Abstract
The 26S proteasome is the molecular machine at the center of the ubiquitin–proteasome system and is responsible for adjusting the concentrations of many cellular proteins. It is a drug target in several human diseases, and assays for the characterization of modulators of its activity are valuable. The 26S proteasome consists of two components: a core particle, which contains the proteolytic sites, and regulatory caps, which contain substrate receptors and substrate processing enzymes, including six ATPases. Current high-throughput assays of proteasome activity use synthetic fluorogenic peptide substrates that report directly on the proteolytic activity of the proteasome, but not on the activities of the proteasome caps that are responsible for protein recognition and unfolding. Here, we describe a simple and robust assay for the activity of the entire 26S proteasome using fluorescence anisotropy to follow the degradation of fluorescently labeled protein substrates. We describe two implementations of the assay in a high-throughput format and show that it meets the expected requirement of ATP hydrolysis and the presence of a canonical degradation signal or degron in the target protein.
Keywords: 26S proteasome, Ubiquitin proteasome system (UPS), Protein degradation, Fluorescence anisotropy, High-throughput degradation assay
1. Introduction
The 26S proteasome is a 2.5 MDa molecular machine, composed of at least 33 different proteins, that recognizes and hydrolyzes proteins targeted for degradation by polyubiquitin modifications [1,2]. The proteasome consists of a 20S proteolytic core particle capped by one or two 19S regulatory particles. The core particle contains six proteolytic sites, which are only accessible through gated pores at either end of the particle [3–5]. The caps contains receptors that recognize ubiquitin chains, six ATPase subunits that unfold and translocate substrates through a central channel to the proteolytic sites in the core particle, and enzymatic subunits that remove the ubiquitin chains from substrates as they are trans-located to degradation [6]. There are at least three different caps that can associate with the core particle, but the 19S regulatory particle is thought to be the cap primarily responsible for ubiquitin-dependent degradation [1].
The proteasome degrades a large number of regulatory proteins, such as cell cycle regulators and transcription factors; it removes misfolded and damaged proteins from cells; and it produces some of the peptides displayed by MHC complexes [7]. The proteasome has been a target for drug design in several human diseases, and proteasome inhibitors are used successfully in the treatment of cancers, particularly multiple myeloma and mantle cell lymphoma [8–10]. Cancer drugs currently used in the clinic target the proteolytic active sites in 20S core particles to inhibit proteasomal activity. Recently, proteasome inhibitors targeting the 19S regulatory particle have been developed, including inhibitors of the deubiquitinating activities associated with the proteasome (Usp14/Ubp6 and Uch37/UCHL5) and molecules that disrupt ubiquitin binding to the proteasome [11]. One inhibitor that targets both Usp14 and Uch37 results in the accumulation of polyubiquitinated proteins, which leads to cell apoptosis and has been shown to inhibit tumor growth in mice [12,13].
In several neurodegenerative disorders, accumulation of aggregation-prone proteins has been implicated in disease progression, and in some cases the proteasome's activity appears to be impaired in affected cells [14–17]. Thus, it may be beneficial to develop drugs that can activate proteasome activity to improve the clearance of toxic proteins and protein aggregates. Indeed, an inhibitor of the deubiquitinating enzyme Usp14 enhances proteasomal protein degradation and leads to the clearance of protein aggregates in human cells [18].
The search for new modulators of proteasome activity requires assays to measure proteasomal activity that can be carried out in an efficient and reproducible manner. One common proteasomal degradation assay uses peptide substrates that release a fluorescent group upon cleavage, resulting in an increase in fluorescence intensity that is proportional to degradation activity [19,20]. These substrates are convenient and sensitive reporters of the proteolytic activity of the 20S core of the proteasome and can be used in a high-throughput format. However, the peptide substrates are less well suited to investigate the activity of the 26S proteasome holoenzyme, as their hydrolysis bypasses the steps of substrate recognition by the ubiquitin receptors and does not require unfolding by the ATPase subunits. 26S proteasome activity is generally monitored by following the degradation of ubiquitinated proteins [21,22]. Ubiquitinated proteins can be produced in vitro using purified enzymes, cell lysates, chemical strategies, or engineered bacteria [21,23–30]. The assays typically follow protein degradation by sampling the reaction at discrete time points and measuring the amount of the substrate protein remaining by SDS-PAGE and protein staining, Western blotting, or autoradiography (e.g., [21,22,31–35]). These methods provide information on protein size and thus on the ubiquitination state and can detect the formation of partially degraded protein fragments, but can be time-, labor-, and reagent-consuming and difficult to implement in a high-throughput format. Incorporating a fluorescent protein such as green fluorescent protein (GFP) into the model protein makes it possible to follow protein degradation by measuring fluorescence using in-gel or high-throughput assays. For example, substrates containing a degradation signal (degron)-fused GFP have been used to monitor proteasome activity by following the decay of fluorescence intensity resulting from degradation of the GFP moiety (e.g., [29,36,37]). Fluorescent proteins often have complicated maturation kinetics and can be difficult to unfold so that they resist degradation by some ATP-dependent proteases [38]. However, easier-to-unfold circular permutants of GFP have been developed that make it possible to monitor degradation even by proteases less robust than the proteasome [29,38–41].
Here, we present a 26S proteasome activity assay based on the measurement of the fluorescence anisotropy of a small molecule dye attached to a substrate protein (Fig. 1a). The fluorescence anisotropy signal depends on the lifetime of the fluorophore and its rotational relaxation time [42,43], which in turn depends on the size of the dye-labeled molecule. Thus, fluorescence anisotropy can interpret the changes in both the abundance and the molecular size of substrates in the reaction. Indeed, anisotropy has been applied in the studies of proteolysis catalyzed by conventional proteases such as thrombin, enterokinase, Factor Xa, chymotrypsin, trypsin, or more specialized enzymes such as calpain II or botulism neurotoxin metalloproteases in a high-throughput format [44–47]. The assay described here measures fluorescence anisotropy to monitor 26S proteasome activity continuously (Fig. 1a). It follows the degradation of a proteasome-targeted protein into short polypeptides and thus reflects all the steps that occur within the 19S cap: substrate binding to the substrate receptors, initiation of degradation, unfolding, translocation into the core particle, and proteolysis. We describe two implementations of the assay in which an Alexa Fluor 546 dye is conjugated to the protein through a SNAP-tag. In one implementation, the substrate is targeted to the proteasome via a polyubiquitin modification that is attached to the protein through an in vitro ubiquitination reaction with purified E1, E2, and E3 enzymes. In the other implementation, the substrate is targeted to the proteasome by a ubiquitin-like domain encoded in the substrate's gene. The ubiquitin-like domain is recognized by the proteasome, thus bypassing the need for ubiquitination. In both cases, the dye is attached to a folded protein that must be unfolded and translocated into the core particle in order to be degraded, which allows probing 26S proteasomal activity. We anticipate that this assay will be a useful tool for high-throughput experiments to probe the effect of inhibitors and activators on proteasome activity, especially those that may act upstream of peptide hydrolysis.
