Abstract
Background:
We identified auranofin as an antimicrobial compound utilizing a high-throughput screen using a Caenorhabditis elegans–Staphylococcus aureus infection model.
Results/methodology:
Treatment of infected nematodes with auranofin resulted in a prolonged survival rate of 95%, reached with 0.78 μg/ml. Further investigation of the antimicrobial activity of auranofin found inhibition against S. aureus, Enterococcus faecium and Enterococcus faecalis. Importantly, the fungal pathogens Cryptococcus neoformans was also effectively inhibited with an MIC at 0.5 μg/ml. Auranofin appears to target the thioredoxin system.
Conclusion:
This work provides extensive additional data on the antibacterial effects of auranofin that includes both reference and clinical isolates and reports a novel inhibition of fungal pathogens by this compound.
Keywords: : antimicrobial, auranofin, Bucillus subtilis, Candida albicans, Cryptococcus neoformans, Enterococcus faecalis, Enterococcus faecium, Staphylococcus aureus
With the rising emergence of drug-resistant pathogens, both bacterial and fungal, there is a need for new antimicrobial compounds. Staphylococcus aureus has emerged as a significant Gram-positive bacterial pathogen, presenting drug-resistant strains such as methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus. In 2005, 94,000 life-threating infections were attributed to S. aureus [1]. The increased prevalence of antimicrobial resistance is not just restricted to bacterial pathogens; it can also be found among fungal microbes. Common medications provided for fungal infections are the azole group of drugs. In a study conducted between 2010 and 2012, 496 clinical isolates of fungi were collected and evaluated for antifungal resistance. Among the pathogens evaluated fluconazole resistance was found among Candida glabrata (6.8%), Candida parapsilosis (5.7%) and Candida tropicalis (3.6%) [2]. For example, in a 5-year study between 2008 and 2013 conducted in Atlanta, Georgia and Baltimore, Maryland, researchers identified 3848 candidemia blood stream infection cases. The study identified an increasing trend in the number of cases with echinocandins resistance, increasing from 1.2 to 2.9% (147% increase) in Atlanta, and increasing from 2.0 to 3.5% (77% increase) over the course of the study in Baltimore [3].
There is an increasing concern for resistance for other pathogens. Especially, for the infectious fungal pathogen Cryptococcus neoformans, treatment is largely dependent upon amphotericin B and azole drugs. In particular, fluconazole is provided as maintenance therapy. In a survey performed in China that included 426 clinical isolates, 2% of the C. neoformans strains were found to be fluconazole resistant [4]. In a report from Kenya, 3% of a collection of 67 C. neoformans isolates exhibited MICs equal to 16 μg/ml. Within the same study, 10% of the isolates had an MIC to 5-flucytosine between 8 and 16 μg/ml [5]. In a 2007 surveillance study in Kenya, fluconazole resistance was reported at 11.2% with MICs ≥64 μg/ml [6]. Among a group of 27 patients from South Africa that experienced cryptococcal meningitis relapse, 76% presented with fluconazole resistance and experienced a high mortality rate of 54% within 6 months of follow-up [7].
In an effort to discover new antimicrobial drugs we screened a collection of US FDA-approved compounds using a Caenorhabditis elegans–S. aureus infection model [8]. The anti-inflammatory, auranofin, was identified to have the ability to inhibit an infection within the nematode. As an anti-inflammatory compound, auranofin has been used to treat arthritic conditions and previous groups indicate that auranofin can exhibit antimicrobial activities. Bacterial pathogens found to be susceptible to auranofin include Clostridium difficile, Treponema denticola and Pseudomonas putida [9–11]. Both C. difficile and T. denticola had a reduction in selenoproteins, as a result of auranofin exposure and investigation of microbial inhibition by auranofin has provided evidence that the drug binds to hydrogen selenide, blocking selenium utilization by bacteria, preventing selenoprotein synthesis [9]. The function of blocking selenoproteins appears to also be conserved in parasitic worms as well and x-ray crystallography demonstrates the binding of thioredoxin-glutathione reductase with auranofin. We found that the antimicrobial capacity of auranofin extends beyond S. aureus and bacteria, inhibiting fungi and we report our findings on the most important resistant bacterial pathogens (often represented with the acronym ESKAPE for: pathogens: Enterococcus faecium, S. aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.) and two important fungal pathogens C. neoformans and Candida spp.
Materials & methods
Strains & culture conditions
Bacterial and fungal reference strains used in the described studies are listed in Table 1. Bacterial strains were grown at 37°C and fungal cultures were grown at 30°C, unless otherwise stated. All bacterial and fungal cells were stored at -80°C until needed. The clinical isolates were derived from the USA (Massachusetts General Hospital, MA, USA) and China (made available by BEI Resources).
Table 1. . Microbial strains used in this study.
| Name | Strain |
|---|---|
|
Bacteria | |
|
Staphylococcus aureus |
MW2 |
|
Enterococcus faecium |
2421 |
|
Klebsiella pneumoniae |
77326 |
|
Acinetobacter baumannii |
ATCC 17978 |
|
Pseudomonas aeruginosa |
PA14 |
|
Enterobacter sp. |
KCTC 2625 |
|
Bacillus subtilis |
PY 79 |
|
Enterococcus faecalis |
MMH 594 |
|
Fungi | |
|
Candida albicans |
SC5314 (CAN14) |
|
Candida glabrata |
ATCC 90030 |
|
Candida parapsilosis |
ATCC 22019 |
|
Candida tropicalis |
ATCC 13803 |
| Cryptococcus neoformans | KN99α |
Infection assay
The screening methodology has been described previously [8]. Our high-throughput screening (HTS) assay sought to identify compounds that exhibited prolonged survival of worms infected with the MRSA strain MW2. In brief, each well of a 384-well plate (Corning no. 3712) received 0.1 μl of compound at a concentration of 2 mg/ml in dimethyl sulfoxide (DMSO) for a final evaluated concentration of 2.86 μg/ml for the assay.
The Biomol 4 library (Enzo Life Sciences, NY, USA), consisting of 640 FDA-approved drugs, was screened in search of compounds that improved the survival of S. aureus-infected worms. The positive control for the assay was vancomycin, at 10 μg/ml, while 1% DMSO was included as the negative control. Plates were seeded with MW2 S. aureus that was grown overnight in tryptic soy broth (TSB) under aerobic conditions then seeded into fresh TSB media for additional overnight growth at 37°C under static growth conditions by adding 100 μl of the agitated culture to 10 ml of fresh TSB (Figure 1). The MW2 culture was added to 384-well plates at a final concentration of 0.15 OD600 at a volume of 35 μl.
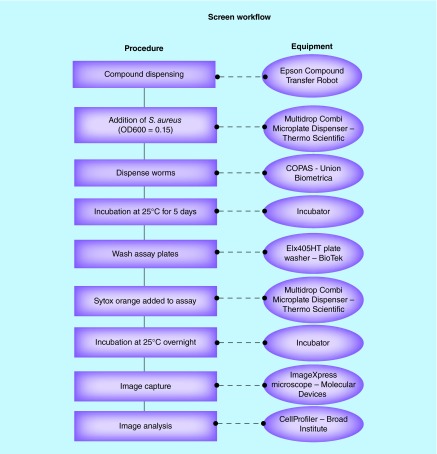
Figure 1. . Workflow chart of the screening methodology.
