessed by a colony formation assay in soft agar. Data are presented as means±s.d. and significance is **P<0.005 (n=4 independent experiments).
Also, an increase in P c-MycS62 and, to less extent, in total c-Myc, characterizes Ambra1-defective tumours (Fig. 6b), together with an higher number of proliferating cells (Fig. 6c).
We then analysed whether Ambra1 allelic dosage and expression levels (Supplementary Fig. 5a-b), were responsible for spontaneous tumor insurgence in wild-type and Ambra1+/gt mice between 12 and 17 months of age. Indeed, as shown in Fig. 6d, a monoallelic deletion of Ambra1 is sufficient to increase the occurrence of tumors which primarily affected liver and lungs, the latter with areas of pulmonary parenchyma, in which the spongy organization typical of the alveoli was no longer recognisable and the tissue had a solid appearance (Fig. 6e, left panels). These lesions appear as circumscribed round masses, with a papillary architecture and glandular aspects, in the absence of a well-structured fibrous capsule, this suggesting tumour aggressiveness (Fig. 6f); In fact, they express the thyroid transcription factor-1 (TTF-1),31 a bona fide marker of lung carcinomas, normally expressed also in bronchial and type II alveolar epithelial cells of the healthy tissue (Fig. 6e, middle/right panels). Also, the presence of Ambra1 in the Ambra1+/gt tumours demonstrates that they are not characterised by loss of heterozygousity (Supplementary Fig. 5c). Indeed, Ambra1 levels appear to be augmented in the hemizygous samples, most likely due to a feedback attempt of the cells to enhance the activity of their autophagy machinery. Last, Ambra1+/gt apparently healthy bronchioli show a high positivity to Ki-67, a cell proliferation marker, whilst cell proliferation is usually extremely low in normal (wild-type) lung epithelia, with only ≈0.5% of airway cells actively dividing32–34 (Fig. 6g, IHC upper panels). However, tumour-bearing lung sections also show a focal positivity to the proliferation marker (Fig. 6g, IHC lower panels), suggesting that tumour growth is also sustained by cell proliferation. In the light of Ambra1’s role in the positive regulation of autophagy, we monitored the autophagy levels in Ambra1-deficient tumours. An inhibition of the autophagy pathway was found, as expected, in Ambra1+/gt tumours, as proven by analysis of the autophagy marker p62 (Supplementary Fig. 5d-e). This is in line with what reported upon inactivation of other autophagy regulators.35–39
Importantly, we also found AMBRA1 mutations in human tumours (in the endomethrium, large intestine and urinary tract, with 2.2%, 2.9% and 2.94% frequence of mutation, respectively) and a clear inverse correlation between AMBRA1 and P C-MYCS62 levels in a number of lung adenocarcinoma cell lines and a breast carcinoma cell line (Fig. 6g-h, Supplementary Fig. 2e and S6). Interestingly, when overexpressing AMBRA1 in cancer cell lines characterised by the lowest endogenous AMBRA1 levels, we were able to decrease C-MYCS62 phosphorylation (right panels in Figs. 6h-i, and Supplementary Fig. 2a-b, 6a-b), whilst, in the same cell lines, AMBRA1 over-expression induced lower proliferation and decreased tumorigenicity (Fig. 6j-k and Supplementary Fig. 6c-d). Based on our results, we thus demonstrate that disruption of the Ambra1 locus is sufficient to induce c-Myc hyperphosphorylation, hyperproliferation, and tumorigenesis.
Discussion
We previously demonstrated that the upstream autophagy regulator AMBRA1,8, 20, 21 affects cell proliferation both in vitro and in vivo.20, 23 In this study we show that this role may be based on the mTOR-controlled AMBRA1 regulation of the phosphatase PP2A toward its substrate C-MYC, impacting cell proliferation and tumorigenesis.
