Abstract
A complete understanding of the fundamental mechanisms of learning and memory continues to elude neuroscientists. Although many important discoveries have been made, the question of how memories are encoded and maintained at the molecular level remains. So far, this issue has been framed within the context of one of the most dominant concepts in molecular biology, the central dogma, and the result has been a protein-centric view of memory. Here, we discuss the evidence supporting a role for neuroepigenetic mechanisms, which constitute dynamic and reversible, state-dependent modifications at all levels of control over cellular function, and their role in learning and memory. This neuroepigenetic view suggests that DNA, RNA and protein each influence one another to produce a holistic cellular state that contributes to the formation and maintenance of memory, and predicts a parallel and distributed system for the consolidation, storage and retrieval of the engram.
Introduction
Learning is described as a persistent, experience-dependent change in behaviour and memory as the internal representation of this experience, which has traditionally been defined as the engram.1–3 How organisms learn has been a question of interest since before the days of Darwin, who proposed that organisms gain innate adaptation through evolution, or Lamarck, who argued that this must occur in response to current environmental demand and is therefore acquired in a lifetime.4,5 The importance of this question is further echoed by those who have addressed it across a variety of levels of analysis, including animal behavior,6 cognition,7,8 development9 and the physiology underlying synaptic transmission, each domain providing its own important contributions, as well as caveats.10
Most empirical evidence suggests that memory formation has two primary components, one that is protein-synthesis independent, and a second time-dependent phase that relies on activity-induced gene transcription and protein synthesis, which lead to enhanced synaptic efficacy.10,11 This is based on the observation that protein synthesis occurs in a predictable time frame following a behavioural experience through a process known as memory consolidation, and that protein-synthesis inhibitors, when administered within this period, block the formation of memory.2,3 However, several reports call into question the strength of this perspective, including (1) the non-specific effects of protein-synthesis inhibitors (e.g., anisomycin can also influence neurotransmitter release, and its effects can be rescued without affecting protein synthesis per se).12–14 Protein-synthesis inhibitors can also induce phosphorylation of CREB15 and apoptosis;16 (2) the demonstration of memory formation in a variety of tasks even during more than 90% reduction of protein;17–20 (3) the existence of simple and selective forms of cellular memory that are protein-independent, such as protection from viral integration in plants,21 and (4) the fact that it is implicit in many empirical descriptions of this phenomenon that protein synthesis is sufficient for long-term memory.
It is evident that we are still a long way from fully understanding the intimate relationship between protein synthesis and memory, and at the very least questioning of the sufficiency, but not necessity of protein synthesis may be beneficial. In fact, a re-evaluation of the protein-synthesis hypothesis of memory may open up new opportunities for understanding, such as determining whether memory is established by serial or parallel processes, as well as questions about the location of the engram.22
The general way we think about memory derives from William James’ distinction between sensory processing (primary memory), and a more permanent trace of this processing (memory proper), that is intuitively linear if we think about storage.23 Unfortunately, this linear trajectory does not follow for less well-known memory functions, such as re-updating and retrieval.24,25 In addition, we know from the seminal work of Brenda Milner with patient H.M. who suffered severe memory impairment following a bilateral temporal lobectomy, that sensory processing and storage of experience can be dissociated for particular types of memories, presumably by disrupting this linear sequence. Much like early observations on the use of protein-synthesis inhibitors in memory, this work demonstrated that the hippocampus is necessary, but not sufficient, for all memory.26 These findings elicited further work, which suggested that particular types of memories are dependent on certain regions of the brain27–29 but also rely on parallel communication with other regions, in some cases competitively,30,31 leading to the conclusion that no one region is sufficient to consolidate, store and retrieve all memory. In comparison, if one assumes that protein synthesis is sufficient for memory, it naturally follows that memory must follow a serial path that terminates in the production of protein. This view derives from the central dogma, which dominated molecular biology for many years and proposed a linear trajectory from DNA to RNA to protein. This view is so entrenched in molecular biology that Crick himself put out an explanatory paper describing how his work had often been misinterpreted as an oversimplification of multiple possibilities. However, despite this, it has remained as an implicit bias in molecular neuroscience.32 Thus given the questioning of a serial view of memory at the molecular and cognitive levels, an alternative and parallel view warrants exploration.
By the same logic, protein sufficiency and its consequences for the location of information storage can be questioned. Following a protein-centric view, the end goal of all molecular changes is primarily a change in protein level, and the structural and change in synaptic efficacy that follows must serve a general storage function. This is akin to Lashley’s equipotentiality argument, which stated that one brain region could serve the same general function as any other for memory.22 However, this idea of systems equipotentiality was challenged long ago by the discovery of discrete brain regions for language production and understanding, such as Broca’s and Wernike’s areas,33 although we have not questioned this at the molecular level.