Screening chemical compound collections involved co-incubation of Caenorhabditis elegans with Staphylocuccus aureus and US FDA-approved compounds. Screening equipment was used to facilitate evaluation of the compounds in a 384-well format.
During the infection process, synchronized worms at the early adult stage of development were dispended into MW2 seeded 384-plates, depositing 15 worms per well using a Union Biometrica Complex Object Parametric Analyzer (COPAS Biosort). glp4::sek1 worms were employed for this assay because the glp4 mutation renders the worms susceptible to pathogen infection, and the sek1 mutation provides the convenience of not producing progeny when incubated at 25°C. Prior to being dispensed into the assay plates, worms were maintained on SK media plates and were provided Escherichia coli HB101 as a food source. Worms were harvested and washed with M9 buffer.
The total volume for the HTS assay plate was 70 μl, comprised of 70% M9 buffer, 19% sheath solution (Union Biometrica Part no. 300-5101-000), 10% TSB and 1% DMSO or compound dissolved in DMSO. Plates were incubated at 25°C with 80–85% humidity for 5 days before being washed six-times with a microplate washer (BioTek ELx405) to remove bacteria from the wells. A volume of 10 μl of M9 was left in the wells after the washing process and 60 μl of 1.4 μM Sytox Orange (Life Technologies), which stains the nucleic acids in cells with compromised membranes, indicating a dead worm, was added to each well. The stained plates were covered with a Breathe-Easy membrane (Diversified Biotech) and further incubated overnight at 25°C with 80–85% humidity.
Stained worms were imaged with an ImageXpress Microscope (Molecular Devices, Sunnyvale, CA, USA), capturing both bright field and TRITC (535 nm excitation, 610 nm emission) fluorescent images. Collected images were analyzed with CellProfiler analysis software [12] from the Broad Institute. The software facilitated the interrogation of the stained and unstained worms to determine the ratio Sytox stained worm area observed in fluorescent imaging compared with the worm area captured in the bright field image. The Z score for each of the wells was calculated using the formula Z = (x-μ)/σ where x is the raw score (generated in CellProfiler), μ is the mean of the population and σ is the standard deviation of the population.
Disc-clearing assay
The disc diffusion test was performed on yeast extract, peptone, dextrose (YPD) for fungal cultures. The assay was repeated three times. Discs were soaked in 10 μl of either DMSO or 10 mg/ml compound stock solution and air dried. Three hundred microliters of an overnight culture of fungi were spread on plates. After completely drying the agar plate in a laminar flow hood, DMSO or compound impregnated discs were overlaid on the plate and incubated at 35°C for 18 h. Antimicrobial susceptibility was determined by presence of a zone of inhibition.
Minimal inhibitory concentrations
Compounds (10 mg/ml stock solution in DMSO) were tested by broth microdilution, in triplicate, in 96-well plates. To test the bacterial MICs for compounds, the total volume in each well was 100 μl of test compounds and bacterial cells in MH broth. Two-fold serial dilutions were carried out to get compounds in the concentration range 0.0625–64 µg/ml. The bacterial concentration was adjusted to an initial OD600 of 0.03. After incubation at 35°C for 18 h, the absorbance was measured at 595 nm in accordance with CLSI document M07-A8 [13].
To interrogate the compound MIC against fungal cultures, colonies of C. neoformans (strain KN99α) and C. albicans (strain CAN14) were inoculated in 5 ml of YPD media overnight at 30°C. The cells were harvested by centrifugation at 4000 RPM for 5 min and washed with phosphate buffered saline (PBS); the cell pellets were resuspended in RPMI 1640 medium. The cell count was calculated with a hemocytometer and adjusted to be between 5.0 × 104 to 2.5 × 106 cells/ml. Compounds were prepared and added to RPMI 1640 medium at 2X the final concentration. MIC was determined in 96-well plates, working volume per well was 100 μl (50 µl compound + 50 µl cells). For C. albicans, OD595 was read after incubating the plate at 35°C for 24 h. For KN99α, OD595 was read after incubating the plate at 35°C for 70 h in accordance with CLSI document M27-A2 [14].
Static versus cidal assessment
The minimum bactericidal concentration was determined as follows. In total, 10 μl of bacterial culture from each microwell of the MIC assay was plated on MH agar and incubated at 35°C overnight. The lowest compound concentration at which there was no bacterial colony growth was considered the minimum bactericidal concentration.
The minimum fungicidal concentration was determined as follows. In total, 10 μl of yeast culture from each microwell of the MIC assay was plated on YPD agar and incubated at 35°C overnight. The lowest compound concentration at which there was no fungal colony growth was considered the minimum fungicidal concentration [15].
Checkerboard assays
The antimicrobial activity of a combination of auranofin with other agents was determined through a checkerboard assay. Briefly, the compounds whose combinations were being tested were arrayed in serial concentrations, vertically for one compound and horizontally for the other compound in the same 96-well microplate. The rest of the procedure involving addition of bacteria and measurement of growth was carried out as described in the previous section for measurement of MIC.
The fractional inhibitory concentration (FIC) index for two compounds, A and B, is defined by the following equation: FIC = ([A/MICA] + [B/MICB]) [16]. MICA and MICB are MICs of compound A or B, respectively. A is the lowest concentration of compound A in combination with compound B that inhibits bacterial growth and B is the lowest concentration of compound B that inhibits bacteria growth. An FIC <0.5 indicates synergism between the compounds being tested, greater than 2 suggest antagonism.
Results
Compound identification
We identified the antimicrobial activity of auranofin against S. aureus using a whole-animal infection model. During this screen, a collection of 640 FDA-approved compounds (provided by Harvard Medical School Institute of Chemistry and Cell Biology Screening Facility) was screened to identify compounds that improve the survival of the nematode C. elegans infected by S. aureus, normally responsible for a lethal infection [8,17].
