Abstract
The epidermal growth factor receptor (EGFR) is activated through binding to specific ligands and generates signals for proliferation, differentiation, migration, and cell survival. Recent data show the role of nuclear EGFR in tumors. Although many EGFR ligands are upregulated in cancers, little is known about their effects on EGFR nuclear translocation. We have compared the effects of six EGFR ligands (EGF, HB-EGF, TGF-α, β-Cellulin, amphiregulin, and epiregulin) on nuclear translocation of EGFR, receptor phosphorylation, migration, and proliferation. Cell fractionation and confocal immunofluorescence detected EGFR in the nucleus after EGF, HB-EGF, TGF-α and β-Cellulin stimulation in a dose-dependent manner. In contrast, amphiregulin and epiregulin did not generate nuclear translocation of EGFR. EGF, HB-EGF, TGF-α and β-Cellulin showed correlations between a higher rate of wound closure and increased phosphorylation of residues in the carboxy-terminus of EGFR, compared to amphiregulin and epiregulin. The data indicate that EGFR is translocated to the nucleus after stimulation with EGF, HB-EGF, TGF-α and β-Cellulin, and that these ligands are related to increased phosphorylation of EGFR tyrosine residues, inducing migration of SkHep-1 cells.
Keywords: EGFR ligands, epidermal growth factor receptor, nuclear translocation, tyrosine phosphorylation sites, cell migration, proliferation
1. Introduction
The epidermal growth factor receptor (EGFR) is one of four members of the HER family of receptor tyrosine kinases [1]. Eight members of the epidermal growth factor (EGF) family ligands have been described: epidermal growth factor (EGF), heparin-binding EGF-like growth factor (HB-EGF), transforming growth factor alpha (TGF-α), betacellulin, amphiregulin, epiregulin, epigen and neuregulin 2 beta (NRG2β) [2]. Endogenously, these ligands are found in the form of transmembrane pro-ligands, which are proteolytically cleaved to release the mature soluble form that interacts with EGF receptors [3]. In several in vitro and animal model systems, distinct EGF family ligands can promote divergent biological outcomes from EGFR activation [2, 4]. Ligand-mediated EGFR activation is achieved by a conformational change in the extracellular domain of the receptor upon ligand binding, resulting in receptor dimerization and internalization [5]. Active EGFR dimers undergo autophosphorylation of tyrosine residues in the cytoplasmic tail of the receptor (carboxy-terminal), recruiting phosphotyrosine-binding proteins and activating multiple signal transduction pathways.
Apart from the prominent role in signal transduction, EGFR also has some non-canonical functions, such as transcriptional regulation, DNA synthesis, and DNA repair in the nucleus [6–8]. Nuclear translocation of EGFR is mediated by a number of ligands, but little is known about their individual roles. Here, we have investigated the effects of six ligands on of EGFR nuclear translocation and SKHep-1 cell migration.
2. Materials and Methods
2.1. Cell culture and EGFR ligand stimulation
SkHep-1, a human liver cancer cell line (ATCC, VA, USA), was cultured at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (Life Technologies, NY, USA) containing 10% fetal bovine serum, 1 mM sodium pyruvate, 50 units/ml penicillin, and 50 g/ml streptomycin (Life Technologies). For stimulation, EGFR ligands (1, 5, 10, 50 and 100 ng/ml) were incubated to previously serum-starved (12–16 hours) cells. The EGFR ligands used were a human recombinant epidermal growth factor (EGF, Life Technologies), Heparin-binding EGF-like growth factor (HB-EGF, R&D Systems, MN, USA), Transforming growth factor-α (TGF-α, R&D Systems), Betacellulin (BTC, R&D Systems), Epiregulin (EPR, R&D Systems), Ampiregulin (AR, R&D Systems).
2.2. Subcellular fractionation
Subcellular fractionation was performed as described previously [9, 10]. Briefly, cells were harvested by scraping, and lysed in a lysis buffer. The homogenate was centrifuged at 1,500g for 5 min to sediment the nuclei. The supernatant was centrifuged at 16,100g for 20 min, and the resulting supernatant formed the non-nuclear fraction. The nuclear pellet was resuspended in NETN buffer to extract nuclear proteins. Nuclear lysates were collected after centrifugation at 16,100g for 20 min at 4°C. Protease and phosphatase inhibitors were added to all buffers. Protein concentrations were quantified by Bradford assay. All reagents were purchased from Sigma-Aldrich, MO, USA.