Notably, protein-protein interactions between PP2A and cellular scaffolds other than its canonical subunits contribute to the specificity of PP2A signalling.40 In this scenario, at least two possible roles for AMBRA1 can be hypothesized: AMBRA1 could be either a regulatory subunit of the complex or a member of the heterotrimeric complex, formed by PP2A-C, PR65A and a regulatory subunit that we have yet to identify. As a PP2A interactor, we found that AMBRA1 may facilitate C-MYC dephosphorylation in a specific manner, since other PP2A substrates resulted unaffected by AMBRA1 dosage. On the other hand, we also found a hyperphosphorylation of the transcription factor p107, another PP2A substrate,25 in Ambra1-defective systems. However, the dephosphorylation of p107 is carried out by both PP2A and the phosphatase PP1,41 although it is difficult to distinguish the two activities, since specific antibodies for the different phosphorylation sites of the two phosphatases are not available.
As for the biochemistry of AMBRA1-PP2A interaction, a recently published structure of the PP2A holoenzyme suggests that the structural arrangement of the PP2A-C/B subunit interface and the conformational flexibility of the PR65 subunit make the PP2A complex susceptible to structural changes that may affect substrate hydrolysis by PP2A-C or alter the substrate specificity.42–44 It is possible that AMBRA1 binding, mediated by a PP2A consensus site, identified in this work, results in allosteric regulation of the PP2A complex structure that could mediate enhancement of the recruitment of the substrate C-MYC and increase PP2A activity on C-MYC.
Also, we recently characterized AMBRA1 as a target of mTOR in the autophagy process;8 we now show here that the AMBRA1-PP2A-mediated regulation of C-MYC is also under mTOR control. This is of the highest importance, given the key role of mTOR in regulating a cell fate by interfering with its metabolic choices. It remains to be unveiled how does mTOR exert this control on AMBRA1, whether this is due to direct or undirect mechanisms.
Based on this network regulation of cell proliferation, we also demonstrate that heterozygous disruption of the Ambra1 locus, is sufficient to induce c-Myc hyperphosphorylation and hyperproliferation, thus suggesting a correlation with tumour insurgence. In fact, AMBRA1 mutations are reported in human tumor samples and increased tumorigenesis affects Ambra1-mutated mice. Indeed, a well-established link between autophagy and tumorigenesis does exist.35 The role of the autophagy core machinery components (ATG7, ATG4C) has been, indeed, dissected in tumorigenesis and in tumour progression.36–39 In particular, a double role for autophagy has emerged in two different steps: tumour formation and maintenance. First, autophagy represents a preventive mechanism against the accumulation of damaged organelles and genome, so counteracting tumour formation.35–38 Second, autophagy supplies fuel essential for established tumour growth.45–47 Considering AMBRA1’s place in the cascade of events leading to autophagy regulation,8, 20, 21 we propose that AMBRA1 plays a key role in this context, finely regulating both molecules involved in cell proliferation and in autophagy initiation. Interestingly, the tumor suppressor function of BECLIN 1,48, 49 another autophagy upstream signaller and AMBRA1 interactor,1, 20 has also been associated to alternative mitogenic signalling pathways and cytokinesis which, if deregulated, also contribute to cancer development.50
In sum, considering the roles of the upstream autophagy signallers in tumorigenesis, we believe that great care should be taken when choosing the appropriate molecular target for clinical anticancer-trials focussed on autophagy inhibition.51
Furthermore, a large body of evidence has demonstrated that C-MYC is relevant in different physiological contexts other than cancer, such as stem cell renewal and cellular differentiation.52 Identification of the AMBRA1’s physiological role and its potential involvement in all the above-mentioned processes will pose an interesting challenge.