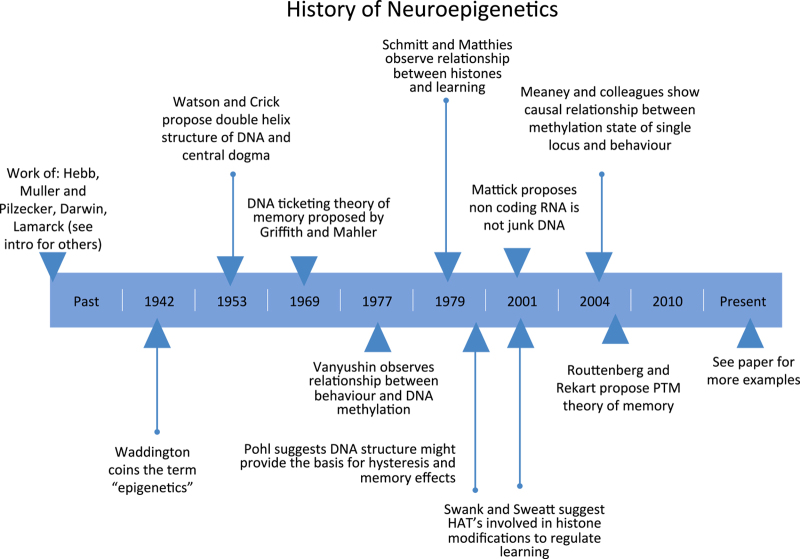
An emerging, and potentially complementary, view is that neuroepigenetic mechanisms, which constitute bidirectional and reversible changes in nucleic acids and proteins, occur both before and after transcription. These effects proceed within the same time frame as learning, and therefore represent an attractive alternative to the extreme interpretations of adaptation and heritability by Lamarck and Darwin.34 Contrary to contemporary perspectives, the concept of epigenetics within the context of learning and memory, or cognitive neuroepigenetics as it is now known, is not new (Figure 1). For example, more than 40 years ago, Griffith and Mahler35 proposed the DNA ticketing theory of memory, which postulated that the source for memory lay in the modification of nucleic acids, and suggested that the engram could extend beyond changes in neuronal function that are the result of protein synthesis alone.
Figure 1.
Timeline of significant discoveries in the field of cognitive neuroepigenetics.
Although this model was not directly testable due to technical limitations at the time, certain predictions have held true. For example, Vanyushin provided early experimental evidence that DNA methylation is associated with both active avoidance and food-seeking in rats.36 These pioneering findings were later rediscovered by Sweatt and colleagues, who showed an association between gene-specific DNA methylation, histone modification and memory formation.18–21 These studies have led to other intriguing advances, including the demonstration that molecular substrates classically associated with learning and memory, such as CREB-binding protein, also possess histone acetyltransferase activity, which accounts, in part, for some of their ability to modulate memory.37,38 Thus, the field has been primed to interrogate epigenetic mechanisms of DNA methylation and histone acetylation to establish their generality, and elucidate their complementary effects.
Meaney and colleagues discovered a causal role for epigenetic mechanisms in behavioural regulation by showing that variations in maternal care lead to reversible changes in DNA methylation within the glucocorticoid receptor promoter, resulting in significant effects on reactivity to stress later in life.39,40 This seminal discovery set the stage for the investigation of new epigenetic marks that are both behaviourally induced, and gene locus-specific. For example, Kumar et al.41 found that histone modification is associated with the progression from acute to chronic drug use, whereas Bredy et al.42 provided evidence to suggest that patterns of histone acetylation around a single gene promoter can be influenced in different ways depending on the type of learning. Many of these initial studies have been overlooked based on the fact that neuroepigenetic mechanisms initially did not appear to be persistent, and thus could not serve a role in maintaining the memory trace. This issue has been discussed by pioneers in the field, including Crick,43 Lisman,44 and Sweatt,45 as a key criterion for any molecular substrate to be affiliated with molecular memory processes. However, more recent studies have reported long-term persistent changes in epigenetic mechanisms, as well as demonstrating that some initial epigenetic marks may transition to others.46,47
Taken together, the findings suggest that neuroepigenetic mechanisms could provide a foundation for testable hypotheses of changes on nucleic acids as well as protein in relationship with a memory code.27–29 In addition, the strength of a neuroepigenetic view of learning and memory is that it diverges from traditional molecular neuroscience, which itself stems from the central dogma of molecular biology.28 This bias within molecular biology has most notably led to the labelling of non-coding regions of DNA as genetic noise or ‘junk DNA’, a conclusion that has since proven to be incorrect.48 As an extension of this, we argue that the unstated assumption of protein as the final goal of transcription has led to just as critical an oversight in the search for the fundamental molecular mechanisms of learning and memory, and that neuroepigenetic mechanisms offer an alternative explanation of the molecular underpinnings that lead to the engram, which are bidirectional, parallel and mechanistically dispersed.
Influence of DNA: old player, new roles in the adult brain?
DNA modification
There are a variety of DNA modifications; however, relatively few have been studied in the context of learning and memory. So far, the canonical modification, 5-methylcytosine (5mC) has mostly been associated with gene repression.49 However, the accumulation of 5mC is dynamic and has also been shown to enhance gene expression. Therefore, it has a far more functionally relevant role in the regulation of activity-dependent gene expression than previously assumed.50 This epigenetic mark also has oxidative derivatives, including 5-hydroxymethylation (5hmC), which has been shown to regulate gene expression within the context of learning and memory.47,51 Moreover, a recent study has demonstrated that the DNA glycosylases Ogg1 and MutY, which target the base modification 8-oxoG, have a role in adaptive behaviour, which implies a physiologically relevant role for 8-oxoG in the adult brain.52 Further, in a series of preliminary experiments, we have discovered that the accumulation of N6-methyladenosine (m6A) on DNA increases following extinction learning, and that knockdown of the putative m6A methyltransferase N6AMT1 blocks the consolidation of extinction memory (Xiang Li et al., unpublished). Contrary to early studies showing an inverse relationship between 5mC and gene expression in the brain, we found that the accumulation of m6A is required for the recruitment of the transcriptional machinery and serves to drive activity-dependent gene expression. In addition, emerging biochemical evidence suggests that there are many modified bases beyond cytosine that are theoretically functional.53,54 Mutagenesis and epileptogenesis studies have also shown that some of these base modifications can be induced under physiologically relevant conditions, similar to the way in which some histone modifications were recognized.54–56 These findings suggest the existence of a diverse and potentially functional repertoire of reversible DNA modifications on all four bases that could contribute to learning and memory processes.