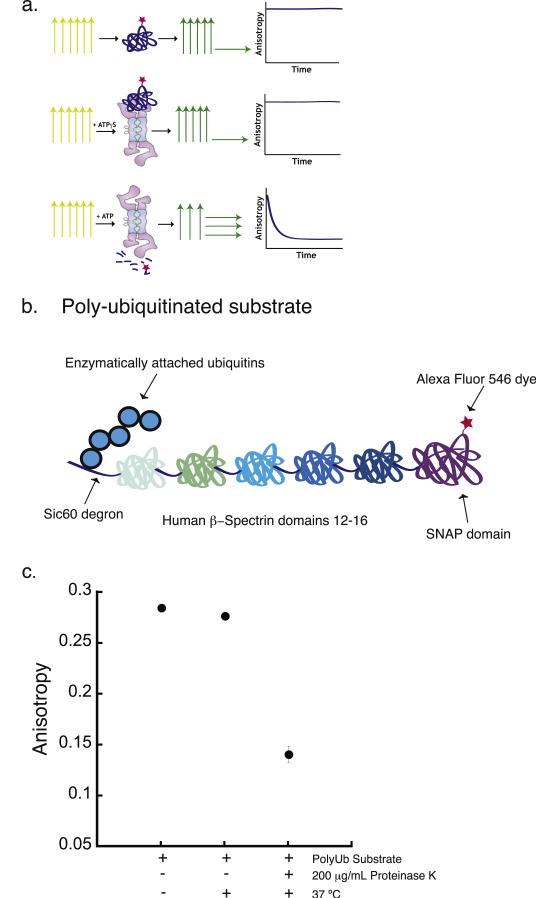
Fig. 1.
Proteasomal degradation assay based on fluorescence anisotropy measurement. (a) Cartoon of the experimental design of the proteasomal degradation assay based on the measurement of bulk steady-state fluorescence anisotropy. The three rows represent three different assay conditions. The top row shows the dye-labeled substrate alone, the middle row shows dye-labeled substrate in the presence of protea-some and the nonhydrolyzed ATP analogue ATPγS, and the bottom row shows dye-labeled substrate in the presence of proteasome and ATP. Under each of these conditions, the sample is excited with polarized light, represented by the yellow vertical arrows. The fluorescence emission from the sample and its polarization are represented by the green and yellow horizontal and vertical arrows. The graphical representations in the rightmost column indicate the expected experimental results when the anisotropy of the fluorescence signal emitted from the substrate is monitored over the time course of the degradation reaction. (b) Schematic representation of a polyubiquitinated substrate used for the degradation assay. The substrate is composed of a 60-amino acid unstructured region derived from yeast Sic1 at the N terminus (Sic60), followed by five domains from the human β-spectrin protein and finally a SNAP-tag domain at the C terminus. The Sic60 sequence acts as the degron and provides the polyubiquitination signal as well as the initiation region for proteasomal degradation. The SNAP-tag domain allows site-specific labeling of the substrate with a fluorescent dye. (c) Fluorescence anisotropy of the polyubiquitinated substrate before and after 2 h incubation at 37 °C in the absence or presence of 200 μg/mL proteinase K. Error bars show the standard error of the mean anisotropy obtained in four separate experiments.
2. Materials and methods
2.1. Substrate construction
We designed two types of model substrates for 26S proteasome degradation assays: One set of proteins contained a Sic60 tag to allow polyubiquitination and the other set contained a ubiquitin-like (UbL) domain encoded in their primary sequences. The Sic60 tag is derived from the first 60 amino acids of the yeast Sic1 protein and contains a four-amino acid motif (PPPY) that is recognized by the yeast ubiquitin ligase Rsp5 [21,33]. We placed the Sic60 tag at the N terminus of the substrate protein, followed by domains 12–16 of human β-spectrin [48,49], a SNAP-tag domain (New England Biolabs) for dye labeling, and finally a hexahistidine tag at the C terminus (Sic60PY-Spectrin12–16-SNAP-His6). The UbL substrates were composed of a UbL domain, which consisted of residues 1–77 of Saccharomyces cerevisiae Rad23 at the N terminus, followed by either a VDGGSGGGS or GSGGSGSG linker, a SNAP-tag domain, an unstructured region, and finally a C-terminal hexahistidine tag (UbL-SNAP-40-His6). The unstructured region was derived from the first 35 amino acids of S. cerevisiae cytochrome b2 (which are part of the mitochondrial targeting sequence) with the lysine residues mutated to arginine or glutamine [33,50]. In a derivative of this protein, the unstructured region was omitted (UbL-SNAP-His6). The annotated sequences can be found in Supplementary Materials Table S1.
2.2. Protein expression and purification
The substrate for polyubiquitination (Sic60PY-Spectrin12–16-SNAP-His6) and the UbL substrates (UbL-SNAP-40-His6 and UbL-SNAP-His6) were overexpressed in and purified from the Escherichia coli strain Rosetta (DE3)pLysS (Novagen). Constructs were cloned into a pETDuet vector (Novagen) and expressed from the T7 lac promoter. Bacterial strains were grown in 1–1.5 L of LB Medium at 37 °C to an optical density at 600 nm of 0.4–0.6. Protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and incubation continued for an additional 2.5–3 h at 28 °C. Cells were harvested by centrifugation, resuspended in NPI-10 buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, and 1 mM DTT, pH 8.0) or PBS (phosphate buffered saline with 1 mM DTT, pH 7.5). Protease Inhibitor Cocktail Set V, EDTA-free was added (Calbiochem 539137) and homogenized using a high-pressure homogenizer (EmulsiFlex C3, Avestin) with 1–2 sample passes at 15,000 psi. The lysate was clarified by centrifugation, and the supernatant was filtered through a 0.22 μm filter and then loaded onto a 5 mL Ni-NTA column (Qiagen). His-tagged protein was eluted with four column volumes of NPI-250 (50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, and 1 mM DTT, pH 8.0) and the fractions containing the target protein were added either to a dye-labeling reaction (UbL substrates, see below) or to a combined ubiquitination and dye-labeling reaction (ubiquitination substrate, see below). The ubiquitinated and labeled substrate was purified further using an S300 16/60 Sephacryl size exclusion column (GE Healthcare) to separate the dye-labeled, ubiquitinated substrate from all other components of the reaction. Protein concentration was estimated by measuring the absorbance at 280 nm using an extinction coefficient calculated from the protein's sequence (ExPASy's ProtParam). For ubiquitinated proteins, we estimated the approximate number of ubiquitins added by the conjugation reaction from the molecular weight of the dye-labeled, ubiquitinated bands observed by SDS-PAGE. Protein purity was assessed by SDS-PAGE and we estimated that at least 12 ubiquitin moieties were attached to the protein in a typical ubiquitination reaction.