Methods
Cell culture and reagents
HEK293, MEFs and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma Aldrich) supplemented with 10% fetal bovine serum (FBS, GIBCO), 2mM L-glutamine, 1% penicillin/streptomycin solution at 37°C. Human lung and breast carcinoma cell lines, with respective control cells, were purchased from American Type Culture Collection (ATCC) and cultured according to the supplier’s instructions. For autophagy induction and for amino acid starvation, cells were washed with PBS and cultured for 1-3hr in Earle's Balanced Salt Solution (EBSS; Sigma-Aldrich) or in medium without amino acids (GIBCO), respectively. When indicated, cells were incubated in the presence of 50μg/ml cycloheximide (CHX, Sigma-Aldrich), 2µM Rapamycin (Sigma Aldrich), 1,5µM Torin1 (Tocris). HeLa and HEK293 cells were transiently transfected with expression vectors using Lipofectamine 2000 (Invitrogen) as indicated by the supplier. Viral infection of A549 and MCF-7 cells with the retroviral AMBRA1-coding construct was performed as previously described.8
Plasmid constructs
Constructs coding for AMBRA1 and its deletion mutants (Flag-AMBRA1ΔWD40, Myc-F1, Myc-F2, Myc-F3, Myc-F1ΔWD40, Myc-F3a, Myc-F3b) or AMBRA1 site-specific mutants (AMBRA1NT-PXP, AMBRA1CT-PXP, AMBRA1PXP) were cloned in pLPCX vector8, 19, 20 (Clontech). Plasmid encoding for Flag-PR65A was kindly provided by Dr. A.C. Gingras. C-MYC construct was cloned in pLPCX vector by using the V5-Flag-C-MYC as a template (kindly provided by Dr. R. Sears). C-MYC encoding sequence was amplified by PCR and cloned in the acceptor vector by using EcoRI and NotI restriction sites. C-MYCS62D mutant and AMBRA1PXP mutants have been created by site-directed mutagenesis (Agilent), using C-MYC and AMBRA1 as templates, respectively.
Antibodies
All the antibodies were used at a dilution of 1:1000 unless specified otherwise. Anti-Flag (cat. num. F3165; 1:2000), Beta Tubulin (cat. num. T4026; 1:3000) and anti-ACTIN (cat. num. A2066; 1:2000) were from Sigma Aldrich. Anti-C-MYC (9E10, cat. num. sc-40), anti-C-MYC (N272, cat. num. sc-764), anti C-MYC (C33, cat. num. sc-42), anti-ULK1 (cat. num. sc-33182), anti-BECLIN 1 (cat. num. sc-10086 and sc-11427), anti-PR65A (cat. num. sc-15355), anti-P AKT (cat. num. sc-16646), anti-AKT (cat. num. sc-8312), anti-GSK3β (cat. num. 377213), anti-P GSK3β (cat. num. sc-11757), anti-ERK1/2 (cat. num. 292838), anti-P ERK1/2 (cat. num. sc-16982), anti-Cyclin D (cat. num. sc-182), anti-Cyclin E (cat. num. sc-20684), anti-Cyclin B (cat. num. sc-245), anti-Cyclin A (cat. num. sc-596), anti-P62 (cat. num. sc-28359; 1:3000) and anti-HSP 90 (cat. num. sc-7947; 1:3000) were from Santa Cruz. Anti-AMBRA1 (cat. num. 26190002; 1:3000) used on human extracts was from Novus. Anti-Ambra1 (used on mouse extracts, cat. num. AB131) and anti-PP2A-C (1D6, cat. num. 05-421) were from MERCK Millipore. Anti-P C-MYCS62 (cat. num. ab78318; 1:500) was from Abcam. Anti-P RPS6 (cat. num. 5364; 1:3000), anti-LC3 (cat. num. 3868; 1:6000) and anti-RAPTOR (cat. num. 2280) were from Cell Signalling. For immunohistochemistry analysis, anti-P62 (ct. num. PM045; 1:2000) from MBL was used. Anti-Ki67 (SP6, cat num. RM-9106; 1:2000) was from Thermo Scientific. Anti TTF-1 (cat num. 8G7GG3/1; 1:100) was from Dako.
RNA interference
RNA interference was performed using the AMBRA1 and control siRNA oligonucleotides that were previously described.8, 20 RAPTOR (cat. num. M-004107-01) and C-MYC (cat. num. AM16704) siRNAs were purchased from Dharmacon and LifeTechnologies, respectively. 2x105 cells/well were transfected with 100pmol siRNA in 6 well plates by Lipofectamine 2000 (Invitrogen), as suggested by the supplier. Where indicated, 24hrs after the AMBRA1-siRNA treatment, AMBRA1 constructs (AMBRA1WT or AMBRA1PXP) or βGal or C-MYC constructs (C-MYCWT or C-MYCS62D) have been transiently expressed. Where indicated, 24hrs after the AMBRA1-siRNA treatment, RAPTOR- or CTRL-siRNA were transfected in depleted cells.