DNA structure
It is often assumed that DNA only encodes information in its nucleotide sequence. This is partly because when Watson and Crick proposed their double-helix model of DNA they described a right handed form of DNA now called B-DNA, and implied a static conformation. However, Pohl57 found that changes in the conformational state of DNA can be detected based on previous changes in DNA structure. This not only suggests that DNA structure influences downstream effects such as protein binding, but also indicates that there is a molecular representation of a previous experience that can persist over time and is reflected in the current conformation of DNA. Pohl therefore suggested that, ‘DNA might provide the basis for hysteresis and memory effects…[and thus] should at least be considered as a possibility in biological systems.’ Indeed, much like its RNA and protein products, DNA can encode information through dynamic changes in its secondary structure.58 In fact, DNA can adopt at least 20 different conformations, which can act in concert with or independently of the sequence to regulate, among other functions, the recognition specificity of binding proteins.41–44
There also appears to be an interaction between DNA modification and structure, as specific conformations can be recognized by DNA-modifying enzymes, and changes in structure can modulate the ability of these enzymes to modify DNA.59,60 Furthermore, chromatin states can also interact and be influenced by DNA structure.61 Despite this, the prevailing view is that changes in DNA structure likely represent transcriptional and translational noise and are the by-product of transcription, adopted only in the absence of chromatin compaction during nucleosome remodelling.62 However, based on the current evidence, it is plausible that there are both stable and transient DNA conformational changes. These conformational changes may occur briefly as a result of transcription or local cellular state change, or be bound by base modifications and potentially rendered stable. If confirmed, this would represent yet another example of a parallel and bidirectional mechanism distributed across the genome, which may encode part of the engram.
DNA editing
The possibility of DNA editing is contentious in light of the current conceptualizations of the genome and its functional readout, in large part because stability is assumed. DNA editing refers to any collection of mechanisms that are predictably engaged to modify the underlying sequence of DNA in an experience-dependent manner. The most well characterized example of this can be found in the immune response, which is an exquisite memory system. However, beyond the variable diversity joining (VDJ) recombination process and somatic hypermutation associated with diversification of the immune system, there are emerging reports of other forms of functionally relevant DNA editing, including retrotransposon insertion and dynamic single-nucleotide variants (SNVs), which can occur anywhere in the neuronal genome. Although it was previously assumed that retrotransposition only occurred during early embryogenesis,63 it has now been demonstrated that the expression of the long interspersed nuclear element 1 (L1) retrotransposon continues during adulthood and is elevated in the brain.64 L1 retrotransposition occurs in response to a range of environmental stimuli, including voluntary exercise and chronic cocaine exposure.65,66 Indeed, single-cell retrotransposon sequencing analysis has recently revealed pervasive L1 mobilization in human hippocampal neurons.67
With respect to SNVs, it is becoming increasingly evident that each post-mitotic neuron in the human cortex can have a distinct genome, with conservative estimates of around 1,500 somatic SNVs per neuron,68 whereas others suggest that up to 10,000 SNVs may accumulate in healthy differentiated neurons across the lifespan.69 Pena de Ortiz and colleagues have for many years reported on the profound DNA recombinase activity that occurs in the brain in response to experience.70–72 Finally, DNA double-strand breaks have recently been shown to be necessary to regulate the expression of immediate early response genes, which are known to be important for learning and memory.73 Together, the evidence suggests that DNA editing may serve as a critically important source of functional diversification in post-mitotic neurons, enabling them to optimize their transcriptional responses to rapidly changing environmental signals by destabilizing and actively changing underlying genomic code. This is a significant departure from the concept of DNA as a static carrier of heritable information; however, its role in the regulation of gene expression related to learning and memory remains to be explored further.
Influence of RNA: ancient mechanisms, new neuroepigenetic player?
Despite possessing superior cognitive processes, ‘higher order’ organisms share approximately the same number of protein-coding genes with lower eukaryotes. Non-coding DNA-derived transcripts that possess no protein-coding capacity (non-coding RNA) have instead increased in the mammalian genome across evolution.48 Various classes of non-coding RNAs have been shown to participate as modular scaffolds and decoys, in cellular localization, and importantly, in activity-dependent cellular processes independent of protein, such as protection from viral infection.21,74,75 Recent evidence also indicates that different classes of non-coding RNA are directly involved in learning and memory. For example, microRNAs are critically involved in various forms of fear-related learning and memory,76,77 and recent studies have shown that long non-coding RNAs are also regulated by experience and appear to play a role in behavioural adaptation.78,79 Moreover, much like DNA, the post-transcriptional regulation of RNA is influenced by dynamic changes in chemical modification, structure, and editing.
RNA modifications
One recently emerging mechanism for how RNA is epigenetically regulated is through chemical modification. To date, there are at least 140 ‘epitranscriptomic’ modifications that are known to occur in RNA. Although little is currently known about their function in the context of learning and memory,80 one can postulate about their role. Pseudouridine (pseudoU) has been shown to affect RNA decay, potentially maintaining RNA involved in a short-term trace, or prolonging that associated with the long-term processes of memory consolidation.81,82 In addition, an enzyme which promotes the accumulation of pseudoU has recently been shown to be associated with cognitive dysfunction.83 These chemical modifications are also targeted to RNA via small nucleolar RNAs, which themselves have been shown to be involved in behavioural adaptation.84,85 RNA modifications have been demonstrated to affect the qualitative nature of RNA translation, thus constituting yet another potential way to alter protein function in an activity- or experience-dependent manner.86,87 In recent work, we have discovered that the RNA modification m6A is highly dynamic in the brain and critically involved in the formation of fear memory.88 We predict that, like DNA modification, RNA modification will come to be appreciated as an important epigenetic mechanism associated with behavioural adaptation that occurs both bidirectionally and in parallel to other neuroepigenetic mechanisms.