2.3. Protein ubiquitination and dye labeling
Ubiquitination of Sic60PY-Spectrin12–16-SNAP-His6 was carried out as described [21], but with some modifications that included a simultaneous dye-labeling reaction. Ubiquitination occurred in 1–2 mL reactions consisting of ubiquitination buffer (25 mM Tris–HCl, pH 7.5, 50 mM NaCl, 4 mM MgCl2, 1 mM DTT), 200 nM purified recombinant Schizosaccharomyces pombe E1, 2.5 μM recombinant, purified yeast UbcH7 (cloned into pGEX-6P-1 and purified via affinity chromatography followed by cleavage with PreScission Protease (GE)), ~2 μM purified yeast Rsp5 [21], 0.35 mM ubiquitin, and 4 mM ATP (as described in Ref. [33]), together with ~2 μM Sic60PY-Spectrin12–16-SNAP-His6 substrate. The specific activity of Rsp5 can vary, and we conducted small-scale (~50 μL) test ubiquitination reactions with each Rsp5 preparation to determine the most effective Rsp5 concentration. For dye labeling, 2 mM DTT and 7 μM SNAP-Surface Alexa Fluor 546 (New England Biolabs) were also included in the combined ubiquitination/labeling reaction. Conjugation and dye labeling were allowed to proceed for 110 min at 25 °C. The reaction was then centrifuged at 4 °C and 9000g to pellet any aggregates and the ubiquitinated substrates in the supernatant were purified on a Sephacryl S300 16/60 column (GE Healthcare) connected to an Akta Prime FPLC (GE Healthcare). Fractions containing the dye-labeled ubiquitinated substrate were identified by SDS-PAGE and detection of Alexa Fluor 546 fluorescence using a laser gel scanner (Typhoon 9400; GE Healthcare; excitation at 532 nm and emission filter 580 BP 30). After fluorescence imaging, the gels were also stained by Coomassie to determine the overall purity of the protein preparation. The number of ubiquitin moieties attached to a substrate was determined by comparing the molecular weight of the diffuse ubiquitinated protein bands to molecular weight standards (PageRuler Prestained Protein Ladder; ThermoFisher) in SDS-PAGE analysis. The purified ubiquitinated dye-labeled proteins were aliquoted, flash frozen in PBS with 10% glycerol, and stored at −80 °C.
A 1 l bacterial culture yielded approximately 1.8 mg (20 nmoles) of Sic60-Spectrin12–16-SNAP after the His purification. Each combined ubiquitination and dye-labeling reaction with ~3 nmoles of His purified Sic60PY-Spectrin12–16-SNAP-His6 yielded approximately 300–500 pmol of ubiquitinated labeled substrate after the final size-exclusion chromatography, assuming that on average 12 ubiquitin moieties were attached to each Sic60PY-Spectrin12–16-SNAP-His6 molecule. SNAP domains have only one dye-labeling site, so a single fluorophore can be attached to each protein molecule, and labeling efficiencies were 40–60%. The single-turnover proteasomal degradation assays used 0.8 pmol of substrate per reaction, so that each labeling reaction yielded sufficient substrate for at least 300 assays.
2.4. Dye-labeling reaction for UbL substrates
UbL-SNAP substrates were purified by Ni-NTA affinity chromatography, and 5 μM substrate were combined with 2 mM DTT and 10 μM SNAP-Surface Alexa Fluor 546 (New England Biolabs) in PBS for 60 min at 30 °C. After the labeling reaction, large aggregates were removed by centrifugation at 4 °C and 9000g, while free dye was removed by size-exclusion chromatography (Superdex S75 16/60, GE). Fractions containing the protein were identified by SDS-PAGE and dye-labeled protein was detected by in-gel fluorescence imaging using a laser gel scanner (Typhoon 9400, GE). After fluorescence imaging, the gels were stained by Coomassie to detect unlabeled contaminants. The protein was then aliquoted, flash frozen, and stored with 10% glycerol at −80 °C.
A 1 l bacterial culture yielded approximately 5 mg (150 nmoles) of UbL-SNAP-40-His6 protein after His purification. Each dye-labeling reaction with 5 nmol of His purified UbL-SNAP-40-His6 yielded approximately 2 nmol of labeled substrate after the final size-exclusion chromatography. Labeling efficiency varied between protein preparations from 35 to 50% for the UbL-SNAP-40-His6 protein to approximately 75% for the UbL-SNAP-His6 protein.
2.5. 26S proteasome purification
Yeast proteasome was purified from S. cerevisiae strain YYS40 (MATa rpn11::RPN113×FLAG-HIS3 leu2 his3 ura3 trp1 ade2 can1 ssd1) by affinity chromatography using FLAG antibodies (M2 agarose affinity beads, Sigma) as described previously [21], except that the lysis buffer consisted of 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10% glycerol, 1 mM DTT, and 5 mM ATP and cells were lysed using a homogenizer at 25,000 psi (EmulsiFlex Avestin C3). Proteasome preparations were analyzed by SDS-PAGE and native gel electrophoresis and compared with the published compositions [21,22]. Proteasome concentration was determined using the Pierce 660 nm protein assay reagent from ThermoFisher. Each proteasome preparation was checked for activity by testing degradation of polyubiquitinated Sic60PY-Spectrin12–16-SNAP-His6.
2.6. Proteinase K digestion
A portion of 10 nM Alexa Fluor 546-labeled substrate (Sic60PY-Spectrin12–16-SNAP-His6) in 1X degradation buffer (50 mM Tris–Cl pH 7.5, 5 mM MgCl2, 2% glycerol, 1 mM DTT, and 0.2 mg/mL BSA) was mixed with Proteinase K (100–500 μg/mL, as indicated) in proteinase K degradation buffer (50 mM Tris–Cl pH 7.5, 5 mM CaCl2, and 0.2 mg/mL BSA) and incubated at room temperature or 37 °C for 60–120 min. Reaction products were analyzed by SDS-PAGE and fluorescence imaging or by measuring fluorescence anisotropy as described above.
2.7. Gel-based proteasomal degradation assays
A portion of 50 nM purified yeast proteasome was mixed with 20 nM Alexa Fluor 546-labeled substrate and either ATP or ATPγS at the indicated concentration in 1X degradation buffer (50 mM Tris–Cl pH 7.5, 5 mM MgCl2, 2% glycerol, 1 mM DTT, and 0.2 mg/mL BSA). Samples were taken at the designated time points and added to a stop buffer containing SDS to quench the reaction. Samples were then analyzed by SDS-PAGE on 4–20% SDS-Tris-Glycine gradient gels (Lonza Biosciences) and imaged on a laser gel scanner (Typhoon 9400, GE) to detect the fluorescence emission from the Alexa Fluor 546 dye attached to the substrate (excitation 532 nm; emission filter 580 BP 30). Fluorescence intensity was quantified for each time point using ImageJ by taking the total fluorescence intensity of the bands representing full-length unmodified and ubiquitinated protein together. The decay of fluorescence intensity over time was analyzed by fitting the data to an equation describing single-exponential decay to a constant offset using Kaleidagraph software (version 4.1, Synergy Software).
2.8. Proteasome anisotropy degradation assays
Proteasome at the concentration indicated was mixed with approximately 20 nM Alexa Fluor 546-labeled substrate with ATP, ATPγS, or ADP at the indicated concentration, as well as DMSO only or epoxomycin dissolved in DMSO. Reactions were carried out in 1X degradation buffer (50 mM Tris–Cl pH 7.5, 5 mM MgCl2, 2% glycerol, 1 mM DTT, and 0.2 mg/mL BSA) at room temperature, and fluorescence anisotropy was measured as described below. Degradation assays carried out for the Michaelis–Menten analysis were conducted at the substrate and proteasome concentrations indicated. BSA (bovine serum albumin) was used in all degradation buffers to minimize nonspecific binding of the proteasome and fluorescent substrates to the 384-well plate. We confirmed that the 0.2 mg/mL BSA does not significantly affect the proteasome activity in the bulk assays.
2.9. Fluorescence anisotropy measurements and data analysis
Fluorescence polarization measurements were performed on an Analyst GT multimode reader (Molecular Devices; G-factor = 1.05) in 384-well plates (Corning or Greiner) with reaction volumes between 36 and 40 μL (excitation at 535 nm/50 nm bandwidth, emission at 580 nm/20 nm bandwidth). A polarization reading was taken every 0.5–1 min for the Michaelis–Menten experiments or every minute for all other experiments for a total experiment time course of 15–25 min. Anisotropy values (r) were calculated from the polarization readings (P) using the equation r = (2P)/(3 – P) [42]. To analyze the anisotropy decay, reaction curves measured under single-turnover conditions (pre-steady state experiments) were fitted to an equation describing single exponential decay to a constant offset using Kaleidagraph software (version 4.1, Synergy Software). The decay rate constants were interpreted as the rate constants of the entire protein degradation reaction and agreed well with the decay constants obtained in the gel-based proteasomal degradation assays.