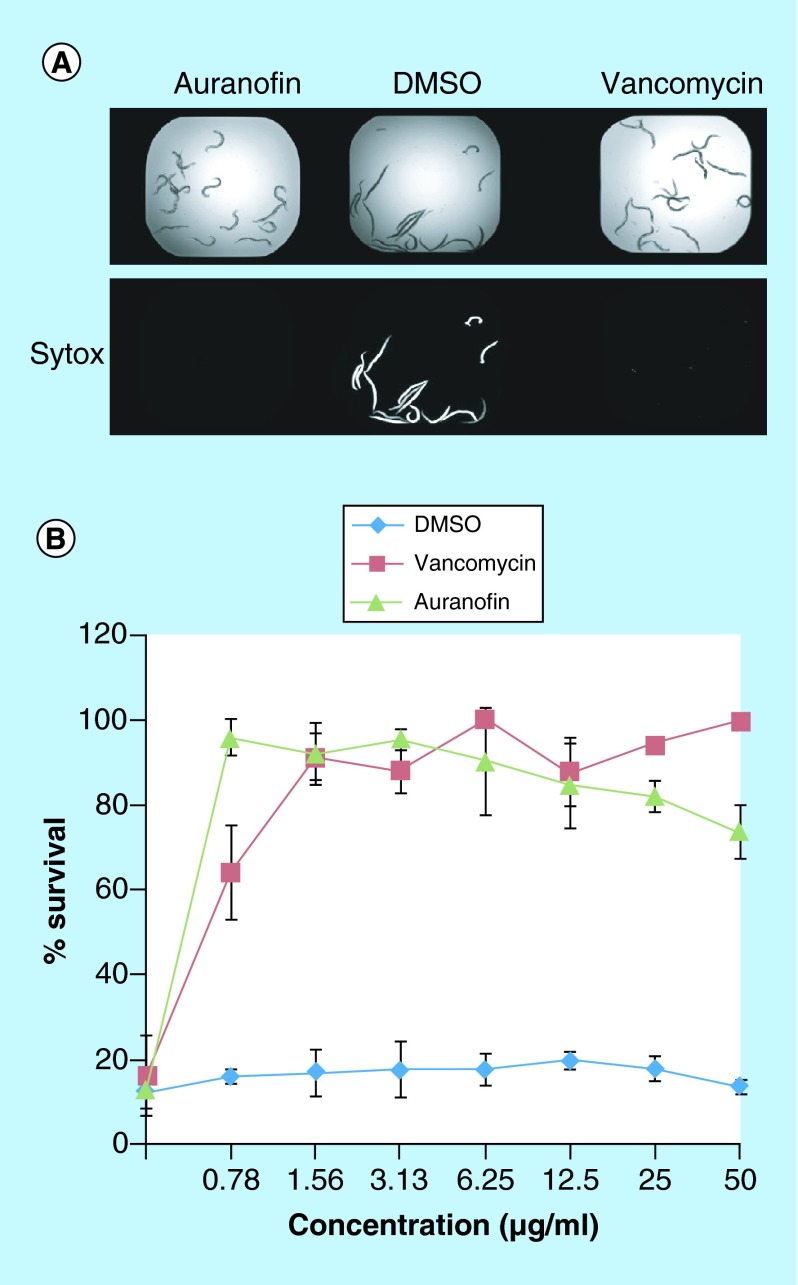
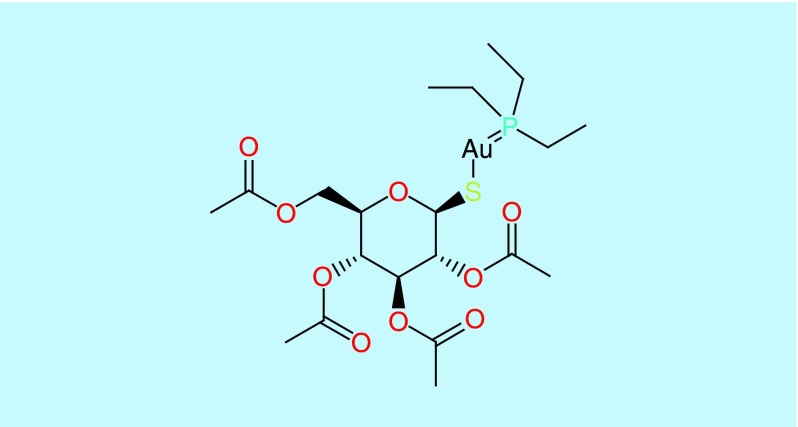
After a 5-day incubation period, worms were stained with Sytox orange, a nucleic acid stain. The detection of the fluorescent worms indicated cell damage and was a marker for dead worms and was compared with treatment with vancomycin (10 μg/ml) as a control [8,18,19] (Figure 2A). Auranofin, 2,3,4,6-tetra-o-acetyl-1-thio-D-glucanpyranosato-S-(triethyl-phosphine), a monomeric gold(I) species where the triethylphosphine group stabilizes the gold thiol complex [20] (Figure 3), was identified with a Z score of 18.06 and 13.06 in two replicates, a Z score greater than 3 was considered a hit within our screen. The lack of staining and prolonged survival of the worms (Figure 2B) suggests that auranofin could inhibit the S. aureus infection in vivo either by means of inhibiting the bacteria or promoting immune responses in the host.
Figure 2. . Worms survival was improved through exposure to auranofin.
(A) Brightfield and fluorescent images of worms infected with Staphylococcus aureus and incubated with auranofin, dimethyl sulfoxide or vancomycin. Both auranofin and vancomycin show a lack of staining with Sytox indicating worm survival. (B) When a series of drug concentrations were interrogated for worm survival, it was found that Caenorhabditis elegans survived after S. aureus infection at even very low concentrations of auranofin, comparable to vancomycin.
DMSO: Dimethyl sulfoxide.
Figure 3. . Chemical structure of auranofin.
A range of drug concentrations were evaluated to monitor the effects on prolonging the survival of S. aureus infected C. elegans. Both vancomycin and auranofin improved the survival of infected worms (p < 0.01), auranofin reaching 95.9% nematode survival at 0.78 μg/ml. A 95% nematode survival was achieved for vancomycin at 1.56 μg/ml.
Antibacterial activity of auranofin
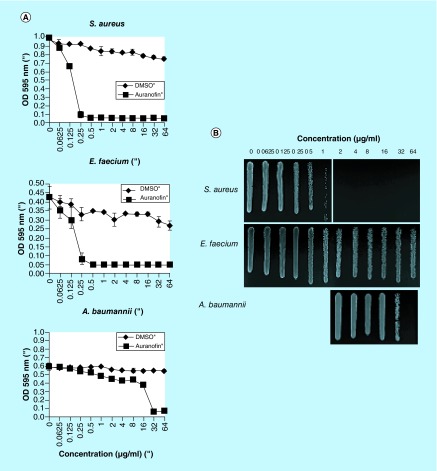
Since the antistaphylococcal activity of auranofin has been described [21], we investigated some of the most medically relevant pathogens, testing the ESKAPE bacterial pathogens (E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and Enterobacter) for susceptibility to auranofin. We found that auranofin was able to inhibit S. aureus and E. faecium and A. baumannii, affecting both Gram-positive and a Gram-negative bacteria. The MICs against these pathogens were 0.25 μg/ml, 0.5 μg/ml and 32 μg/ml, respectively, (Figure 4A) and the inhibition of the bacterial strains was bacteriostatic at the MIC concentration but bactericidal at higher concentrations (Figure 4B). These findings indicate that auranofin is effective against other medically important bacteria, in addition to S. aureus, and the MICs are lower for Gram-positive pathogens.
Figure 4. . Auranofin inhibits additional bacterial pathogens.