RNA structure
Chemical modifications are also known to affect the folding of RNA and are critical for determining its secondary and tertiary structures, which can impact the function of RNA inside the cell.74,89 This is interesting because changes in RNA structure have been linked to learning and memory. A stem-loop structure in the 3′ untranslated region of brain-derived neurotrophic factor (BDNF), a key neurotrophic factor for learning and memory, has been shown to be calcium-dependent and necessary for RNA stabilization.90 Furthermore, the G-quadruplex RNA structure is required for calcium/calmodulin-dependent protein kinase type II alpha chain and postsynaptic density protein 95 localization to neurites, both key factors in plasticity required for learning.91 It has also been shown that alternative splicing of exon 10 of tau, a protein that is strongly linked to neurodegenerative processes that lead to deficits in learning and memory, is regulated by a stem loop induced by a particular RNA helicase.92 Thus, dynamic changes in RNA structure may represent a novel mechanism for how RNA is co-opted for memory processes, without the need for new protein synthesis.
RNA editing
RNA editing is a process whereby an organism can increase the complexity and repertoire of transcripts that are able to be produced without changes in the genetic code.48,93 Specifically, two major classes of enzymes, including the ADAR family of adenosine deaminases and the APOBEC family of cytosine deaminases, mediate RNA editing. In what are now classic examples of the functional relevance of RNA editing, ADAR1 and two have been shown to promote the editing of the 5-hydroxytryptamine 2c receptor and the GluR2 subunit of the AMPA receptor, and can even modify synaptic structure, all of which are known to affect learning and memory.94 In addition, other RNA editing enzymes such as ADAR3 are only expressed in the brain of higher order vertebrates, which further suggests unexplored roles for RNA editing in cognition.95,96 It has also been shown that RNA editing can be altered by RNA structure, suggesting that RNA editing might represent a complementary and parallel process that can act on the qualitative state of a protein but is dependent on other protein synthesis-independent pathways in the cell.97
Influence of protein: old player, new tune
The conceptualization of protein synthesis-dependent learning and memory dates back to some of the first studies performed to understand biological processes contributing to memory, in which ‘protein-synthesis inhibitors’ were used to demonstrate the necessity of protein for memory storage (reviewed in refs 2,3). Unfortunately, as outlined above, these drugs have been shown to act by impairing nascent RNA, as well as a plethora of off-target processes, including phosphorylation of CREB, modifying adrenergic release, and potentially generating state dependency, all of which complicate the interpretation of their effect.12–14,98,99 However, classic examples of the necessity of protein synthesis for memory should not be overlooked. These include many different examples involving a variety of intracellular signalling cascades, as outlined by Kandel and others, including simple habituation in Aplysia, as well as fear learning in mice.11 In addition, histone modifications seem to act in part by altering gene expression and protein levels.100 Moreover, another way in which proteins may overcome molecular turnover is via prion-like conformational state changes, which can by definition self-perpetuate.101 Similarly, Routtenberg and Rekart102 have proposed the post-translational modification of protein hypothesis, which states that instead of relying only on static protein machinery for the formation and maintenance of a memory, memory can also be quasi-stored in dynamic, but consistently similar, post-translational modifications of proteins, which can affect subsequent processing of stimuli without the need to be in an active state. This idea is intriguing, and has been expanded within this paper to include DNA and RNA alongside traditional protein-synthesis views.
Neuroepigenetics: dynamic integration across all levels
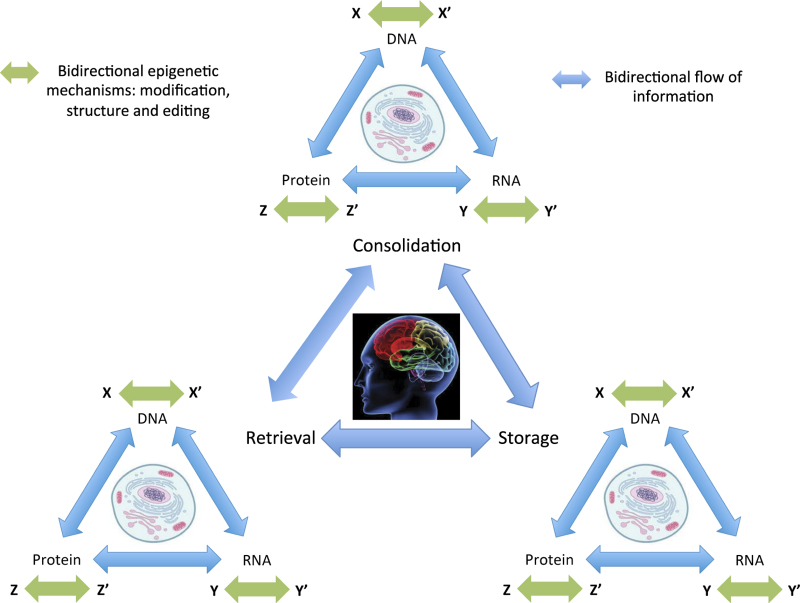
It is likely that the mnemonic state of an organism is encoded by changes in both nucleic acids and proteins, which can be temporarily biased or bounded by modifications, editing and structural changes to produce memory. Coming back to the question of serial or parallel systems and the location of memory storage, we can integrate what is known about protein synthesis, post-translational modifications and neuroepigenetic mechanisms into a more holistic view of the molecular basis of learning and memory (Figure 2).
Figure 2.