The anisotropy data can be converted to the fraction of substrate remaining (X) as a function of time, using the equation
| (1) |
where r = measured anisotropy, rp = anisotropy of peptides after cleavage, rf = anisotropy of full-length protein, and g = fluorescence enhancement factor (i.e., the ratio of the total fluorescence intensity of the full-length dye-labeled substrate to the total fluorescence intensity of the cleaved dye-labeled peptide) [51,52]. For some fluorophores, the total fluorescence intensity does not change significantly over the course of the reaction, in which case the correction factor (g – 1)(rf – r) is small and can be neglected [51]. However, we observed a fluorescence enhancement of approximately 50% upon proteasomal degradation for both the Alexa-Fluor546-labeled polyubiquitinated and UbL substrates and thus corrected for this in our data when converting anisotropy values to the fraction of substrate remaining. The conversion also assumes that the degradation reaction proceeds to completion, so that rp represents the anisotropy after 100% of substrate has been converted to peptides. However, enzymatic reactions are often attenuated when reconstituted in vitro [47], and we find that roughly 15% of substrate typically escapes degradation at the end of the assay described here (see below). Therefore, the converted progression curves represent the decay of the 85% of the substrate that is degraded by the proteasome by the end of the reaction (see the Supplementary Materials). It is theoretically possible to introduce a correction factor into the equation above to account for the attenuation by multiplying by the observed extent of reaction (e.g., 0.85 here) and adding a constant offset representing unreacted protein (e.g., 0.15 here).
For Michaelis–Menten analysis, the anisotropy measurements were first converted to fraction of substrate remaining over time using Eq. (1). The linear part of the decay curves was then fitted to a straight line and the slope of the line was converted to an initial reaction rate, V, by multiplying by the initial substrate concentration. We did not correct for the fact that the degradation reactions may not have proceeded to completion, which likely introduced some error in Vmax, but not in KM [47].
3. Results and discussion
3.1. An assay based on fluorescence anisotropy
We set out to develop an assay to monitor protein degradation by the 26S proteasome continuously that could be implemented in a high-throughput format. Modern fluorophores, such as Alexa Fluor dyes, are photostable and their high quantum yields allow the detection of molecules at the nanomolar level with ease. A range of different strategies make it easy to label proteins with these dyes. We chose a dye (Alexa Fluor 546) with a fluorescence lifetime such that the anisotropy of the fluorescence signal of the dye-labeled protein differs significantly from that of the dye-labeled short peptides (Fig. 1a).
3.2. Substrate design
To test whether an assay based on fluorescence anisotropy measurements is able to monitor protein degradation by the 26S proteasome, we designed a proteasome substrate derived from domains 12 to 16 of the human cytoskeletal protein β-spectrin [48,49]. We attached the 20 kDa SNAP-tag domain to the C terminus of the spectrin fragment to allow us to label the protein at a single position. We also attached a 60-amino-acid sequence derived from the yeast protein Sic1 (Sic60) [21] to the N terminus of the spectrin fragment to serve as a degradation signal (degron) (Fig. 1b). The Sic60 sequence contains a PPXY motif (we use PPPY), which is recognized by the ubiquitin ligase Rsp5 and thus makes it possible to polyubiquitinate the protein in vitro [21]. The Sic60 sequence is also long enough to allow the proteasome to engage the protein and to initiate degradation [33]. We expect that degradation will proceed from the N-terminal Sic60 degron toward the C terminus of the protein where the dye-labeled SNAP-tag domain is located [53]. Finally, we added a hexahistidine tag to the very C terminus of the protein for purification. We refer to this substrate as the polyubiquitinated substrate.
3.3. Protein expression and labeling
We purified the substrate by overexpression in E. coli followed by Ni-NTA affinity chromatography. The protein was then ubiquitinated by incubation with Sch. pombe E1, E2 (UbcH7), and E3 (Rsp5) enzymes in vitro and simultaneously dye-labeled using a benzyl-guanine derivative of Alexa Fluor 546 (SNAP-surface Alexa Fluor 546). After the reaction, the substrate was purified by size-exclusion chromatography. The final full-length dye-labeled polyubiquitinated substrate had average molecular weight ~200 kDa, which suggests that about 12 ubiquitin moieties were added to a substrate molecule by the ubiquitination reaction. The molecular weight of the final protein, and thus the number of ubiquitin moieties attached, varied slightly from preparation to preparation.
3.4. Anisotropy signal
We first determined the difference in fluorescence anisotropy between a full-length dye-labeled protein and dye-labeled peptides generated by proteolysis. The fluorescence anisotropy of the polyubiquitinated substrate protein described above was 0.28, which is within a reasonable range of the value expected for a 200 kDa protein labeled with a dye with a 4 ns lifetime (Fig. 1c) [42]. We then digested the protein with the nonspecific protease Proteinase K at 37 °C for 2 h and observed that the anisotropy decreased to around 0.1 (Fig. 1c). SDS-PAGE analysis of the reaction product demonstrated that the substrate was digested completely into short peptides smaller than 5 kDa, and no protein fragments of intermediate-size could be detected (Fig. S1). We conclude that full-length and digested substrates can be detected and distinguished from each other by measuring the fluorescence anisotropy of a covalently attached dye in our assay, as expected [44–46] (Fig. 1c).
3.5. Proteasomal degradation monitored by SDS-PAGE analysis
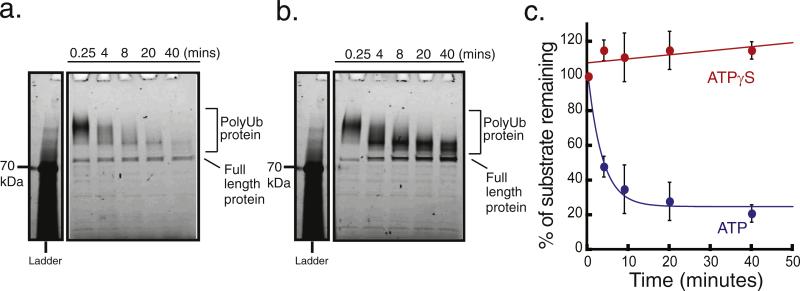
Next, we asked whether it would be possible to use fluorescence anisotropy measurements to follow proteasome activity. We first confirmed that the dye-labeled, ubiquitinated substrate could be degraded by the proteasome in the presence of ATP by the classic SDS-PAGE gel based assay. Portions of 10–20 nM polyubiquitinated dye-labeled substrate were incubated with 50 nM purified yeast proteasome in the presence of 2 mM ATP at room temperature to carry out the degradation reaction with excess proteasome, near single-turnover conditions. Samples were taken at five time points over the course of an hour and immediately added to a stop buffer that contained SDS to disassemble the proteasome and inhibit any further proteolysis. We analyzed the samples by SDS-PAGE and quantified the amount of substrate remaining at each time point by imaging the Alexa Fluor 546 fluorescence on a laser gel scanner (Typhoon 9400; GE) (Fig. 2a). The ubiquitinated protein ran as a diffuse high-molecular-weight band on the gel, centered at ~200 kDa, whereas protein lacking the ubiquitin modification showed a well-defined band at the expected molecular weight of 90 kDa (Fig. 2a). We estimated the total amount of undigested substrate protein by measuring the total fluorescence intensity of the region of a gel lane from the high-molecular-weight end of the band representing ubiquitinated protein to just below the band representing full-length unmodified protein. The total fluorescence in this region decreased with time (Fig. 2a). At most 15% of full-length protein remained at the end of the assay, and no lower-molecular-weight bands appeared over time, suggesting that the substrate was degraded progressively and almost completely to short peptides (Fig. 2a).