(A) MICs were determined for the bacteria susceptible to auranofin: Staphylococcus aureus, Enterococcus faecium and Acinetobacter baumannii. (B) The static versus cidal nature of the inhibition was tested by plating out cells that were exposed to the various concentrations of auranofin. Growth at the MIC or higher indicated that auranofin was bacteriostatic. Lack of growth indicated that auranofin was bactericidal.
We investigated further the effects of auranofin against Gram-positive bacteria. First, we investigated if auranofin inhibits an array of clinical isolates, particularly MRSA strains, or whether the inhibition we observed was restricted to the reference strain MW2. We found that the MIC for auranofin for all 11 isolates was less than or equal to 0.5 μg/ml (Table 2).
Table 2. . Drug MIC for Staphylococcus aureus clinical isolates.
| Isolate | Vancomycin | Oxacillin | Auranofin |
|---|---|---|---|
| BF1 |
2 |
>64 |
0.25 |
| BF2 |
2 |
>64 |
0.25 |
| BF3 |
4 |
32 |
0.25 |
| BF4 |
2 |
16 |
0.25 |
| BF5 |
2 |
>64 |
0.5 |
| BF6 |
2 |
1 |
0.25 |
| BF7 |
2 |
>64 |
0.25 |
| BF8 |
2 |
>64 |
0.5 |
| BF9 |
2 |
0.25 |
0.25 |
| BF10 |
2 |
>64 |
0.5 |
| BF11 | 2 | >64 | 0.25 |
Concentrations are in μg/ml.
Since the MIC against E. faecium was also low, 0.5 μg/ml, we examined if auranofin could successfully inhibit clinical isolates of this pathogen as well [22]. Indeed, we found that auranofin had a low MIC of 1 μg/ml against E. faecium clinical isolates, including strains that exhibited resistance to chloramphenicol and vancomycin (Table 3). These findings suggest that auranofin is able to inhibit multiple bacterial pathogens and the activity is not restricted to laboratory reference strains.
Table 3. . Drug MIC for Enterococcus faecium clinical isolates.
| Isolate | Chloramphenicol | Vancomycin | Auranofin |
|---|---|---|---|
| C68 |
16 |
>64 |
1 |
| D14 |
8 |
2 |
1 |
| D24 |
4 |
1 |
1 |
| D25 |
8 |
1 |
1 |
| D29 |
64 |
2 |
1 |
| W312 |
8 |
>64 |
1 |
| WC176 | 16 | >64 | 1 |
Concentrations are in μg/ml.
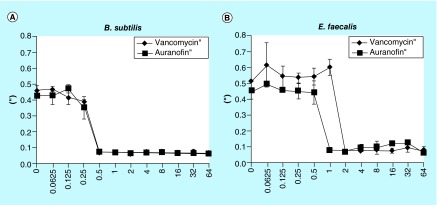
With such low MICs against two Gram-positive bacteria within the ESKAPE collection, we expanded our interrogation of bacterial strains to E. faecalis (and included Bacillus subtilis as a control). Both B. subtilis and E. faecalis indicated clearing around the auranofin disc (Figure 5). The MIC of auranofin again B. subtilis is 0.5 μg/ml (Figure 5A), and 1 μg/ml against E. faecalis (Figure 5B). Both bacteria are highly susceptible to auranofin.
Figure 5. . Gram-positive bacteria inhibited by auranofin.
(A) The MIC was determined to be 0.5 μg/ml against Bacillus subtilis and (B) 1 μg/ml against Enterococcus faecalis.
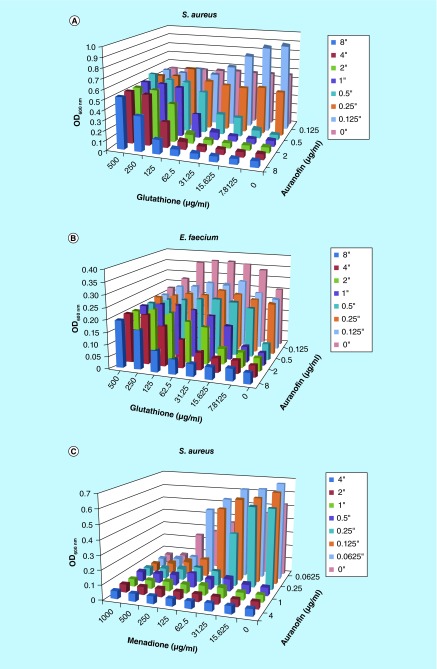
The target of auranofin was investigated based on known molecular interactions between auranofin and thioredoxin reductase where it functions as a thioredoxin system inhibitor, binding directly to thrioredoxin-glutathione reductase of Schistosoma mansoni, Giardia lamblia and Echinococcus granulosus [23–25]. Interestingly, we found that auranofin is more active against Gram-positive bacteria that have a thioredoxin system, but are glutathione deficient [26]. Thus we introduced glutathione to see if it would antagonize the inhibition of auranofin. In both S. aureus and E. faecium, glutathione reduced the inhibitory effect of auranofin and resulted in higher MICs (Figure 6A). More specifically, in this series of experiments, the MIC of auranofin against S. aureus was 0.5 μg/ml but was increased to 1 μg/ml in the presence of 31.25 μg/ml of glutathione and even 8 μg/ml in the presence of 125 μg/ml glutathione. E. faecium exhibited similar findings, exhibiting an MIC of 0.5 μg/ml is this assay in the absence of glutathione. However, the MIC increased to 8 μg/ml with the antagonistic effect of 125 μg/ml glutathione (Figure 6B). Glutathione alone did not inhibit either of the bacteria strains, thus suggesting antogonsitic activity specific to auranofin. Therefore, suggesting that auranofin is also targeting the thioredoxin system of S. aureus and E. faecium.
Figure 6. . Glutathione antagonizes auranofin inhibition and oxide stress is synergistic.
(A) Staphylococcus aureus is inhibited by auranofin. However, the MIC is increased in the presence of glutathione. (B) The same antagonism is found against auranofin that was found to inhibit Enterococcus faecium. The color chart provided indicates that concentration of auranofin that was interrogated. (C) In the presence of menadione, Staphylococcus aureus exhibited increased susceptibility to auranofin, lowering the MIC to 0.125 μg/ml.
Since the thioredoxin system plays a role in protecting cells from oxide stresses experienced in the host environment [26], we exposed S. aureus to various oxide stresses in combination with auranofin. We found that hydrogen peroxide did not provide any synergistic activity to auranofin mediated inhibition of S. aureus. The addition of diamide marginally reduced the MIC of auranofin from 0.5 μg/ml to 0.25 μg/ml in the presence of 1000 μg/ml diamide, a thiol oxidizing agent. However, menadione, which generates superoxides, demonstrated synergistic activity with auranofin, with an FIC of 0.5, reducing the auranofin MIC from 0.5 to 0.125 μg/ml (Figure 6C). Thus, suggesting that the thioredoxin system plays a role in defending against superoxide stressors. It also indicates that compounds that elicit increases in superoxides could function synergistically with auranofin.