Immunoblotting
Cell lysates were prepared with RIPA Buffer. Mouse tissues were homogenized mechanically in tissue lysis buffer (50mM Tris HCl pH 7.5, 320mM Sucrose, 50mM NaCl, 1% Triton X-100 and protease and phosphatase inhibitors). Homogenates were centrifuged at 13,000xrpm at 4°C for 30min and supernatants were collected. Protein extracts were quantified by Lowry protein assay (Biorad), and denatured by adding Laemmli Buffer (0,1% Bromophenol blue, 10% Glycerol, 2% SDS, 5% ß-mercaptoethanol, 270mM Tris HCl pH.6.8). A total of 30-50μg of proteins was analysed by SDS-polyacrylamide gel electrophoresis and immunoblotting. Proteins were separated on acrylamide gel (Biorad) and electroblotted onto nitrocellulose membranes (Protran, Schleicher & Schuell). Blots were incubated with primary antibodies in 5% non-fat dry milk in PBS plus 0,1% Tween-20 overnight at 4°C. Detection was achieved using horseradish peroxidase-conjugate secondary antibody (Biorad) and visualized with ECL plus (Amersham Bioscience). For the Western Blot analysis of immunoprecipitated C-MYC, TrueBlot@ secondary antibody (EBioscience) was used, in order to minimize detection of Immunoglobulines in the immunoprecipitation samples.
Immunoprecipitation
Cells were lysed in CHAPS buffer (40mM Hepes pH 7.4, 150mM NaCl, 2mM EDTA, 10mM β-glycerophosphate, 0,3% CHAPS and protease and phosphatase inhibitors). Lysates (1-3mg) were then incubated at 4°C for 30min, following a centrifugation at 4°C for 15min at 13000rpm to remove insoluble debris, equal amounts of proteins were incubated with 1-2μg of monoclonal anti-C-MYC or anti PP2A-C antibodies for at least 2-3 hours. In the case of Flag-PR65A immunoprecipitation, anti-Flag conjugated agarose beads have been used (Sigma Aldrich). Then, protein G conjugated sepharose beads (Roche) were added to anti-C-MYC (C-33) and ati-PP2A-C primary antibodies, and incubated with rotation at 4°C for 1hr. The beads were finally collected by centrifugation and washed four times with the lysis buffer plus 150mM NaCl. Proteins bound to the beads were eluted with 20μl of SDS-PAGE sample buffer and boiled at 95°C for 10min.
Phosphatase assay
For phosphatase assay, P C-MYCS62-enriched54 HeLa cells were suspended in lysis buffer (50mM Tris-HCl, pH 7.4, 7.5% Glycerol, 1mM EDTA, 150mM NaCl, 0.5% Nonidet P-40, 1mM Na3VO4, Complete Protease Inhibitor), incubated for 30min in ice and cleared from debris by centrifugation and incubated with C-MYC antibody (C-33, mouse monoclonal, Santa Cruz) followed by incubation with G Protein Sepharose Beads (Roche). The obtained immunocomplex was washed in lysis buffer without Na3VO4. Wild-type or Ambra1gt/gt MEFs were lysed in NET buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 1% Nonidet P-40, 15mM EDTA, Complete Protease Inhibitor). The catalytic subunit of the PP2A phosphatase (PP2A-C) was immunoprecipitated from wild-type or Ambra1gt/gt protein extracts by using anti PP2A-C antibody (1D6, mouse monoclonal, Millipore). The same amount of C-MYC was added to PP2A-C coming from wild-type or Ambra1gt/gt cells and the phosphatase reaction was carried out in vitro, for 15min at 37°C, in the “phosphatase reaction” buffer (50mM Tris-HCl, pH 7.4, and 2mM DTT). As a negative control, C-MYC was incubated with an immunoprecipitation carried out with mouse Immunoglobulin mix (SIGMA), and treated as PP2A-C immunocomplexes. The products of the phosphatase reactions have been boiled for 10min at 95°C in Laemmli Buffer and subsequently analysed by western blot, by using antibodies against P C-MYCS62, total C-MYC and PP2A-C. A similar PP2A phosphatase assay protocol was already reported.52 TrueBlot@ secondary antibody (EBioscience) was used in order to minimize the detection of the immunoglobulines in the immunoprecipitation samples.