A neuroepigenetic model of memory suggests dynamic and reversible, state-dependent, modifications on DNA, RNA and protein that occur during consolidation, storage and retrieval. Much like Roberson and Sweatt’s45 model: X indicates cytosine and X′ 5-mC, Y adenosine and Y′ N6-methyladenosine, Z is acetylated histone and Z′ deacetylated histone. Although more evidence is required to establish the generalized nature of this model across all phases of learning and memory, it is well established that epigenetic mechanisms influence memory storage, and emerging findings suggest an influence on consolidation102 and retrieval. 104–106,111–123
As mentioned above, the process of encoding and memory storage is intuitively linear. However, when speaking about memory it is important to keep in mind that, much like the fact that transcription does not always lead to protein expression, encoding does not always result in storage, which in turn does not always lead to retrieval. Memories can be state-dependent, inaccessible at one point in time but, with a change of internal state, immediately retrieved. For example, Gisquet-verrier et al.103 have shown that impairment by protein-synthesis inhibitors can be reversed if they are delivered both post-training and pre-test. Is this because of alterations to existing proteins or has the epigenetic state of DNA, RNA and protein been changed? The evidence remains to be seen, but the question is important. Put more simply, experience could be encoded across a spectrum of epigenetic modifications to DNA, RNA and protein, and over time this reversibility may become less probable and a similar cellular state may be more easily reproducible, such as occurs in the case of metaplasticity and epigenetic priming. The strength of this complementary view is that this molecular system is parallel and distributed, instead of serial and equipotential, which is also in line with the emerging evidence in neuroepigenetics. This view also leads to the prediction that most memories will be metastable and cannot by definition be an exact replication at the molecular or cognitive level of the past; instances when this does occur would necessitate moving a chemical equilibrium so far in one direction that its reversibility becomes biochemically unfeasible, including the chemical stability of protein formation.
What is on the horizon?
The question of where cognitive neuroepigenetics as a field is going requires an appreciation of the precision of emerging technology and clarity in how current technologies are applied. Currently, we know that there are a variety of ways in which nucleic acid can alter memory and learning, but there is a paucity of data to definitively link all of the pieces. More explicitly, there are three requirements that need to be met in order to tackle this issue: (1) specific measurement, (2) specific manipulation and (3) functional validation. Specific measurement refers to the need to extend fundamental data about established brain regions and molecular pathways, based on cell-type and locus-specific analyses. A promising approach to achieve this resolution is single-cell sequencing, which has already begun to reveal a level of precision often missed in population studies.68 In addition, we have developed methodology to enrich and profile epigenetic mechanisms in specific cell types, including those that are selectively involved in a memory trace.104 Furthermore, others have developed ‘click DNA labelling’, which uses chemically specific interactions with selected targets such as DNA modification, to produce genome-wide profiling of this epigenetic mark at single base resolution.105,106 Together, these technologies can be applied to the analysis of particular active cells, loci and nucleotide modifications that potentially make up Lashley’s dynamic engram at the molecular level.22
With an exceptional level of detail in the measurement of epigenetic changes on the horizon, this begs the question as to what is required to best use the information generated from these studies. Thankfully, there already exists a technique to locus-specifically edit and direct putative modifications in the brain, namely clustered regularly interspaced palindromic repeat (CRISPR)-Cas9. This new technology uses the enzymatic machinery from bacteria, which normally functions to target and destroy viruses, and instead exchanges the RNA targeting sequence to virus for one of an experimenter’s choice to selectively target regions of DNA for excision.107,108 Furthermore, and more excitingly for the field of neuroepigenetics, this technique can also use a deactivated version of the cutting enzyme (dCas9) to site-selectively direct proteins, or epigenetic modifiers in awake behaving animals to probe learning and memory processes. For example Heller et al.109 have used a similar technology called transcription activator-like effector nuclease (TALEN), which uses a cutting enzyme and direction sequence, to functionally validate the sufficiency of modifications around one locus to modulate a behavioural effect. This technique has also been paired with light activation to reversibly modulate epigenetic modifiers in vivo in a rapid and discrete temporal window.110 In the future, these kinds of studies should serve as the standard. In particular, as techniques for measurement and manipulation evolve, so too should it become less acceptable to simply relate bulk protein or mRNA levels to behavioural change, as the data no longer support the 1:1 relationship as outlined in central dogma.
Conclusions
Sufficient evidence now exists to support the concept of reversibility as a common thread in our understanding of the molecular mechanisms of learning and memory, which extends well beyond the traditional protein-centric model. It is evident that nucleic acids and related epigenetic mechanisms contribute to learning and memory in a variety of ways, and can bidirectionally impact each stage of the cognitive process. A neuroepigenetic view predicts a parallel and distributed system for the consolidation, storage and retrieval of the engram based on dynamic and reversible changes to DNA, RNA and protein in the brain. This view may also help to explain the increased complexity of higher order organisms and how they have evolved to maintain their capacity to learn and store information despite constant changes in the environment.
Acknowledgments
We gratefully acknowledge grant support from the NIH (5R01MH105398-TWB) and the Australian Research Council (SR120300015-TWB). We would also like to thank Ms Rowan Tweedale for helpful editing of the manuscript and Dr James McGaugh for his thoughtful comments, which helped organize earlier drafts of this manuscript.
Footnotes
The authors declare no conflict of interest.
References
- Hebb, D. O. The Organization of Behavior 335 (Wiley, 1949). [Google Scholar]
- McGaugh, J. L. Memory—a century of consolidation. Science 287, 248–251 (2000). [DOI] [PubMed] [Google Scholar]
- Lechner, H. A., Squire, L. R. & Byrne, J. H. 100 years of consolidation—remembering Müller and Pilzecker. Learn. Mem. 6, 77–87 (1999). [PubMed] [Google Scholar]
- Zirkle, C. The Inheritance of acquired characters and the provisional hypothesis of pangenesis. Press Am. Soc. Nat. 69, 417–445 (1935). [Google Scholar]
- Darwin, C. Darwin. 386 (John Murray, London, UK, 1859).