Antifungal activity
As detailed in the previous section, our investigation of the molecular target of auranofin found that the thioredoxin system is the likely inhibited target of Gram-positive bacteria. The thrioredoxin system is conserved in both prokaryotic and eukaryotic organisms with developed differences. We investigated the effect of auranofin against fungi that rely on thioredoxin reductase for oxidative stress resistance. In C. neoformans, thioredoxin reductase (TRR1) is essential for viability [27,28]. Thus, there was the potential that fungi could be inhibited by auranofin if it did indeed target the thioredoxin system.
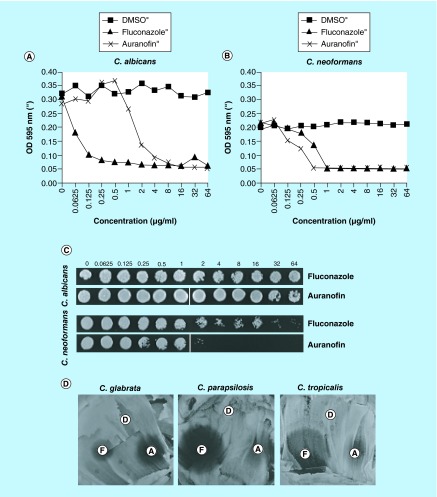
By testing the microdilutions of the compounds in a liquid assay, we determined that the MIC of auranofin was 8 μg/ml against C. albicans (Figure 7A) and 0.5 μg/ml, against C. neoformans (Figure 7B). The compounds exhibited fungistatic activity against C. albicans, however fungicidal activity was reached against C. neoformans at concentrations greater than 2 μg/ml (Figure 7C).
Figure 7. . Auranofin inhibits fungal pathogens.
(A) The MIC was determined for Candida albicans and (B) Cryptococcus neoformans fungi. (C) The static versus cidal nature of the inhibition was examined for the various auranofin concentrations. Growth at concentration equivalent or higher than the MIC indicated that aurnaofin was fungistatic and inhibition of growth indicated fungicidal activity. (D) Additional non-albicans Candida strains were tested for susceptibility to auranofin. A zone of inhibition was identified for each of the Candida strains (A: auranofin, D: DMSO, F: fluconazole).
DMSO: Dimethyl sulfoxide.
Although C. albicans was not as susceptible to auranofin as C. neoformans, we investigated if non-albicans Candida spp. were susceptible to the compound, testing C. glabrata, C. parapsilosis and C. tropicalis (Table 1), some of the most medically relevant fungal non-albicans Candida spp. Indeed, we were able to find zones of inhibition when tested against Candida glabrata. (Figure 7D). Further investigation of the reference strain and three C. tropicalis clinical isolates demonstrated MICs that ranged from 0.125 to 1 μg/ml, demonstrating the antifungal effect against this non-albicans strain (Table 4). Interrogation of C. glabrata clinical isolates found an inhibition at 0.25–32 μg/ml (Table 5). Out of the 15 clinical isolates examined, 13 exhibited sensitivity to auranofin, but 2 isolates were resistant. Thus our findings indicate that auranofin is effective at inhibiting fungal pathogens as well as bacterial pathogens, inhibiting both fungal clinical isolates and reference strains.
Table 4. . Drug MIC for Candida tropicalis isolates.
| Isolate | Isolate | Fluconazole | Amphotericin B | Auranofin |
|---|---|---|---|---|
| ATCC 13803 |
Reference |
8 |
1 |
1 |
| 11 |
Clinical |
0.5 |
0.5 |
0.125 |
| 85-S |
Clinical |
1 |
0.125 |
0.125 |
| 172-S | Clinical | 2 | 0.5 | 0.5 |
Concentrations are in μg/ml.
Table 5. . Drug MIC for Candida glabrata isolates.
| Isolate | Isolate | Fluconazole | Amphotericin B | Auranofin |
|---|---|---|---|---|
| ATCC 90030 |
Reference |
8 |
1 |
0.25 |
| 6891 |
Clinical |
2 |
0.5 |
0.5 |
| 6922 |
Clinical |
2 |
0.5 |
1 |
| 6927 |
Clinical |
2 |
1 |
0.5 |
| 6930 |
Clinical |
1 |
1 |
0.5 |
| 6931 |
Clinical |
0.5 |
0.5 |
0.5 |
| 6932 |
Clinical |
0.5 |
0.5 |
32 |
| 6943 |
Clinical |
8 |
1 |
1 |
| 7110 |
Clinical |
1 |
0.5 |
1 |
| 7117 |
Clinical |
2 |
0.5 |
0.5 |
| 7221 |
Clinical |
1 |
0.5 |
0.5 |
| 7255 |
Clinical |
2 |
1 |
0.5 |
| 7815 |
Clinical |
2 |
0.5 |
1 |
| 7869 |
Clinical |
4 |
1 |
16 |
| 7871 |
Clinical |
2 |
1 |
1 |
| 8066 | Clinical | 2 | 0.5 | 1 |
Concentrations are in μg/ml.
Since there was some inhibition found against C. albicans, we also interrogated the ability of auranofin to inhibit biofilm. We found that C. albicans biofilm formation was not significantly reduced by auranofin. There was only reduction achieved with the addition of 32 μg/ml (Supplementary Figure 1).
Among the fungal pathogens examined, more significant inhibition was found against the fungal pathogen C. neoformans. Interrogation of C. neoformans clinical isolates indicates that the inhibitory activity of auranofin was conserved among the 11 isolates tested, ranging from 2 to 8 μg/ml (Table 6).
Table 6. . Drug MIC for Cryptococcus neoformans clinical isolates.
| Isolate | Amphotericin B | Fluconazole | Auranofin |
|---|---|---|---|
| KN99α |
0.125 |
2 |
2 |
| BF113 |
0.125 |
16 |
8 |
| BF114 |
<0.0625 |
64 |
4 |
| 41291 |
0.125 |
8 |
4 |
| 41292 |
0.125 |
64 |
4 |
| 41294 |
0.125 |
8 |
2 |
| 41295 |
0.125 |
64 |
8 |
| 41296 |
0.125 |
>64 |
4 |
| 41297 |
0.125 |
8 |
2 |
| 41298 |
0.125 |
8 |
2 |
| 41299 |
0.125 |
4 |
4 |
| 41300 |
0.125 |
>64 |
4 |
| C31 |
0.125 |
4 |
1 |
| F10 |
0.25 |
4 |
4 |
| RN01 |
0.25 |
4 |
2 |
| WP |
0.5 |
8 |
2 |
| 27JF |
0.25 |
8 |
2 |
| 28JF |
0.25 |
8 |
2 |
| 5396 |
0.25 |
8 |
1 |
| 90896 |
0.25 |
32 |
1 |
| 93 |
0.25 |
4 |
1 |
| 94 |
0.125 |
8 |
2 |
| 646B |
0.125 |
1 |
2 |
| 975 |
0.125 |
2 |
1 |
| 9220 |
0.125 |
2 |
0.5 |
| 9272 |
0.125 |
8 |
2 |
| 9273 |
0.125 |
2 |
1 |
| 10131 |
0.125 |
8 |
1 |
| 10211 |
0.125 |
4 |
2 |
| 10264 |
0.125 |
4 |
2 |
| 10287 |
0.25 |
1 |
1 |
| 10335 |
0.125 |
8 |
1 |
| 10379 |
0.125 |
1 |
1 |
| 92868 | 0.25 | 8 | 1 |
Concentrations are in μg/ml.