2.3. Western blot
Western blotting was performed and detected as described previously [9, 11]. Primary antibodies used were polyclonal anti-EGFR (Santa Cruz Biotechnology, TX, USA), monoclonal anti-α-tubulin (Sigma-Aldrich), anti-Lamin B1 (Abcam, MA, USA), Histone H3 (Sigma-Aldrich), p-Histone H3 (S10) (Milipore, MA, USA), Erk1/2, p-Erk1/2 (T202/Y204), AKT, p-AKT (T308), p-EGFR (T669), p-EGFR (Y845), p-EGFR (Y992), p-EGFR (Y998), p-EGFR (Y1045), p-EGFR (S1046/1047), p-EGFR (Y1086), p-EGFR (Y1148), p-EGFR (Y1173) (Cell Signaling Technology, MA, USA). The PDVF membranes were developed using enhanced chemiluminescence (ECL Plus; GE Healthcare Life Sciences, NJ, USA). Subsequently, the films were scanned and analyzed using Image J software (http://rsbweb.nih.gov/ij/).
2.4. Confocal fluorescence microscopy
Cells were double-labeled with mouse monoclonal anti-EGFR (Millipore) and a rabbit polyclonal antibody against the nuclear membrane marker Lamin-B1 (Abcam, MA, USA), and then incubated with secondary antibodies conjugated to anti-mouse Alexa 555 or anti-rabbit Alexa 488 (Life Technologies), respectively. Hoechst 33342 (Life Technologies) was used as marker for the nuclear compartment. Images were collected using a Zeiss LSM 880 with Airyscan with a 63X, 1.4 NA objective lens.
2.5. Proliferation assays
Serum starved cells were stimulated with EGFR ligands (100ng/ml) for 24, 48 and 72 hours. Over time, cells were trypsinized, stained by trypan blue to mark unviable cells. Only viable cells were counted. The percentage of DNA-synthesizing cells were analyzed after 24 h of ligand stimulation using Click-iT® Edu Alexa Fluor® 488 Imaging Kit (Life Technologies) according to the manufacturer’s instructions. The images were obtained and analyzed using Cytatyon™ 5 Cell Imaging Multi-Mode Reader managed by Gen5™ software (BioTek, VT, USA). The proportion of Edu (5-ethynyl-2′-deoxyuridine)-positive cells was quantified related to nuclear marker Hoechst. Three independent experiments and nine fields were counted per treatment.
2.6. Measurement of Histone 3 phosphorylation by flow cytometry
The percentage of cells positive for phospho-Histone H3 (Ser10) were determined by flow cytometry. After ice-cold 70% ethanol fixation, cells were incubated in blocking solution (1% BSA in PBS) and with Alexa Fluor 488-conjugated with phospho-H3 (S10) antibody (Cell Signaling Technology) at a 1:100 dilution for 1 hour. Quantification of phospho-H3-positive cells were performed in a Guava Easycyte 6L flow cytometer (Millipore) and data was analyzed using FlowJo software (version 7.2.5) (FlowJo, OR, USA)
2.7. Scratch assay
Cell migration was assessed using a wound healing assay. Previously, cells were serum starved for 24 hours and then a “wound” was made by displacing cells using a 1 mL pipette tip (Axygene, CA, USA). After wounding, cells were washed with PBS and incubated with growth media supplemented with EGFR ligands (100ng/ml) described above. The wounded area was imaged using a 10X Objective and a QIClick camera (QImaging, BC, Canada) installed on IX70 Olympus inverted microscope (Olympus, Japan) via Image-Pro Plus software (Media Cybernetics, MD, USA) immediately after wounding (0 hour), 12, 24 and 36 hours later. Quantification of cell migration was determined by calculating the coverage of the wounded area with cells after 36 hours with or without ligands using ImageJ software (http://rsbweb.nih.gov/ij/).
2.8. Data and statistical analysis
Semi-log plots were generated using densitometric data from nuclear EGFR accumulation upon ligand stimulation. The EC50 were estimates for nuclear EGFR detection using the nonlinear regression fit by the software Prism 6 (GraphPad software, CA, USA). Significance of changes in treatment groups was determined by Student’s t test or one-way analysis of variance and Bonferroni’s multiple comparison test, using Prism 6 software. Data are represented as mean ± S.E.