- Skinner, B. F. Operant behavior. Am. Psychol. 18, 503–515. (1963). [Google Scholar]
- McClelland, J. L., McNaughton, B. L. & O’Reilly, R. C. Why there are complementary learning systems in the hippocampus and neortex: Insights from the successes and failures of connectionist models of learning and memory. Psychol. Rev. 3, 419–457 (1995). [DOI] [PubMed] [Google Scholar]
- Neisser, U. Cognitive Psychology. East Norwalk, CT, US Applet Century Crofts. Available at http://www.amazon.ca/exec/obidos/redirect?tag=citeulike09-20&path=ASIN/049550629X (1967).
- Wurtz, R. H. Recounting the impact of Hubel and Wiesel. J. Physiol. 587, 2817–2823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmoen, I. A. & Apuzzo, M. L. J. The brain on itself: nobel laureates and the history of fundemental nervous system function. Neurosurgery 61, 891–908 (2007). [DOI] [PubMed] [Google Scholar]
- Kandel, E. R. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 5, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J. L., Jensen, R. A. & McGaugh, J. L. Attenuation of experimentally-induced amnesia. Prog. Neurobiol. 16, 155–186 (1981). [DOI] [PubMed] [Google Scholar]
- Roberts, R. B., Flexner, J. B. & Flexner, L. B. Some evidence for the involvement of andrenergic sites in the memory trace. Proc. Natl Acad. Sci. USA 66, 310–313 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serota, R. G., Roberts, R. B. & Flexner, L. B. Acetoxycycloheximide-induced transient amnesia: protective effects of adrenergic stimulants. Proc. Natl Acad. Sci. USA 69, 340–342 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer, L. M. & Slice, L. W. Anisomycin induces COX-2 mRNA expression through p38MAPK and CREB independent of small GTPases in intestinal epithelial cells. Biochim. Biophys. Acta Mol. Cell. Res. 1745, 393–400 (2005). [DOI] [PubMed] [Google Scholar]
- Rudy, J. W., Biedenkapp, J. C., Moineau, J. & Bolding, K. Anisomycin and the reconsolidation hypothesis. Learn. Mem. 13, 1–3 (2006). [DOI] [PubMed] [Google Scholar]
- Wittstock, S. & Menzel, R. Color learning and memory in honey bees are not affected by protein synthesis inhibition. Behav. Neural. Biol. 3, 224–229 (1994). [DOI] [PubMed] [Google Scholar]
- Wittstock, S., Kaatz, H. H. & Menzel, R. Inhibition of brain protein synthesis by cycloheximide does not affect formation of long-term memory in honeybees after olfactory conditioning. J. Neurosci. 13, 1379–1386 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubli, U., Faraday, R. & Lynch, G. Pharmacological dissociation of memory: anisomycin, a protein synthesis inhibitor, and leupeptin, a protease inhibitor, block different learning tasks. Behav. Neural. Biol. 43, 287–297 (1985). [DOI] [PubMed] [Google Scholar]
- Lattal, K. M. & Abel, T. Different requirements for protein synthesis in acquisition and extinction of spatial preferences and context-evoked fear. J. Neurosci. 21, 5773–5780 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo, J. A. & Dougherty, W. G. Plant pathology and RNAi: a brief history. Annu. Rev. Phytopathol. 43, 191–204 (2005). [DOI] [PubMed] [Google Scholar]
- Lashley, K. in Society of Experimental Biology Symposium No. 4: Physiological Mechanisms in Animal Behaviour (eds Danielli J. F. & Brown R.) 454–482 (Academic Press, 1950).
- James, W. The Principles of Psychology Vols. 1 & 2, 688 (Holt, 1890).
- Nader, K. A single standard for memory; the case for reconsolidation. Debates Neurosci. 1, 2–16 (2007). [DOI] [PubMed] [Google Scholar]
- Misanin, J. R., Miller, R. R. & Lewis, D. J. Retrograde amnesia produced by electroconvulsive shock after reactivation of a consolidated memory trace. Science 160, 554–555 (1968). [DOI] [PubMed] [Google Scholar]
- Scoville, W. B. & Milner, B. Loss of recent memory after bilateral hippocampal lesions. 1957. J. Neuropsychiatry Clin. Neurosci. 12, 103–113 (2000). [DOI] [PubMed] [Google Scholar]
- Squire, L. R. Nondeclarative memory: multiple brain systems supporting learning. J. Cogn. Neurosci. 4, 232–243 (1992). [DOI] [PubMed] [Google Scholar]
- Squire, L. R. Memory systems of the brain: a brief history and current perspective. Neurobiol. Learn. Mem. 82, 171–177 (2004). [DOI] [PubMed] [Google Scholar]
- Packard, M. G. & McGaugh, J. L. Inactivation of hippocampus or caudate nucleus with lidocaine differentially affects expression of place and response learning. Neurobiol. Learn. Mem. 65, 65–72 (1996). [DOI] [PubMed] [Google Scholar]
- Yassa, M. A. & Reagh, Z. M. Competitive trace theory: a role for the hippocampus in contextual interference during retrieval. Front. Behav. Neurosci. 7, 107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, R. J. & White, N. M. A triple dissociation of memory systems: hippocampus, amygdala, and dorsal striatum. Behav. Neurosci. 107, 3–22 (1993). [DOI] [PubMed] [Google Scholar]
- Crick, F. Central dogma of molecular biology. Nature 227, 561–563 (1970). [DOI] [PubMed] [Google Scholar]
- Geschwind, N. The organization of language and the brain. Science 170, 940–944 (1970). [DOI] [PubMed] [Google Scholar]
- Waddington, C. H. The epigenotype. Endeavour 1, 18–20 (1942). [Google Scholar]
- Griffith, J. S. & Mahler, H. R. DNA ticketing theory of memory. Nature 223, 580–582 (1969). [DOI] [PubMed] [Google Scholar]
- Ashapkin, V. V., Romanov, G. A., Tushmalova, N. A. & Vanyushin, B. F. Selective DNA synthesis in the rate brain induced by learning Biokhimija 48, 355–362 (1982).