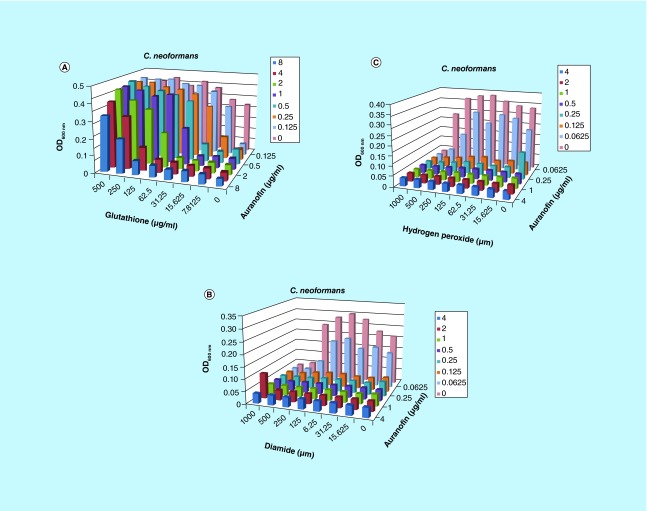
Much like the bacterial pathogens, we found that glutathione is able to antagonize the inhibition by auranofin. The MIC to auranofin was increased from 0.125 μg/ml in our assay to 4 μg/ml with 62.5 μg/ml glutathione (Figure 8). The inhibition concentration was increased to 16 μg/ml with auranofin in the presence of 250 μg/ml glutathione. Glutathione alone did not inhibit the fungal cells at concentrations ≤500 μg/ml. Again, the targeted mechanism of auranofin inhibition in fungal cells is suggested to be the thioredoxin system.
Figure 8. . Glutathione antagonizes auranofin inhibition of Cryptococcus neoformans.
(A) The auranofin-driven inhibition of Cryptococcus neoformans is reduced by the addition of glutathione. The MIC increases in a dose-dependent manner with increasing concentrations of the compound. The color chart provided indicates that concentration of auranofin that was interrogated. (B) The MIC was reduced in combination with diamide and (C) menadion.
The sensitivity of C. neoformans to oxidizing agents was tested in combination with auranofin to evaluate if thioredoxin can inhibit oxide stress. We found that the MIC of auranofin was reduced in the presence of diamide to 0.0625 μg/ml, and in combination with hydrogen peroxide to 0.0325 μg/ml, but it did not constitute the level of synergism, only an additive effect. There was no change in the presence of menadione.
The most common antifungal compounds utilized against fungal infections are amphotericin B and fluconazole. Therefore, we investigated whether any synergistic activity could be found with auranofin. The ∑FICs for amphotericin and fluconazole in combination with auranofin were 1 and 2, respectively (Supplementary Figure 2). Thus, there were additive effects but not synergism.
Discussion
Auranofin has been shown to inhibit S. aureus [21,29], Mycobatcerium tuberculosis [29] and the parasites Schistosoma mansoni, Entamaeba histolytica and Giardia lamblia have also demonstrated susceptibility to auranofin, both in vitro and in vivo [23,24,30]. The platyhelminth Echinococcus granulosus are susceptible to auranofin, inhibiting larval worms at 2.5 μM in vitro. In the case of all of the parasitic inhibition, auranofin was found to disrupt the thioredoxin–glutathione system where thioredoxin glutathione reductase acts as a key enzyme [24,25,30]. In this report, we demonstrate the novel antifungal activity that is associated with targeting the thioredoxin system. Also, we expand what is known about the antibacterial activity of auranofin. We report that auranofin is active against S. aureus in the in vivo C. elegans model and that this compound is active against other Gram-positive bacteria, such as E. faecium and E. faecalis, that are not inhibited by the full array of available antimicrobial agents, and B. subtilis. Also, we extend these findings by demonstrating that it inhibits not only laboratory reference strains, but also clinical isolates of bacteria and fungi.
Jackson-Rosario et al. demonstrated that auranofin binds selenium, a catalyst in energy metabolism that affects redox balance [9,11]. Investigation of microbial inhibition by auranofin has provided evidence that the drug binds to hydrogen selenide, blocking selenium utilization by bacteria, preventing selenoprotein synthesis [9]. The function of blocking selenoproteins appears to also be conserved in parasitic worms as well, x-ray crystallography demonstrates the binding of thioredoxin–glutathione reductase with auranofin. The gold from the compound appears to be the inhibiting portion of the molecule [23].
Our investigation into the bacteria target was prompted by the previous findings that it inhibits the thioredoxin system. The thioredoxin system is comprised of NADPH, thioredoxin reductase (TrxR) and thioredoxin (Trx). In some systems the thioredoxin system is backed up by the glutathione–glutaredoxin (GSH) system. However, for many Gram-positive bacteria and some Gram-negative bacteria, the systems are not redundant, leaving the thioredoxin system to serve the essential task of defending against oxidative stresses through disulfide reductase activity. Indeed, we found that for both S. aureus and E. faecium, auranofin inhibitory activity was diminished with increasing concentrations of glutathione. GSH is absent in S. aureus and produced at very low levels for E. faecium [31,32]. The thioredoxin system protects the cell against oxygen species and maintains the intracellular thiol-disulfide balance. Recent work by Harbut et al. demonstrate that auranofin appears to target the thioredoxin system of bacteria [29].
Oxide stressors threaten the bacteria, generated as part of normal metabolic and physiological conditions. Uziel et al. found that trxA and trxB both experienced increased transcription in the presence of menadione, reduced by thioredoxin in the presence of NADPH [33], or diamide but were not responsive to hydrogen peroxide [34], mirroring the indications we found in our checkerboard assay that indicate auranofin acts synergistically with menadione and showed some additive effect with diamide when tested with S. aureus.
As noted above, we present a finding that auranofin inhibits C. albicans, C. tropicalis, C. glabrata and C. neoformans. A previous report has suggested that C. albicans and C. glabrata have susceptibility to auranofin [35]. Notably, C. neoformans was inhibited at a low MIC that was reduced even further by oxide stressors. C. neoformans is a significant fungal pathogen to susceptible patients, mostly immunocompromised individuals. The finding that this particular fungal pathogen is inhibited by auranofin is novel. Although there are drugs to treat C. neoformans, they can require prolonged use and can lead to toxic effects [36,37]. Importantly, auranofin was effective at inhibiting not only laboratory reference strains but also clinical isolates for the fungi to which it exhibited the lowest MICs, C. tropicalis, C. glabrata and C. neoformans, extending the breadth of pathogens to which it exhibits inhibition. Further, our findings that glutathione antagonizes auranofin suggests the thioredoxin system is the likely target.