Fig. 2.
Proteasomal degradation of the polyubiquitinated substrate monitored by SDS-PAGE. Degradation of the polyubiquitinated substrate by the proteasome in the presence of ATP or ATPγS, analyzed by SDS-PAGE and protein detection by a laser gel scanner (Typhoon 9400, GE). 20 nM substrate was incubated with 50 nM proteasome at 30 °C in the presence of (a) 2 mM ATP or (b) 2 mM ATPγS. Aliquots were taken at the times indicated and added to a stop buffer that contained SDS in tris-glycine buffer. The first time point was taken at 0.25 min. The first lane shows molecular weight standards, with the 70 kDa band being visualized using the Typhoon Imager. (c) Quantification of polyubiquitinated substrate degradation in the presence of ATP (blue circles) or ATPγS (red circles). The graph plots the amount of protein remaining as the percentage of protein present at 0.25 min. Protein amounts were estimated by integrating dye fluorescence intensity in the region from the top of the diffuse band representing ubiquitinated protein to the bottom of the band representing full-length unmodified protein. Error bars show the standard error of the percentage of fluorescence intensity remaining in three separate experiments.
Protein degradation by the 26S proteasome is driven by ATP hydrolysis and is inhibited if ATP is replaced with the poorly hydrolyzed ATP analogue ATPγS [54,55]. When we presented polyubiquitinated dye-labeled substrate to the proteasome in the presence of 2 mM ATPγS, the center of the diffuse band representing polyubiquitinated protein moved toward decreasing molecular weights, while the band at 90 kDa representing full-length unmodified protein increased in intensity (Fig. 2b). Presumably, these changes were caused by the deubiquitinases on the protea-some removing ubiquitin moieties from the substrate in an ATP-independent manner [56,57]. No protein degradation occurred in the presence of ATPγS because the total combined fluorescence intensity of the ubiquitinated band and the full-length band remained constant (Fig. 2b). The total fluorescence intensity over time from both the ATP and the ATPγS experiments was quantified and normalized to the first time point, shown in the graph in Fig. 2c.
The fluorescence-based gel imaging detected no partially degraded products labeled with the fluorophore, suggesting that the substrate was degraded progressively. Most likely, degradation proceeded from the N-terminal Sic60 degron toward the C terminus, as the proteasome initiates degradation at unstructured regions close to the ubiquitination site [21,50,53,58]. This assay, however, relies on detection of peptides containing the fluorescent tag, so that a single cleavage could remove the tag, rendering the remainder of the protein invisible. Therefore, we labeled a closely related substrate that lacked the fluorescent moiety (Sic60PY-Spectrin12–16-His6) radioactively at 14 methionine residues distributed throughout the polypeptide chain, ubiquitinated it, and presented the protein to 50 nM purified yeast proteasome (Fig. S2 in Supplementary materials). Analysis of the degradation reaction by SDS-PAGE and electronic autoradiography showed that no partially degraded protein fragments accumulated as the full-length protein disappeared. We conclude that Sic60-Spectrin12–16-His6 was degraded completely and progressively [33,40,53,59,60]. We expect that the fluorescently tagged substrate behaves similarly, as observed by the fluorescence-based SDS-PAGE analysis of degradation by the proteasome.
3.6. Assay reports on 26S proteasome activity
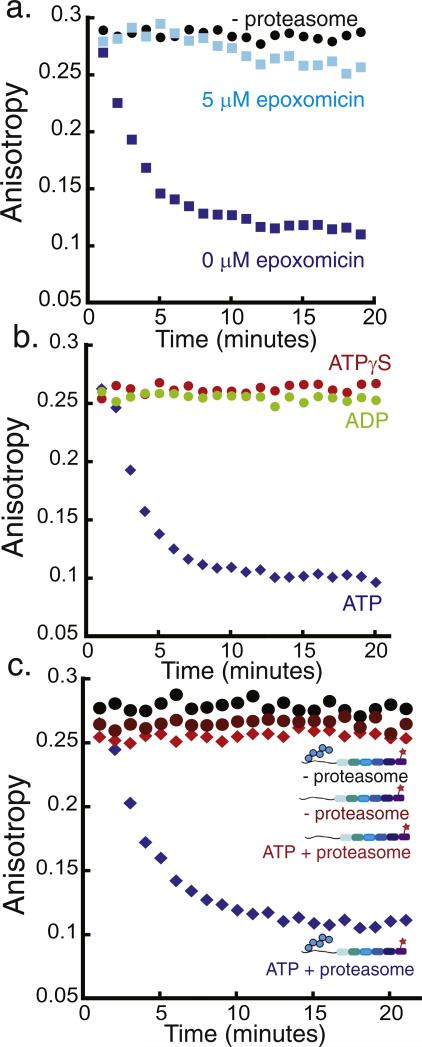
We then measured the fluorescence anisotropy over time under the same assay conditions as described above, where proteasome was in excess of substrate (Fig. 3a), though much lower proteasome concentrations can also effectively be used to conduct degradation reactions (Fig. S3 in Supplementary materials). In the presence of ATP and proteasome, the anisotropy decreased exponentially from the value expected for intact polyubiquitinated protein and plateaued at the value expected for short peptides by the end of the time course (Fig. 3a). The anisotropy values obtained after proteasomal digestion of the substrate were comparable to the values obtained from Proteinase K digestion of the substrate (compare Figs. 1c and 3a). The progression curves were well described by a two-state model in which the dye-labeled full-length intact protein is converted to the species in which the dye is attached to small peptides, just as observed when proteasomal degradation is monitored by SDS-PAGE (Fig. 2). The degradation rate constant obtained by anisotropy measurement agrees well with the decay rate constant obtained by SDS-PAGE analysis (Supplementary materials Table S2). Anisotropy progression curves can be converted to represent the fraction of substrate protein degraded over time explicitly, which requires correction factors for the change in fluorescence intensity upon degradation and for the extent to which the reaction progresses (see Materials and methods and Fig. S4). The two approaches yielded very similar rate constants for proteasomal protein degradation (Figs. S4 and S5). In the absence of proteasome, the anisotropy values stayed constant over the assay time course (Fig. 3a). The decrease in fluorescence anisotropy was caused by proteolysis by the proteasome because a specific inhibitor of the proteasome's proteolytic sites, epoxomicin, prevented the decrease in fluorescence anisotropy (Fig. 3a). We conclude that the anisotropy assay allows us to follow the kinetics of proteasomal degradation.
Fig. 3.