- Wood, M. a. et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 12, 111–119 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey, C. G. et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 27, 6128–6140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, I. C. G. et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004). [DOI] [PubMed] [Google Scholar]
- Weaver, I. C. G. Epigenetic programming by maternal behavior and pharmacological intervention. Epigenetics 2, 22–28 (2007). [DOI] [PubMed] [Google Scholar]
- Kumar, A. et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314 (2005). [DOI] [PubMed] [Google Scholar]
- Bredy, T. W. et al. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn. Mem. 14, 268–276 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick, F. Neurobiology: memory and molecular turnover. Nature 312, 101–101 (1984). [DOI] [PubMed] [Google Scholar]
- Lisman, J. E. A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc. Natl Acad. Sci. USA 82, 3055–3057 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson, E. D. & Sweatt, J. D. A biochemical blueprint for long-term memory. Learn. Mem. 6, 381–388 (1999). [PMC free article] [PubMed] [Google Scholar]
- Baker-Andresen, D. et al. Persistent variations in neuronal DNA methylation following cocaine self-administration and protracted abstinence in mice. Neuroepigenetics 4, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc. Natl Acad. Sci. USA 111, 7120–7125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J. S. Deconstructing the dogma. Ann. N Y Acad. Sci. 1178, 29–46 (2009). [DOI] [PubMed] [Google Scholar]
- Shin, J., Ming, G.-L. & Song, H. DNA modifications in the mammalian brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco, M. R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2011). [DOI] [PubMed] [Google Scholar]
- Li, X., Wei, W., Ratnu, V. S. & Bredy, T. W. On the potential role of active DNA demethylation in establishing epigenetic states associated with neural plasticity and memory. Neurobiol. Learn. Mem. 105, 125–132 (2013). [DOI] [PubMed] [Google Scholar]
- Bjørge, M. D. et al. Synergistic actions of Ogg1 and mutyh DNA glycosylases modulate anxiety-like behavior in mice. Cell. Rep. 13, 2671–2678 (2015). [DOI] [PubMed] [Google Scholar]
- Bande, O. et al. Base pairing involving artificial bases in vitro and in vivo. Chem. Sci. 7, 995–1010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela, F. & Debelak, H. The N(8)-(2′-deoxyribofuranoside) of 8-aza-7-deazaadenine: a universal nucleoside forming specific hydrogen bonds with the four canonical DNA constituents. Nucleic Acids Res. 28, 3224–3232 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, J., Henshall, D. C., Simon, R. P. & Chen, J. Formation of the base modification 8-hydroxyl-2′-deoxyguanosine and DNA fragmentation following seizures induced by systemic kainic acid in the rat. J. Neurochem. 74, 302–309 (2000). [DOI] [PubMed] [Google Scholar]
- Tsankova, N. M., Kumar, A. & Nestler, E. J. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 24, 5603–5610 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl, F. M. Hysteretic behaviour of a Z-DNA-antibody complex. Biophys. Chem. 26, 385–390 (1987). [DOI] [PubMed] [Google Scholar]
- Harteis, S. & Schneider, S. Making the bend: DNA tertiary structure and protein–DNA interactions. Int. J. Mol. Sci. 15, 12335–12363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. S., Kan, J. L., Baker, D. J., Kaplan, B. E. & Dembek, P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J. Mol. Biol. 217, 39–51 (1991s). [DOI] [PubMed] [Google Scholar]
- Laayoun, A. & Smith, S. S. Methylation of slipped duplexes, snapbacks and cruciforms by human DNA(cytosine-5)methyltransferase. Nucleic Acids Res. 23, 1584–1589 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Mulholland, N., Fu, H. & Zhao, K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell. Biol. 26, 2550–2559 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morange, M. What history tells us. IX. Z-DNA: when nature is not opportunistic. J. Biosci. 32, 657–661 (2007). [DOI] [PubMed] [Google Scholar]
- Kano, H. et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri, A. R. et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature 7322, 443–446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri, A. R., Zhao, C., Marchetto, M. C. N. & Gage, F. H. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 19, 1002–1007 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze, I. et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl Acad. Sci. USA 108, 3035–3040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton, K. R. et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161, 228–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato, M. A. et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, J. L. et al. The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron. 89, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Cesario, M. et al. Behavioral/systems/cognitive an inhibitor of DNA recombination blocks memory consolidation, but not reconsolidation, in context fear conditioning. J. Neurosci. 26, 5524–5533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra-Rodríguez, L. et al. Identification of flap structure-specific endonuclease 1 as a factor involved in long-term memory formation of aversive learning. J. Neurosci. 29, 5726–5737 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Pérez, E. et al. Identification and characterization of the V(D)J recombination activating gene 1 in long-term memory of context fear conditioning. Neural Plast. 2016, 1–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi, R. et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale, R. C. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro, P. A. & Bredy, T. W. Emerging role of non-coding RNA in neural plasticity, cognitive function, and neuro psychiatric disorders. Front Genet. 3, 1–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q. et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 14, 1115–1117 (2011). [DOI] [PubMed] [Google Scholar]
- Bredy, T. W., Lin, Q., Wei, W., Baker-Andresen, D. & Mattick, J. S. MicroRNA regulation of neural plasticity and memory. Neurobiol. Learn. Mem. 96, 89–94 (2011). [DOI] [PubMed] [Google Scholar]