The thioredoxin reductase, TRR1, in C. neoformans is essential for viability [27,28]. Missall and Lodge demonstrated that TRR1 is induced during oxidative stress by hydrogen peroxide [27], the oxidizer that elicited the greater additive effect when tested in combination with auranofin. TRR1 is also found in the C. albicans genome and responds to oxidative stresses [38]. The difference in sensitivity between C. albicans and C. neoformans could be associated with the diploid structure of the C. albicans genome versus the haploid of C. neoformans, thus requiring more drug to inhibit thioredoxin reductase. C. glabrata is also a haploid and expressed sensitivity to auranofin with a lower MIC than that of C. albicans. However, C. tropicalis is diploid but is susceptible to auranofin. In additional to ploidy, the availability of glutathione reductase may also contribute to fungal cell auranofin sensitivity.
The less sensitive nature of C. albicans to auranofin could also be associated with the reactive oxygen species (ROS) detoxification contribution from the four encoded glutathione genes (GRX1, GRX2, GRX3 and GRX5). Further, C. glabrata is also known to have glutathione but with only three copies encoded in the genome [39]. The status of glutathione availability is not known for C. tropicalis or C. parapsilosis, however, our investigations suggest that glutathione is potentially encoded by these genomes.
Although we did not see significant reduction in Candida albicans biofilm with application of auranofin based on our test conditions, Siles et al. found inhibition to effective at 5 μM [40]. Our noncongruent findings may be associated with the fact that we used different assays comparing biofilm mass versus cell viability (XTT) staining and differing medias.
With the continual emergence of drug-resistant strains of bacteria (particularly S. aureus) and fungi, additional therapy options are needed. Historically, gold and silver have been known for antimicrobial activities. Other gold complex compounds, such as gold(I) N-heterocyclic carbene complexes, have demonstrated broad spectrum antimicrobial activity [41]. This report suggests that auranofin could be repositioned as a new drug option for microbial infections, particularly, S. aureus, E. faecium and C. neoformans. Auranofin has the benefit of being FDA approved (since 1985) to make clinical evaluation for the purpose of reducing bacterial and fungal infections more feasible. Further, this drug can be taken orally, a contrast to many of the current therapeutic options that require intravenous delivery and thus could be ideal for uncomplicated cases.
Conclusion
In summation, the thioredoxin reductase targeting compound auranofin not only provides an exciting compound for microbial inhibition, it also presents a new anti-microbial target. Auranofin inhibits Gram-positive bacteria with low MICs and even has the promiscuity of inhibiting fungi. Within our group of prohibited pathogens, we find that reference and clinical strains are susceptible. Further, we find that even drug resistance strains can be inhibited by auranofin.
Future perspective
HTS with an amenable in vivo model has provided a rapid, facile means of identifying new antimicrobial compounds. It is exciting that we now find the compounds being identified are not restricted to inhibiting the single pathogen to which they were discovered. Further, the mining of FDA-approved compounds provides a source of new drugs that can be repositioned as therapeutics with shortened routes to reach patients. Although our screen was directed to identify a compound that inhibits S. aureus we were excited to find a compound with activity against fungal pathogens. Importantly, the identification of this compound directs our attention at developing drugs that can inhibit the thioredoxin system as means to control microbial infections, highlighting a potential new class of antibiotics and antifungals.
Executive summary.
Auranofin has antimicrobial properties inhibiting bacterial and fungal pathogens.
This is the first report demonstrating the antifungal properties of auranofin against C. neoformans.
Auranofin appears to target the thioredoxin system of C. neoformans, essential to viability.
Auranofin appears to target the thioredoxin system of Gram-positive bacteria.
The inhibitory activity of auranofin against C. neoformans is enhanced in the presence of oxide stressors in the form of hydrogen peroxide or a thiol-oxidizing agent.
Auraonfin appears to inhibit a range of microbial pathogens from Gram-positive bacteria to fungi (inclusive of: B. subtilis, E. faecalis, E. faecium, S. aureus, C. albicans, C. tropicalis, C. glabrata and C. neoformans).
The antimicrobial activity of auranofin is not restricted to laboratory reference strains but is retained against clinical isolates.
Supplementary Material
Acknowledgements
Our appreciation is extended to Dr A Lopes Colombo who provided clinical strains. We thank N Kirienko for providing the R Script used to analyze data from the compound screen. Our gratitude is also extended to the Harvard Medical School Institute of Chemistry and Cell Biology for providing small molecule compound libraries.
Footnotes
Financial & competing interests disclosure
This research was supported by a COBRE-Immune-based intervention against infectious diseases pilot grant and a Brown University Dean's Emerging Areas of New Science award to B Burgwyn Fuchs, and a NIH P01 grant to E Mylonakis. Our work was also supported with funding provided by the Brown-Brazil Initiative awarded to B Burgwyn Fuchs and E Mylonakis. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Cuddy SM. Methicillin-resistant Staphylococcus aureus: a new pandemic? Plast. Surg. Nurs. 2008;28(4):168–169. doi: 10.1097/PSN.0b013e31818ea7ca. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller M, Messer S, Jones RN, Castanheira M. Antifungal susceptibilities of Candida Cryptococcus neoformans and Aspergillus fumigatus from the Asia and Western Pacific region: data from the SENTRY antifungal surveillance program (2010–2012) J. Antibiot. (Tokyo) 2015;68(9):556–561. doi: 10.1038/ja.2015.29. [DOI] [PubMed] [Google Scholar]
- 3.Cleveland AA, Harrison LH, Farley MM, et al. Declining Incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008–2013: results from population-based surveillance. PLoS ONE. 2015;10:e0120452. doi: 10.1371/journal.pone.0120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan W, Khayhan K, Hagen F, et al. Resistance of Asian Cryptococcus neoformans serotype a is confined to few microsatellite genotypes. PLoS ONE. 2012;7(3):e32868. doi: 10.1371/journal.pone.0032868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mdodo R, Moser SA, Jaoko W, et al. Antifungal susceptibilities of Cryptococcus neoformans cerebrospinal fluid isolates from AIDS patients in Kenya. 2012;54(5):1–7. doi: 10.1111/j.1439-0507.2010.01946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bii CC, Makimura K, Abe S, et al. Antifungal drug susceptibility of Cryptococcus neoformans from clinical sources in Nairobi, Kenya. Mycoses. 2007;50:25–30. doi: 10.1111/j.1439-0507.2006.01293.x. [DOI] [PubMed] [Google Scholar]