Monitoring 26S proteasome activity by fluorescence anisotropy. Degradation of the polyubiquitinated Alexa Fluor 546-labeled substrate by purified yeast proteasome as monitored by fluorescence anisotropy measurements. The degradation reaction was monitored at room temperature over 20 min by performing anisotropy readings every minute. One representative experiment is shown in each graph, and the decay rate constants obtained from the exponential fits to repeat experiments can be found in Supplementary Materials Table S2. The graphs plot fluorescence anisotropy against time. (a) Degradation of approximately 20 nM polyubiquitinated substrate in the presence of 2 mM ATP and 50 nM proteasome (blue squares), 50 nM proteasome + 5 μM epoxomicin (light blue squares), or no proteasome (black circles). (b) Degradation of approximately 20 nM polyubiquitinated substrate by 50 nM proteasome and 2 mM ATP (blue diamonds), 2 mM ADP (green circles), or 2 mM ATPγS (red circles). (c) Degradation of approximately 20 nM polyubiquitinated substrate in the presence of 2 mM ATP and 50 nM proteasome (blue diamonds), degradation of approximately 20 nM substrate lacking the polyubiquitin modification by 50 nM proteasome (red diamonds), and incubation of approximately 20 nM polyubiquitinated substrate without proteasome (black circles).
The 20S core particle of the proteasome, which contains the pro-tease active sites, can degrade peptides on its own without the 19S caps under certain conditions [19,61,62]. However, to recognize and degrade folded proteins, the 20S core particle has to associate with 19S caps and form the 26S holoproteasome [1]. We asked whether the anisotropy-based degradation assay reflects specifically 26S protea-some activity rather than 20S core activity. We found that protein degradation, as detected by the decrease in fluorescence anisotropy, required ATP hydrolysis, because replacing ATP with the nonhydrolyzable ATP analogue ATPγS completely inhibited the decrease in anisotropy observed in the presence of ATP (Fig. 3b). Similarly, replacing ATP with ADP also stabilized the substrate (Fig. 3b).
Degradation also required the substrate protein to contain a proteasome-binding tag, in the case of the model substrate analyzed here a polyubiquitin modification [63]. Protein lacking a polyubiquitin modification remained stable when presented to proteasome, even in the presence of ATP (Fig. 3c). Both the polyubiquitinated substrate and the substrate lacking the polyubiquitin modification maintained constant anisotropy values over time when proteasome was omitted from the reaction (Fig. 3c). Thus, no change in anisotropy over time was detected for substrates lacking a proteasome-binding tag or when ATP hydrolysis was inhibited, suggesting that the assay monitored 26S proteasome activity specifically.
3.7. Developing model substrates independent of ubiquitin modification
In the experiments described above, we targeted a protein to the proteasome by a polyubiquitination signal. Following the degradation of this type of substrate makes it possible to investigate the role of the polyubiquitin chain in proteasomal degradation. However, the synthesis of ubiquitinated proteins requires reaction of the protein with purified enzymes. Furthermore, it is difficult to determine the exact number of ubiquitin moieties attached to the substrate after the ubiquitination reaction, making precise determination of substrate concentrations difficult. Therefore, we constructed a proteasome substrate that does not require ubiquitination for degradation but still reports on the activity of the 26S proteasome. This substrate was targeted to the proteasome by the ubiquitin-like (UbL) domain of S. cerevisiae Rad23 (Fig. 4a). The UbL domain can be recognized by several receptors on the proteasome [64–67] and can target model proteins for proteasomal degradation in vitro and in vivo [50,68,69]. We fused the UbL domain to the N terminus of a SNAP-tag domain followed by an unstructured region that functions as a proteasomal initiation site (denoted UbL substrate). A hexahistidine tag was added at the C terminus for purification.
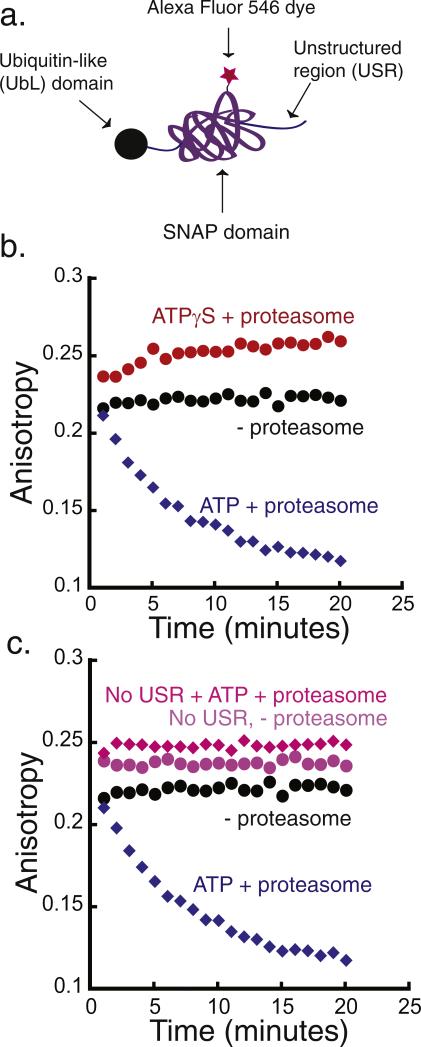
Fig. 4.
Degradation of a simplified model protein. (a) Schematic representation of a model substrate that is targeted to the proteasome by a ubiquitin-like (UbL) domain from S. cerevisiae Rad23 protein. The substrate consists of the UbL domain at its N terminus, followed by the SNAP-tag and a 40-amino acid-long unstructured region at the C terminus that functions as the proteasome initiation site (UbL-SNAP-40-His6). (b) Degradation of UbL-SNAP-40-His6 by purified yeast proteasome monitored by fluorescence anisotropy. The degradation reaction was monitored at room temperature over 20 min, taking an anisotropy reading every minute. Incubation of 20 nM substrate in the presence of 2 mM ATP and 50 nM proteasome (blue diamonds), of 20 nM substrate in the presence of 2 mM ATPγS and 50 nM proteasome (red circles), and of 20 nM substrate alone (black circles). (c) Degradation reaction of UbL substrate with (UbL-SNAP-40-His6) and without (UbL-SNAP-His6) a C-terminal unstructured region. Experiments are as in (b): 20 nM of UbL-SNAP-40-His6 with 50 nM proteasome and 2 mM ATP (blue diamonds), 20 nM UbL-SNAP-His6 with 50 nM proteasome and 2 mM ATP (pink diamonds), 20 nM substrate UbL-SNAP-40-His6 alone (black circles), and 20 nM UbL-SNAP-His6 alone (light purple circles).
We purified the UbL substrates from E. coli, labeled with SNAP-surface Alexa Fluor 546, and conducted the fluorescence anisotropy assay with purified yeast proteasome as described above. We first tested a UbL substrate that had a 40-amino acid-long unstructured region fused to the C terminus of the SNAP domain (Fig. 4b; Fig. S5b in Supplementary Materials). When we incubated this substrate with proteasome in the presence of ATP, the fluorescence anisotropy decreased over time, as expected when the protein is degraded by the proteasome (Fig. 4b and Fig. S5). The decrease in anisotropy depended on the presence of proteasome, and replacing ATP with ATPγS or inhibiting the proteolytic sites with epoxomicin prevented degradation (Fig. 4b, Fig. S5, and Fig. S6). Degradation also depended on a functional proteasome initiation site as a substrate without an unstructured region escaped degradation [50,68] (Fig. 4c, Fig. S5c). Thus, the UbL-SNAP-40-His6 substrate described here is a convenient and sensitive reporter for 26S proteasome activity, and substrates targeted to the proteasome via the UbL domain can be used effectively in the anisotropy assay.