- Spadaro, P. A. et al. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatry. 78, 848–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, G. et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494 (2013). [DOI] [PubMed] [Google Scholar]
- Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D. R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23, 5020–5026 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó, K., Muramatsu, H., Keller, J. M. & Weissman, D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 20, 948–953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen, R. et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 135, 707–713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148 (2002). [DOI] [PubMed] [Google Scholar]
- Bachellerie, J. P., Cavaillé, J. & Hüttenhofer, A. The expanding snoRNA world. Biochimie 84, 775–790 (2002). [DOI] [PubMed] [Google Scholar]
- Hoernes, T. P. et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 530, 441–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo, J. et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 34, 721–733 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi, M. & Tsuda, M. Involvement of the 3′-untranslated region of the brain-derived neurotrophic factor gene in activity-dependent mRNA stabilization. J. Neurochem. 115, 1222–1233 (2010). [DOI] [PubMed] [Google Scholar]
- Subramanian, M. et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 12, 697–704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, A. et al. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell. Biol. 31, 1812–1821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J. S. & Mehler, M. F. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 31, 227–233 (2008). [DOI] [PubMed] [Google Scholar]
- Aktories, K. et al. in Adenosine Deaminases Acting on RNA (ADARs) and A-to-I Editing 1st edn (ed. Samuel C. E.) (Springer, Berlin, 2012).
- Chen, C. X. et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6, 755–767 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C. A., Campbell, S. L. & Sweatt, J. D. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn. Mem. 89, 599–603 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeris, M., Funke, L., Shrestha, J., Rich, A. & Maas, S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 33, 5362–5370 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, A. J., Kogan, J. H., Frankland, P. W. & Kida, S. Creb and memory. Annu. Rev. Neurosci. 21, 127–148 (1998). [DOI] [PubMed] [Google Scholar]
- Lechner, H. A. & Squire, L. R. 100 Years of consolidation— remembering Müller and Pilzecker. Learn. Mem. (Cold Spring Harb. NY) 6, 77–87 (1999). [PubMed] [Google Scholar]
- Lopez-Atalaya, J. P., Ito, S., Valor, L. M., Benito, E. & Barco, A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 41, 8072–8084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si, K., Lindquist, S. & Kandel, E. R. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 115, 879–891 (2003). [DOI] [PubMed] [Google Scholar]
- Routtenberg, A. & Rekart, J. L. Post-translational protein modification as the substrate for long-lasting memory. Trends Neurosci. 28, 12–19 (2005). [DOI] [PubMed] [Google Scholar]
- Gisquet-verrier, X. P. et al. Integration of new information with active memory accounts for retrograde amnesia : a challenge to the consolidation/reconsolidation hypothesis? J. Neurosci. 35, 11623–11633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Baker-Andresen, D., Zhao, Q., Marshall, V. & Bredy, T. W. Methyl CpG binding domain ultra-sequencing: a novel method for identifying inter-individual and cell-type-specific variation in DNA methylation. Genes Brain Behav. 13, 721–731 (2014). [DOI] [PubMed] [Google Scholar]
- Sletten, E. M. & Bertozzi, C. R. From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res. 44, 666–676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. & Chen, P. R. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 12, 129–137 (2016). [DOI] [PubMed] [Google Scholar]
- Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J. A. & Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096–1258096 (2014). [DOI] [PubMed] [Google Scholar]
- Heller, E. A. et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci. 17, 1720–1727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M. & Matthies, H. [Biochemical studies on histones of the central nervous system. III. Incorporation of [14C]-acetate into the histones of different rat brain regions during a learning experiment]. Acta Biol. Med. Ger. 38, 683–689 (1979). [PubMed] [Google Scholar]
- Swank, M. W. & Sweatt, J. D. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J. Neurosci. 21, 3383–3391 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, T. R. et al. Noncoding RNAs in long-term memory formation. Neuroscientist 14, 434–445 (2008). [DOI] [PubMed] [Google Scholar]
- Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002). [DOI] [PubMed] [Google Scholar]
- Watson, J. D. & Crick, F. H. C. Molecular structure of nucleic acids. Nature 171, 737–738 (1953). [DOI] [PubMed] [Google Scholar]
- Ghosh, A. & Bansal, M. A glossary of DNA structures from A to Z. Acta Crystallogr.—Sect. D Biol. Crystallogr. 59, 620–626 (2003). [DOI] [PubMed] [Google Scholar]
- Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, N. et al. Deconvolving the recognition of DNA shape from sequence. Cell 161, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs, R. et al. The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. et al. Reinstatement of long-term memory following erasure of its behavioral and synaptic expression in aplysia. Elife e03896, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtenberg, A. The substrate for long-lasting memory: If not protein synthesis, then what? Neurobiol. Learn. Mem. 89, 225–233 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Z. & Gold, P. E. Intrahippocampal infusions of anisomycin produce amnesia: contribution of increased release of norepinephrine, dopamine, and acetylcholine. Learn. Mem. 16, 308–314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canal, C. E., Chang, Q. & Gold, P. E. Amnesia produced by altered release of neurotransmitters after intraamygdala injections of a protein synthesis inhibitor. Proc. Natl Acad. Sci. USA 104, 12500–12505 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]