- 7.Bicanic T, Harrison T, Niepieklo A, Dyakopu N, Meintjes G. Symptomatic relapse of HIV-associated cryptococcal meningitis after initial fluconazole monotherapy: the role of fluconazole resistance and immune reconstitution. Clin. Infect. Dis. 2006;43:1069–1073. doi: 10.1086/507895. [DOI] [PubMed] [Google Scholar]
- 8.Rajamuthiah R, Fuchs BB, Jayamani E, et al. Whole animal automated platform for drug discovery against multi-drug resistant Staphylococcus aureus . PLoS ONE. 2014;9(2):e89189. doi: 10.1371/journal.pone.0089189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson-Rosario S, Cowart D, Myers A, et al. Auranofin disrupts selenium metabolism in Clostridium difficile by forming a stable Au-Se adduct. J. Biol. Inorg. Chem. 2009;14(4):507–519. doi: 10.1007/s00775-009-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhodes MD, Sadler PJ, Scawen MD SS. Effects of Gold(I) antiarthritic drugs and related compounds on Pseudomonas putida . J. Inorg. Chem. 1992;46:129–142. doi: 10.1016/0162-0134(92)80016-o. [DOI] [PubMed] [Google Scholar]
- 11.Jackson-Rosario S, Self WT. Inhibition of selenium metabolism in the oral pathogen Treponema denticola . J. Bacteriol. 2009;191(12):4035–4040. doi: 10.1128/JB.00164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cell Profiler. www.cellprofiler.org/
- 13.Clinical and Laboratory Standards Institute. Methods For Dilution Antimicrobial Susceptibility Tests For Bacteria That Grow Aerobically; Approved Standard (8th Edition) CLSI; PA, USA: 2009. M07-A8. [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Reference Methods For Broth Dilution Antifungal Susceptibility Testing Of Yeast. CLSI; PA, USA: 2012. [Google Scholar]
- 15.Rajput SB, Karuppayil SM. Small molecules inhibit growth, viability and ergosterol biosynthesis in Candida albicans . SpringerPlus. 2013;2(1):26. doi: 10.1186/2193-1801-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orhan G, Bayram A, Zer Y, Balci I. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis . J. Clin. Microbiol. 2005;43(1):140–143. doi: 10.1128/JCM.43.1.140-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garsin DA, Sifri CD, Mylonakis E, et al. A simple model host for identifying Gram-positive virulence factors. Proc. Natl Acad. Sci. USA. 2001;98(19):10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conery AL, Larkins-Ford J, Ausubel FM, Kirienko NV. High-throughput screening for novel anti-infectives using a C. elegans pathogenesis model. Curr. Protoc. Chem. Biol. 2014;6(1):25–37. doi: 10.1002/9780470559277.ch130160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland TL, Arnold C, Fowler VG. Clinical management of Staphylococcus aureus bacteremia. JAMA. 2014;312(13):1330–1341. doi: 10.1001/jama.2014.9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kean WF, Hart L, Buchanan WW. Auranofin. Br. J. Rheumatol. 1997;36(5):560–572. doi: 10.1093/rheumatology/36.5.560. [DOI] [PubMed] [Google Scholar]
- 21.Cassetta MI, Marzo T, Fallani S, Novelli A, Messori L. Drug repositioning: auranofin as a prospective antimicrobial agent for the treatment of severe staphylococcal infections. Biometals. 2014;27(4):787–791. doi: 10.1007/s10534-014-9743-6. [DOI] [PubMed] [Google Scholar]
- 22.Rice LB, Carias LL, Rudin S, Wood A, Hutton-Thomas R, Laktic V. Enterococcus faecium low-affinity pbp5 is a transferable determinant. Antimicrob. Agents Chemother. 2005;49(12):5007–5012. doi: 10.1128/AAC.49.12.5007-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelucci F, Sayed AA, Williams DL, et al. Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: structural and kinetic aspects. J. Biol. Chem. 2009;284(42):28977–28985. doi: 10.1074/jbc.M109.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tejman-Yarden N, Miyamoto Y, Leitsch D, et al. A reprofiled drug, auranofin, is effective against metronidazole-resistant Giardia lamblia . Antimicrob. Agents Chemother. 2013;57(5):2029–2035. doi: 10.1128/AAC.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonilla M, Denicola A, Novoselov SV, et al. Platyhelminth mitochondrial and cytosolic redox homeostasis is controlled by a single thioredoxin glutathione reductase and dependent on selenium and glutathione. J. Biol. Chem. 2008;283(26):17898–17907. doi: 10.1074/jbc.M710609200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 27.Missall TA, Lodge JK. Thioredoxin reductase is essential for viability in the fungal pathogen Cryptococcus neoformans . Eukaryot. Cell. 2005;4(2):487–489. doi: 10.1128/EC.4.2.487-489.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ianiri G, Indrum AL. Essential gene discovery in the Basidiomycete Cryptococcus . MBio. 2015;6(12):e02334–e02314. doi: 10.1128/mBio.02334-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbut MB, Vilchèze C, Luo X, et al. Auranofin exerts broad-spectrum bactericidal activities by targeting thiol-redox homeostasis. Proc. Natl Acad. Sci. USA. 2015;112(14):4453–4458. doi: 10.1073/pnas.1504022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debnath A, Parsonage D, Andrade RM, et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nature. 2012;18(6):956–960. doi: 10.1038/nm.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Vlamis-Gardikas A, Kandasamy K, et al. Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. FASEB J. 2013;27:1394–1403. doi: 10.1096/fj.12-223305. [DOI] [PubMed] [Google Scholar]
- 32.Newton GL, Arnold K, Price MS, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 1996;178(7):1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmgren A. Reduction of risulfides by thioredoxin. J. Biol. Chem. 1979;254(18):9113–9119. [PubMed] [Google Scholar]
- 34.Uziel O, Borovok I, Schreiber R, Cohen G, Aharonowitz Y. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 2004;186(2):326–334. doi: 10.1128/JB.186.2.326-334.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stylianou M, Kulesskiy E, Lopes JP, Granlund M, Wennerberg K, Urban CF. Antifungal application of nonantifungal drugs. Antimicrob. Agents Chemother. 2014;58(2):1055–1062. doi: 10.1128/AAC.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denning DW. Echinocandin antifungal drugs. Lancet. 2003;362(9390):1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- 37.Dismukes WE. Management of cryptococcosis. Clin. Infect. Dis. 1993;17(Suppl. 2):S507–S512. doi: 10.1093/clinids/17.supplement_2.s507. [DOI] [PubMed] [Google Scholar]
- 38.Enjalbert B. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell. 2002;14(4):1460–1467. doi: 10.1091/mbc.E02-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuellar-Cruz M, Lopez-Romero E, Ruiz-Baca E, Zazueta-Sandoval R. Differential response of Candida albicans and Candida glabrata to oxidative and nitrosative stresses. Curr. Microbiol. 2014;69(5):733–739. doi: 10.1007/s00284-014-0651-3. [DOI] [PubMed] [Google Scholar]
- 40.Siles SA, Srinivasan A, Pierce CG, Lopez-Ribot JL, Ramasubramanian AK. High-throughput screening of a collection of known pharmacologically active small compounds for identification of Candida albicans biofilm inhibitors. Antimicrob. Agents Chemother. 2013;57(8):3681–3687. doi: 10.1128/AAC.00680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozdemir I, Temelli N, Günal S, Demir S. Gold(I) complexes of N-heterocyclic carbene ligands containing benzimidazole: synthesis and antimicrobial activity. Molecules. 2010;15(4):2203–2210. doi: 10.3390/molecules15042203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.