3.8. Michaelis–Menten analysis
Traditional strategies for characterizing enzyme mechanisms often involve reactions under conditions where the enzyme turns over multiple times and the reaction is monitored at steady state. The model substrates described above can be used for the classic Michaelis–Menten analysis of proteasome activity.
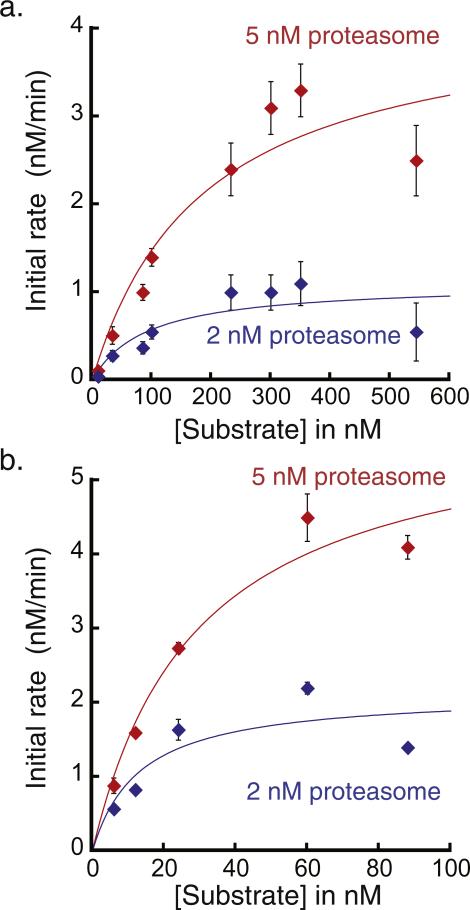
We incubated the UbL-SNAP-40-His6 substrate with purified yeast proteasome in the presence of ATP and followed the fluorescence anisotropy of the labeled substrate over time. We investigated eight substrate concentrations ranging from 10 to 544 nM at two different proteasome concentrations (2 and 5 nM) and measured the initial rates of the fluorescence anisotropy change for each condition (Fig. 5a). The measured initial rates followed classic MichaeliseMenten behavior (Fig. 5a). The Vmax depended linearly on the proteasome concentration, whereas the KM values remained constant within error between two proteasome concentrations.
Fig. 5.
Michaelis–Menten analysis of initial proteasomal protein degradation rates of UbL-SNAP-40-His6 or the polyubiquitinated substrate monitored by fluorescence anisotropy at room temperature. (a) Degradation rates in nM/min were determined in the presence of 2 nM or 5 nM proteasome for eight UbL-SNAP-40-His6 substrate concentrations ranging from 10 to 544 nM. Each point represents the average of initial rates from three to nine separate experiments and error bars represent the standard error of the mean. The kcat values for degradation by 2 and 5 nM proteasome were 0.55 ± 0.14 and 0.85 ± 0.19 min −1, respectively. KM was estimated as 91 ± 76 nM for degradation by 2 nM proteasome and 187 ± 101 nM for degradation by 5 nM proteasome. (b) Degradation rates in nM/min were determined for the polyubiquitinated substrate at five substrate concentrations at 2 nM or 5 nM proteasome concentration. Each point represents average initial rates from at least three experiments and error bars represent the standard error of the mean. The kcat values for degradation by the 2 and 5 nM proteasome were 1.1 ± 0.3 and 1.2 ± 0.2 min −1, respectively. KM was estimated as 13 ± 11 nM at 2 nM proteasome and 29 ± 10 nM at 5 nM proteasome.
We also characterized the polyubiquitinated substrate by Michaelis–Menten analysis (Fig. 5b). These measurements suggest that the polyubiquitinated substrate binds proteasome more tightly than the UbL-SNAP-40-His6 substrate and is degraded at least as rapidly. The ubiquitination method used in this paper is efficient, but leads to attachment of one or more heterogeneous polyubiquitin chains to the substrate, which makes it difficult to know the precise concentration of ubiquitinated substrate and can therefore lead to uncertainty in the determined KM and Vmax values. Other methods allow the synthesis of substrate proteins with precisely defined ubiquitin modifications [24,26,29]. Nevertheless, together, these data demonstrate that the assay described in this paper can easily be used to characterize proteasomal degradation by standard kinetic approaches.
4. Conclusions
In summary, we present a simple and robust assay for 26S proteasome activity based on fluorescence anisotropy. The assay can detect the degradation activity of as little as 2 nM proteasome and only requires 30 μL volume for one reaction. For a standard laboratory operation, the setup time for one assay is less than 30 min and the results can be obtained using a 384-well plate in a high-throughput manner immediately after the course of the assay (for us, 20 min). We designed two different proteasome substrates, each containing a SNAP-tag for site-specific covalent dye labeling. The assay does not depend on the use of SNAP-tags, and other labeling strategies should work just as effectively. One substrate was targeted to the proteasome by an enzymatically attached polyubiquitin tag. Other methods to ubiquitinate proteins exist and are, in principle, compatible with the assay described here. We also targeted a substrate to the proteasome via a UbL domain encoded in the primary sequence of the protein and demonstrated that both types of substrate can be used to study the kinetics of degradation using classical Michaelis–Menten analysis. The advantages of the 26S proteasome activity assay described in this paper are high sensitivity, versatile substrate options, simple operation, and high-throughput detection, and it will be a useful tool to probe for modulators of proteasome activity, particularly those that may impact proteasomal degradation prior to peptide hydrolysis.
Supplementary Material
Acknowledgments
Fluorescence polarization data were acquired in Northwestern University's High-Throughput Analysis lab, which is supported by the Robert H. Lurie Comprehensive Cancer Center. We especially thank Sara Fernandez Dunne and Dr. Chi-Hao Luan for help with the anisotropy experiments. We are also grateful to the members of the Matouschek lab at the University of Texas at Austin for critically reading this manuscript. The work was supported by NSF Grants MCB-1022117 and DMR-1206868 (to JFM), NIH Grants 1U54CA193419 (JFM), 1R01GM105847 (JFM), U54GM105816 (AM), R21 CA 196456 (AM) and R21CA191664 (AM), Welch Foundation Grant F-1817 (AM), and Cancer Prevention and Research Institute of Texas (CPRIT) Grant RP140328 (AM).
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ab.2016.05.026.