Size exclusion chromatography
Four milligrams of HeLa endogenous extract was injected onto a Superose 6 HR 10/30 fast protein liquid chromatography (FPLC) gel filtration column (GE Healthcare), equilibrated with 20mM Tris-HCl (pH 7.5), 200mM NaCl, 12.5% glycerol, 0.2% Nonidet P-40, 1mM EDTA, 1mM EGTA and 1mM DTT, and eluted at a flow rate of 0.25ml/min with the same buffer. Proteins were collected in 250µl fractions, precipitated with 10% trichloroacetic acid (TCA), and subjected to Western blotting. Gel filtration columns were calibrated with the following molecular mass markers: thyroglobulin (669kDa), apoferritin (443kDa), β-amylase (200kDa), alcohol dehydrogenase (150kDa), bovine serum albumin (66kDa), and carbonic anhydrase (29kDa) (Sigma Aldrich).
Immunocytochemistry
Cells were washed in PBS and fixed with 4% paraformaldehyde in PBS for 15min. After permeabilization with 0.2% Triton X-100 in PBS for 5min, cells were blocked in 2% horse serum in PBS and incubated for 1-2hrs at room temperature, with the primary antibodies. We used antibody directed against the P C-MYCS62 or against BrdU. Cells were then washed and incubated for 1h with labeled anti-mouse (Alexa Fluor-488 or -555, Molecular Probes, Eugene, OR) secondary antibodies. Images were examined under Leica TCS SP5 confocal microscope equipped with a x63 oil-immersion objective (Deerfield, IL).
Immunohistochemistry for Ki67 and for p62 was carried out on lung and liver paraffin sections using a monoclonal anti-Ki67 from rabbit (Clone SP6, Thermo Scientific) and a polyclonal rabbit-anti-p62 affinity purified (MBL). Antigen retrival was performed using citrate buffer incubation in the microwave for both antigens. Antibody incubation was carried out overnight at 4°C 1:200 (Ki67) or 1:1000 (p62) and immunoreactions were finally developed and visualized using the Avidin-Biotin-AEC method (Ki67) or the Envision-DAB method (p62). In the case of the xenografts and TTF-1 staining on lung, tissue samples were fixed 4% paraformaldehyde (PFA) and embedded in Tissue-Tek OCT compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan). Sections were prepared and incubated with Ki67 and TTF-1 primary antibodies overnight at 4°C. The day after, labelled Alexa Fluor-488 (Molecular Probes, Eugene, OR) secondary antibody was used. Where indicated, H&E and Trichromic stainings have been carried out, according to standard protocols.
MTS assay
For the MTS assay (Promega), HeLa cells were knocked-down and transfected as previously described. Following 6hrs lipofectamine transfection, cells were splitted and 5000 cells were plated in each well of a 96-well. Transformed MEFs were plated in 96-well at a cellular density of 7000 cells per well. Each sample was analysed in triplicate. MTS assay was performed as indicated by the supplier.