3. Results
3.1. Only EGF, HB-EGF, TGF-α and β-Cellulin mediate EGFR nuclear translocation
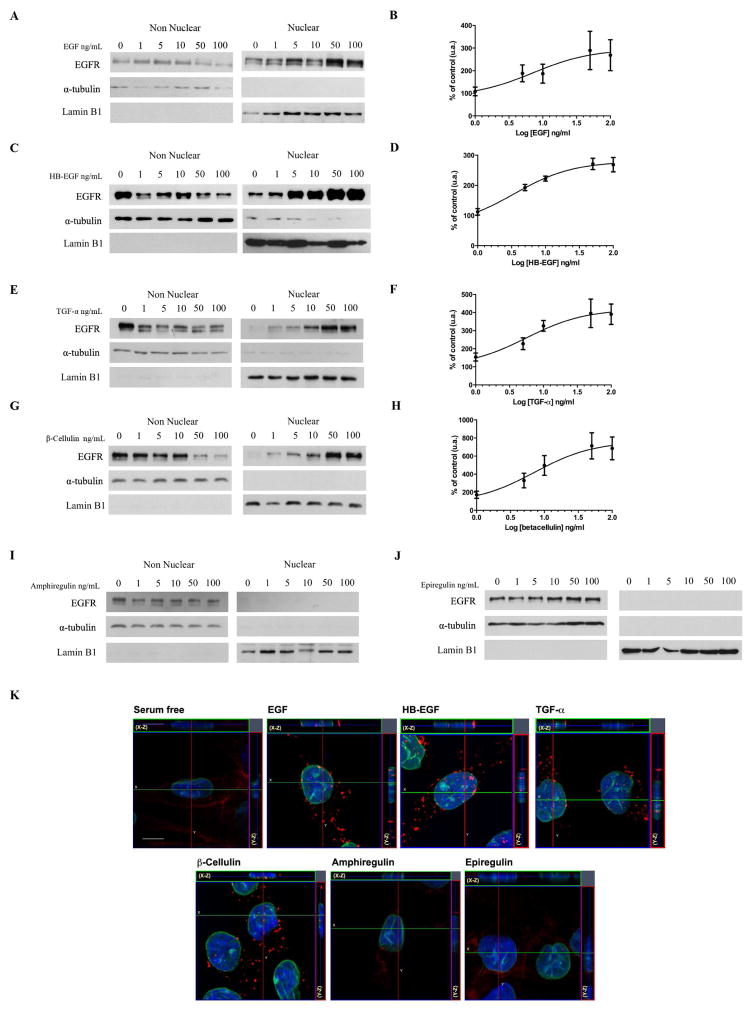
Previous live cell studies using SKHep-1 cells showed that EGFR could be found within the nucleus after 2.5 minutes, reaching a peak within 10 minutes after stimulation with100 ng/ml of EGF [10]. To test the effects of different ligands (EGF, HB-EGF, TGF-α, BTC, AR and EPR) on EGFR nuclear translocation, we stimulated SkHep-1 cells for 10 minutes at increasing ligand concentrations (1, 5, 10, 50 and 100 ng/ml). Nuclear and non-nuclear fractions from non-stimulated (0) and ligand-stimulated cells were analyzed by western blot (Figure 1). Lamin B1 and α-tubulin distributions verified correct fraction separation. In addition to EGF, we found that HB-EGF, TGF-α, and β-Cellulin are able to induce EGFR nuclear translocation in a dose ligand-dependent manner (Figure 1A, C, E and G). In contrast, even at higher concentrations, amphiregulin and epiregulin stimulation, EGFR was not detected in the nucleus (Figure 1I, J).
Figure 1. EGF, HB-EGF, TGF-α, β-Cellulin induce EGFR nuclear translocation in a dose-dependent manner in SkHep-1 cells while amphiregulin and epiregulin do not.
A, C, E, G, I, J, Western blot analysis of total EGFR in non-nuclear and nuclear fractions isolated from resting (0) or EGFR ligands stimulated cells (1, 5,10, 50 and 100 ng/ml) for 10 minutes. α-tubulin and Lamin B1 were used as purity control for nuclear and non-nuclear fractions, respectively. Bands corresponding to EGFR in the nuclear fractions are detected in EGF, HB-EGF, TGF-α, β-Cellulin-stimulated groups (A, C, E, G). EGFR bands are not observed in amphiregulin and epiregulin nuclear fractions (I, J). EGFR Blots are representative of what was observed in three separate experiments. B, D, F, H, EC50 curves from densitometrics analysis confirms the dose-dependent EGFR nuclear translocation upon EGF, HB-EGF, TGF-α, β-Cellulin stimulation. The nuclear detection of EGFR was expressed as percent of EGFR expression in resting cells. Line through EGFR expression is the nonlinear regression fit with Hill equation of ligand dose response. Data are represented as mean ± S.E (N=3). K, confocal images of non-stimulated SkHep-1 cells (serum free) and upon 10 min EGFR ligands stimulation (100ng/ml). Lamin B1, the nuclear membrane marker is showed in green, EGFR in red and the nuclear staining hoechst in blue. Serial optical sections were collected for three-dimensional reconstruction; x–z sections are shown at the top, and y-z sections are shown at the right of each image (bar = 10 μm).