References
- 1.Finley D, Chen X, Walters KJ. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem. Sci. 2015;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 3.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Forster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 5.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. J. Cell. Biol. 2012;199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheridan C. Drug makers target ubiquitin proteasome pathway anew. Nat. Biotechnol. 2015;33:1115–1117. doi: 10.1038/nbt1115-1115. [DOI] [PubMed] [Google Scholar]
- 11.Dou QP, Zonder JA. Overview of proteasome inhibitor-based anti-cancer therapies: perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr. Cancer Drug Targets. 2014;14:517–536. doi: 10.2174/1568009614666140804154511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian Z, D'Arcy P, Wang X, Ray A, Tai YT, Hu Y, Carrasco RD, Richardson P, Linder S, Chauhan D, Anderson KC. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 14.Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp. Mol. Med. 2015;47:e147. doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 16.Dantuma NP, Bott LC. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front. Mol. Neurosci. 2014;7:70. doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang NY, Tang Z, Liu CW. alpha-Synuclein protofibrils inhibit 26 S proteasome-mediated protein degradation: understanding the cytotoxicity of protein protofibrils in neurodegenerative disease pathogenesis. J. Biol. Chem. 2008;283:20288–20298. doi: 10.1074/jbc.M710560200. [DOI] [PubMed] [Google Scholar]
- 18.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 20.Liggett A, Crawford LJ, Walker B, Morris TC, Irvine AE. Methods for measuring proteasome activity: current limitations and future developments. Leuk. Res. 2010;34:1403–1409. doi: 10.1016/j.leukres.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Saeki Y, Isono E, Toh EA. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Tomko RJ, Jr., Hochstrasser M. Proteasomes: isolation and activity assays. Curr. Protoc. Cell Biol. 2015;67:3 43 1–3 43 20. doi: 10.1002/0471143030.cb0343s67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faggiano S, Alfano C, Pastore A. The missing links to link ubiquitin: Methods for the enzymatic production of polyubiquitin chains. Anal. Biochem. 2016;492:82–90. doi: 10.1016/j.ab.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Hemantha HP, Bavikar SN, Herman-Bachinsky Y, Haj-Yahya N, Bondalapati S, Ciechanover A, Brik A. Nonenzymatic polyubiquitination of expressed proteins. J. Am. Chem. Soc. 2014;136:2665–2673. doi: 10.1021/ja412594d. [DOI] [PubMed] [Google Scholar]
- 25.Xu S, Patel P, Abbasian M, Giegel D, Xie W, Mercurio F, Cox S. In vitro SCFbeta-Trcp1-mediated IkappaBalpha ubiquitination assay for high-throughput screen. Methods Enzymol. 2005;399:729–740. doi: 10.1016/S0076-6879(05)99048-4. [DOI] [PubMed] [Google Scholar]
- 26.Bavikar SN, Spasser L, Haj-Yahya M, Karthikeyan SV, Moyal T, Kumar KS, Brik A. Chemical synthesis of ubiquitinated peptides with varying lengths and types of ubiquitin chains to explore the activity of deubiquitinases. Angew. Chem. Int. Ed. Engl. 2012;51:758–763. doi: 10.1002/anie.201106430. [DOI] [PubMed] [Google Scholar]
- 27.McGinty RK, Chatterjee C, Muir TW. Semisynthesis of ubiquitylated proteins. Methods Enzymol. 2009;462:225–243. doi: 10.1016/S0076-6879(09)62011-5. [DOI] [PubMed] [Google Scholar]
- 28.Verma R, Chi Y, Deshaies RJ. Cell-free ubiquitination of cell cycle regulators in budding yeast extracts. Methods Enzymol. 1997;283:366–376. doi: 10.1016/s0076-6879(97)83030-3. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Fonts K, Matouschek A. A rapid and versatile method for generating proteins with defined ubiquitin chains. Biochemistry. 2016;55:1898–1908. doi: 10.1021/acs.biochem.5b01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keren-Kaplan T, Attali I, Motamedchaboki K, Davis BA, Tanner N, Reshef Y, Laudon E, Kolot M, Levin-Kravets O, Kleifeld O, Glickman M, Horazdovsky BF, Wolf DA, Prag G. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J. 2012;31:378–390. doi: 10.1038/emboj.2011.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verma R, McDonald H, Yates JR, 3rd, Deshaies RJ. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Mol. Cell. 2001;8:439–448. doi: 10.1016/s1097-2765(01)00308-2. [DOI] [PubMed] [Google Scholar]
- 32.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 33.Kraut DA, Israeli E, Schrader EK, Patil A, Nakai K, Nanavati D, Inobe T, Matouschek A. Sequence- and species-dependence of proteasomal processivity. ACS Chem. Biol. 2012;7:1444–1453. doi: 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriegenburg F, Seeger M, Saeki Y, Tanaka K, Lauridsen AM, Hartmann-Petersen R, Hendil KB. Mammalian 26S proteasomes remain intact during protein degradation. Cell. 2008;135:355–365. doi: 10.1016/j.cell.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 35.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J. Biol. Chem. 2013;288:29215–29222. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat. Struct. Mol. Biol. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khmelinskii A, Meurer M, Ho CT, Besenbeck B, Fuller J, Lemberg MK, Bukau B, Mogk A, Knop M. Incomplete proteasomal degradation of green fluorescent proteins in the context of tandem fluorescent protein timers. Mol. Biol. Cell. 2015;27:360–370. doi: 10.1091/mbc.E15-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wohlever ML, Nager AR, Baker TA, Sauer RT. Engineering fluorescent protein substrates for the AAA+ Lon protease. Protein Eng. Des. Sel. 2013;26:299–305. doi: 10.1093/protein/gzs105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J. Biol. Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nager AR, Baker TA, Sauer RT. Stepwise unfolding of a beta barrel protein by the AAA+ ClpXP protease. J. Mol. Biol. 2011;413:4–16. doi: 10.1016/j.jmb.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blommel PG, Fox BG. Fluorescence anisotropy assay for proteolysis of specifically labeled fusion proteins. Anal. Biochem. 2005;336:75–86. doi: 10.1016/j.ab.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Cleemann F, Karuso P. Fluorescence anisotropy assay for the traceless kinetic analysis of protein digestion. Anal. Chem. 2008;80:4170–4174. doi: 10.1021/ac7025783. [DOI] [PubMed] [Google Scholar]
- 46.Gilmore MA, Williams D, Okawa Y, Holguin B, James NG, Ross JA, Roger Aoki K, Jameson DM, Steward LE. Depolarization after resonance energy transfer (DARET): a sensitive fluorescence-based assay for botulinum neurotoxin protease activity. Anal. Biochem. 2011;413:36–42. doi: 10.1016/j.ab.2011.01.043. [DOI] [PubMed] [Google Scholar]
- 47.Sem DS, McNeeley PA. Application of fluorescence polarization to the steady-state enzyme kinetic analysis of calpain II. FEBS Lett. 1999;443:17–19. doi: 10.1016/s0014-5793(98)01655-x. [DOI] [PubMed] [Google Scholar]
- 48.Ipsaro JJ, Huang L, Gutierrez L, MacDonald RI. Molecular epitopes of the ankyrin-spectrin interaction. Biochemistry. 2008;47:7452–7464. doi: 10.1021/bi702525z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ipsaro JJ, Huang L, Mondragon A. Structures of the spectrin-ankyrin interaction binding domains. Blood. 2009;113:5385–5393. doi: 10.1182/blood-2008-10-184358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jameson DM, Mocz G. Fluorescence polarization/anisotropy approaches to study protein-ligand interactions: effects of errors and uncertainties. Methods Mol. Biol. 2005;305:301–322. doi: 10.1385/1-59259-912-5:301. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, Chen Y, Fierke CA. A real-time fluorescence polarization activity assay to screen for inhibitors of bacterial ribonuclease P. Nucleic Acids Res. 2014;42:e159. doi: 10.1093/nar/gku850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 54.Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol. Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 57.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 58.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 59.Hoyt MA, Zich J, Takeuchi J, Zhang M, Govaerts C, Coffino P. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daskalogianni C, Apcher S, Candeias MM, Naski N, Calvo F, Fahraeus R. Gly-Ala repeats induce position- and substrate-specific regulation of 26 S proteasome-dependent partial processing. J. Biol. Chem. 2008;283:30090–30100. doi: 10.1074/jbc.M803290200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 62.Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 65.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 66.Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 2002;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 67.Hamazaki J, Hirayama S, Murata S. Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet. 11:e1005401. doi: 10.1371/journal.pgen.1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat. Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM, Matouschek A. Sequence composition of disordered regions fine-tunes protein half-life. Nat. Struct. Mol. Biol. 2015;22:214–221. doi: 10.1038/nsmb.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.