BrdU incorporation assay
Cells were pulsed with BrdU for 1h at final concentration of 100µM. Following incubation with BrdU, cells were fixed in cold methanol for 20min at -20°C and then rehydrated in phosphate saline buffer (PBS). 2M HCl was added to each well and incubated at room temperature for 30min. Cells were washed twice for 5min in 0.1M Sodium Borate (pH 8.5) and washed for 5min in PBS before blocking in 2% bovine serum albumin (BSA/PBS) for 1 h at room temperature. Anti-BrdU was diluted at 1:100 in 2% BSA/PBS and added to each well and incubated at 4 °C overnight. Coverslips were washed three times in PBS for 5min and antibody-antigen complexes were detected with Alexa Fluor 488-conjugated goat anti-mouse antibody diluted 1:500 in 2% BSA/PBS and incubated at room temperature for 1h. Cells were then washed four times with PBS and counterstained with 4’,6-diamidino-2-phenylindol (DAPI) for 5min at room temperature in the dark. Finally cells were mounted in GEL/MOUNT antifade (Biomeda, Corp.), and visualized with Olimpus IX70 microscope using softWoRx and Delta Vision Imaging Workstation. The in vitro experiments were performed on 3-4 clones for each MEFs genotype.
Mice
Ambra1 heterozygous (Ambra1+/gt) mice20 and wild-type littermates were used for the analysis of spontaneous tumorigenesis. Both female and male mice from CD1 and mixed (C57BL/6 x CD1) strain were used. Necropsies were performed on 40 Ambra1gt/+ and 40 wild-type mice, aged 12-17 months and the masses that were present on gross examination were recorded. Experiments were carried out in accordance with the European Community's Council Directive 86/609/EEC. Formal approval of these experiments was obtained from the Italian Ministry of Health (D.L.vo 116/92).
Identification of AMBRA1 genomic alteration in human tumors
Mutations and chromosome alterations in the TCGA dataset of human tumors from the endometrium (sample size: 227), the large Intestine (sample size: 723) and the urinary tract (sample size: 102) are available at cancergenome.nih.gov.
Zebrafish
Zebrafish handling and treatment were approved by the UniPD Ethical Committee on Animal Experimentation (CEASA – Project #25/2012). Cell transplantation experiments were performed as described.55 In brief, embryos were injected, at one-cell stage, with a mixture containing 200pg EGFP mRNA (for in vivo tracking of transplanted cells) and 10ng Biotin-dextran Lysine Fixable (10.000 Mw, Sigma) alone (control donors) or in combination with designated MOs: I) 10ng MO1-ambra1a, II) 20ng MO1-ambra1b, III) 2.5ng MO1-ambra1a + 5ng MO1-ambra1b mixture.
At blastula stage (4 hpf), donor and wild-type host embryos were manually dechorionated. Approximately 30 cells from donor embryos were then transplanted into host embryos. Transplanted embryos were fixed in paraformaldehyde 4% in phosphate buffered saline, for 2 hrs at RT, at 90% epiboly (9 hpf). Biotin injected transplanted cells were detected in hosts using an avidin-biotinylated complex (ABC Kit, Vectastain) and the DAB substrate. Proliferation was analysed by whole mount immunohistochemistry with anti-phosphohistone H3 antibody (polyclonal rabbit Millipore) according to the manufacturer instructions and revealed with NBT/BCIP solution (Roche).
For imaging confocal microscope Leica SP5 were used. Quantitative data are expressed as mean ± SEM. Statistical significance was determined by chi-squared test using the Primer statistical software.
Real-time PCR
RNA was isolated by using RNeasy Micro Kit (Qiagen Inc., Valencia, CA), according to the manufacturer’s instructions. Real-time PCR was performed by using SensiMix Plus SYBR Kit (Quantace Ltd, London, UK). The primer sets used were as follows: mouse Cyclin A forward, 5’-TGCAAACTGTAAGGTTGAAAGC-3’; and reverse, 5’-TGTAGAGAGCCAAGTGGAAGG-3’. Mouse Cyclin B forward, 5’-CTGCAGTGACTACGTGAAGG-3’; and reverse 5’-TGTGGATTAATGGACTGTAAAACC-3’. Mouse Ambra1 forward, 5’-CTACTGGGACCAGCTAAGTGAAA-3’; and reverse 5’- ACGTGGCTCTGCTGGTTC-3’. Real time quantitation was performed by using a Fast-Real Time PCR System (Applied Biosystems, Foster City, CA). Data were normalized to β-actin. Fold-change was determined by using the 2-ΔΔCT method and all reactions were performed in triplicate.