The median effective concentration (EC50 values) was interpolated from the dose response curves of EGFR nuclear quantification (Figure 1B, D, F and H). The EC50 value of HB-EGF, TGF-α, EGF and BTC was 3.4 ± 0.2, 5.6 ± 0.4, 7.1 ± 0.6, and 7.2 ± 0.4 ng/ml, respectively. The results indicate differences in the potency of the different ligands in mediating nuclear accumulation of EGFR.
Corroborating with cell fractionation experiments, three-dimensional reconstruction of serial confocal immunofluorescence showed that EGFR localized to the plasma membrane in AR and EPR stimulated cells, and in non-stimulated SKHep-1 cells (serum free). On the other hand, the receptor was localized in the nucleus after 10 minutes of exposure to EGF, HB-EGF, TGF-α and BTC (Figure 1K). In summary, the results show that EGFR translocates to the nucleus upon 10 minutes of stimulation with EGF, HB-EGF, TGF-α, β-Cellulin, but not with AR and EPR.
3.2. Differential phosphorylation profile upon EGFR stimulation
We hypothesized that different EGFR ligands stimulate EGFR phosphorylation on distinct tyrosine or serine residues. SkHep-1 lysates were subjected to western blot analysis with phosphosite-specific antibodies to determine whether there was differential phosphorylation after stimulation with different EGFR ligands. EGF, HB-EGF, TGF-α, and BTC stimulation phosphorylated more sites than AR, EPR, and control groups (Figure 2). Specifically, EGF, HB-EGF, TGF-α and β-Cellulin stimulated phosphorylation of Tyr845, Tyr 998, Tyr1045, Tyr1086, Tyr1148, and Tyr1173 more efficiently than amphiregulin and epiregulin. Thr669, Tyr992 and Ser1046/47 showed similar phosphorylation levels upon stimulation with the different ligands (Figure 2).
Figure 2. Ligands trigger differential phosphorylation profile in the carboxy-terminal tails of EGFR.
Western blot analysis of whole-cell extracts from non stimulated cells (serum free) and treated with EGFR ligands (100ng/ml) for 10 minutes. The extracts were separated in 10% SDS-PAGE and immunoblotted with p-EGFR (T669), p-EGFR (Y845), p-EGFR (Y992), p-EGFR (Y998), p-EGFR (Y1045), p-EGFR (S1046/1047), p-EGFR (Y1086), p-EGFR (Y1148), p-EGFR (Y1173), EGFR, p-AKT (T308), AKT, p-Erk1/2 (T202/Y204), Erk1/2 antibodies. α-tubulin was used as loading control.
EGFR activation by phosphorylation leads to activation of several downstream intracellular signaling, extracellular-related kinase (ERK) and phosphoinositide 3-kinase (PI3K)-AKT pathways, responsible for a variety of effects [12]. We analyzed the role of EGFR ligands in the activation AKT, an important kinase for cell survival and motility [13]. EGF, HB-EGF, TGF-α, β-Cellulin in SkHep-1 cells showed greater levels of phospho-AKT than AR and EPR. With the exception of amphiregulin, EGFR ligands stimulation resulted in increased phosphorylation of p-ERK 1/2. Our data suggest that specific phosphorylation sites could be involved with the EGFR nuclear translocation. Together, these findings demonstrate that EGFR ligands that mediate nuclear EGFR translocation showed more phosphorylated markers than ligands that did not induce nuclear accumulation.