Tumor xenografts
Wild-type or Ambra1gt/gt MEFs were transformed by pBABE encoding RasV12/E1A oncogenes. Tumors were induced by subcutaneous injection in nude mice of 5x106 cells. Transformed MEFs wild-type, Ambra1gt/gt, Ambra1gt/gt infected with AMBRA1WT or AMBRA1PXP retroviral constructs were suspended in PBS supplemented with 0.1% glucose. βGal retroviral construct was used as a control in wild-type, Ambra1gt/gt MEFs. Tumors were measured with external caliper, and volume was calculated as (4π/3) x (width/2)2 × (length/2). 28 days after injection of the cells, animals were killed and all tumors were excised. Samples from tumors xenografts were dissected and frozen for further analysis.
Colony formation in soft-agar assay
The colony formation assay in soft agar was performed by using the CytoSelect™ 96-Well Cell Transformation Assay, as indicated by the supplier. For AMBRA1-overexpressing A549 and MCF-7 cells, 0.5x104 cells were seeded in 12-wells. The colony growth was monitored day by day and colonies were counted 18 and 12 days after, respectively. Representative images were acquired using bright field microscopy.
Yeast two hybrid assay
The results of the yeast two hybrid assay shown in Figure 2 come from the same data set reported in our previously publication: refer to this reference for the methods.20
Tandem affinity purification assay (TAP)
The results of the TAP shown in Supplementary Table 1 come from the same data set reported in our previously publication: refer to this reference for the methods.8
Statistical analysis
For all experiments shown, n is indicated in the figure legends. Each point value reported in graphs represents the mean±s.d. or ±s.e.m, as indicated in the figure legends. Statistical significance was measured by an unpaired two-tailed t-test. P<0.05 has been considered significant.
No statistical method or inclusion/exclusion criteria were used to predetermine sample size or define the experimental group.
The experiments were not randomized.
The investigators were not blinded to allocation during experiments and outcome assessment.
For the experiments in which not quantification is shown, images representative of at least 3 independent experiments are shown.
Supplementary Material
Acknowledgements
We wish to thank Mark Canney, Valerie Unterkircher, Romina Laricchia and Mara Salomé for excellent technical assistance, and M. Acuña Villa and M.W. Bennett for editorial and secretarial work. We also thank Dr. Silvia Campello for critical reading of the manuscript. We are indebted to Dr. R. Sears (Portland, OR - USA), Dr. A.C. Gingras (Toronto - Canada) and Dr. A. Teleman and K. Dimitriadis (Heidelberg, Germany) for kindly providing us with V5-Flag-C-MYC and Flag-PR65A constructs and Tsc2 MEFs, respectively and with Prof. S. Cannata (Rome) for his precious advices on histopathology. This work was supported by grants from KBVU, Lundbeck Foundation, Novo Nordisk Foundation, AIRC (IG2010 and IG2012 to both FC and MP), and in part from FISM (2009), Telethon Foundation (GGP10225), the Italian Ministry of University and Research (PRIN 2009 and FIRB Accordi di Programma 2011) and the Italian Ministry of Health (RF 2009). VC is supported by Lundbeck Foundation. Also, we are grateful to the Spanish Ministry of Economy and Competitiveness (MINECO) (PS09/01401; PI12/02248, FR2009-0052 and IT2009-0053) and to Fundación Mutua Madrileña (AP101042012) for funding GV’s laboratory.
Footnotes
Author Contribution
V.C. performed most experiments with crucial help from: C.F., M.B. and F.Q. (immunohistochemistry analysis); M.L and M.S. (xenograft assay); P.F.G., M.R. and M.H. (bioinformatic analysis); D.D.Z. (real-time PCR); F.N. and M.A. (mutagenesis and cloning); M.D. (gel-filtration assay); T.S. and L.D.V (experiment in zebrafish); C.G and J.D. (mass-spectrometry analysis); G.M.F. and G.V. provided critical reagents. S.D.B discussed the results and commented on the manuscript; V.C. and F.C. wrote the manuscript, with suggestions from M.P., G.M.F. and G.V.; F.C. and V.C. conceived and designed the research.
Competing financial interests
The authors declare no competing financial interests.
Full Methods and any associated references are available in the Supplementary Material.
References
- 1.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–73. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neufeld TP. Autophagy and cell growth--the yin and yang of nutrient responses. J Cell Sci. 2012;125:2359–68. doi: 10.1242/jcs.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosokawa N, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–9. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 8.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–16. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 9.Banreti A, Lukacsovich T, Csikos G, Erdelyi M, Sass M. PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy. 2012;8:623–36. doi: 10.4161/auto.19081. [DOI] [PubMed] [Google Scholar]
- 10.Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–15. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puustinen P, et al. CIP2A oncoprotein controls cell growth and autophagy through mTORC1 activation. J Cell Biol. 2014;204:713–27. doi: 10.1083/jcb.201304012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–45. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Arroyo JD, Hahn WC. Involvement of PP2A in viral and cellular transformation. Oncogene. 2005;24:7746–55. doi: 10.1038/sj.onc.1209038. [DOI] [PubMed] [Google Scholar]
- 15.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–39. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 19.Junttila MR, Westermarck J. Mechanisms of MYC stabilization in human malignancies. Cell Cycle. 2008;7:592–6. doi: 10.4161/cc.7.5.5492. [DOI] [PubMed] [Google Scholar]
- 20.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 21.Di Bartolomeo S, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benato F, et al. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy. 2013;9:476–95. doi: 10.4161/auto.23278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 25.Jayadeva G, et al. B55alpha PP2A holoenzymes modulate the phosphorylation status of the retinoblastoma-related protein p107 and its activation. J Biol Chem. 2010;285:29863–73. doi: 10.1074/jbc.M110.162354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112:1223–33. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 29.Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–9. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 30.Evans BJ, et al. Physical association of GPR54 C-terminal with protein phosphatase 2A. Biochem Biophys Res Commun. 2008;377:1067–71. doi: 10.1016/j.bbrc.2008.10.108. [DOI] [PubMed] [Google Scholar]
- 31.Holzinger A, et al. Monoclonal antibody to thyroid transcription factor-1: production, characterization, and usefulness in tumor diagnosis. Hybridoma. 1996;15:49–53. doi: 10.1089/hyb.1996.15.49. [DOI] [PubMed] [Google Scholar]
- 32.Szabo E. Lung epithelial proliferation: a biomarker for chemoprevention trials? J Natl Cancer Inst. 2001;93:1042–3. doi: 10.1093/jnci/93.14.1042. [DOI] [PubMed] [Google Scholar]
- 33.Kauffman SL. Cell proliferation in the mammalian lung. Int Rev Exp Pathol. 1980;22:131–91. [PubMed] [Google Scholar]
- 34.Ayers MM, Jeffery PK. Proliferation and differentiation in mammalian airway epithelium. Eur Respir J. 1988;1:58–80. [PubMed] [Google Scholar]
- 35.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marino G, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 38.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inami Y, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sontag E. Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal. 2001;13:7–16. doi: 10.1016/s0898-6568(00)00123-6. [DOI] [PubMed] [Google Scholar]
- 41.Kolupaeva V, Janssens V. PP1 and PP2A phosphatases--cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2013;280:627–43. doi: 10.1111/j.1742-4658.2012.08511.x. [DOI] [PubMed] [Google Scholar]
- 42.Millward TA, Zolnierowicz S, Hemmings BA. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–91. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- 43.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–7. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–51. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 45.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113–9. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 50.Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010;316:3368–78. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 51.Helgason GV, Holyoake TL, Ryan KM. Role of autophagy in cancer prevention, development and therapy. Essays Biochem. 2013;55:133–51. doi: 10.1042/bse0550133. [DOI] [PubMed] [Google Scholar]
- 52.Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5:947–59. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgess A, et al. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A. 2010;107:12564–9. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junttila MR, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130:51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 55.Nusslein-Volhard C, Zebrafish Dahm R. A Practical Approach. Oxford Univ. Press; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.