3.2. Effects of different ligands on SkHep-1 cell migration and proliferation
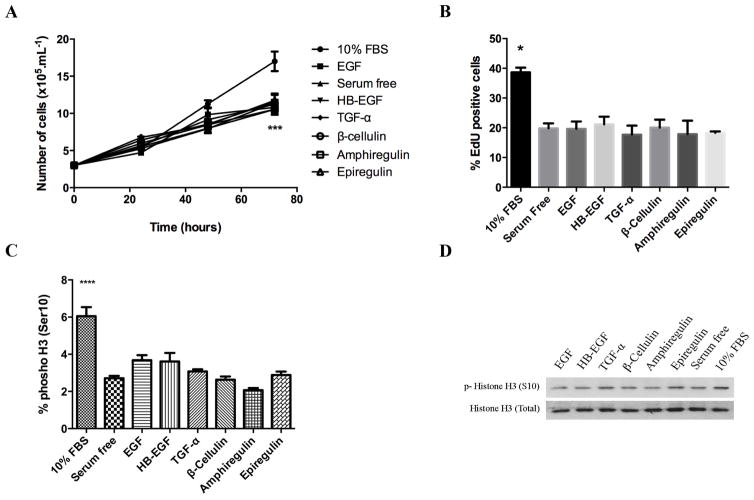
We next investigated the effects of the ligands on SKHep-1 cell migration and proliferation. The six EGFR ligands alone did not show induction of SkHep-1 cells proliferation (Figure 3A). Similar results were obtained using Cell Titer Blue reagent (data not shown). Furthermore, cell proliferation was assessed by EdU incorporation in a population of EGFR ligand-stimulated cells that showed no difference between stimulated and serum free treated cells (Figure 3B). We also tested the capacity of the ligands to induce the Histone H3 phosphorylation occurring at Ser10 that is tightly correlated with chromosome condensation and segregation during mitosis [14]. The fraction of cells in mitosis was determined by quantification of this marker by flow cytometry and Western blot (Figure 3C and D). Similarly, to the other proliferation analysis, the amount of phospho-H3 marker in ligand-stimulated cells was similar to the control group (serum free cells). Migration of untreated and cells treated with EGFR ligands were compared by wound healing assay. SkHep-1 cells were previously cultured in serum free medium to minimize the cell proliferation effects. Representative images were taken at time points of 0, 12, 24 and 36 hours of the identical locations (Figure 4A). Quantification of the results are shown in Figure 4B. A significant enhancement in wound closure was detected in cells exposed to EGF, HB-EGF, TGF-α and β-Cellulin compared with serum free cells (p <0.0001). In addition, epiregulin showed higher wound closure induction than serum free group (p<0.01). In contrast, amphiregulin failed to mimic the effect of others EGFR ligands on wound closure, showing no difference compared to control cells. Together, the results show that EGF, HB-EGF, TGF-α and β-Cellulin mediated migration, but not proliferation of SKHep-1 cells.
Figure 3. EGFR ligands are unable to trigger cell proliferation in SkHep-1.
Serum starved cells were stimulated with EGF, HB-EGF, TGF-α, β-Cellulin, amphiregulin and epiregulin (100ng/ml) for 24 hours A, growth curve of controls (serum free and 10%FBS) and cells treated with EGFR ligands. The cell count data were determined by counting cells on a standard grid-patterned hemocytometer every 24 hours. N=3, ***p<0.001. B, Quantification of EdU-positives numbers cells after 24 hour of EGFR ligands treatment. The average number of EdU-positive cells was quantified as a ratio of the total number of nucleus (hoechst stain) as described in materials and methods. N=3 *p<0.05. C, Quantification of SkHep-1 cells stained with Alexa Fluor 488-conjugated phosphorylated H3 (Ser10) antibody by flow cytometry. N=3, ****p<0.0001. D, western blot of whole cell lysates were probed against antibodies for phosphorylated histone H3 (top) and total histone H3 (bottom). Histone H3 was used as internal control.
Figure 4. EGF, HB-EGF, TGF-α, β-Cellulin and epiregulin induce cell migration while amphiregulin does not.
A, serum starved cells were wounded and then stimulated with EGFR ligands (100ng/ml). Representative phase-contrast images of the cell monolayer were acquired using IX70 inverted microscope (Olympus America) fitted with a 4X objective. The bar in the upper image indicates 200μm. B, Cell migration rates quantification. Wound areas were measured using ImageJ software, and the percentage wound closure was calculated and expressed related to serum free group (N=3, *p<0.05, **p<0.01, ****p<0.0001).
4. Discussion
Similar to hepatocyte growth factor receptor (HGFR, c-Met) and insulin receptor, EGF/EGFR complexes translocate to the nucleus [10, 11, 15]. Nuclear translocation of EGFR family promotes cell survival, and is associated with enhanced resistance to radiation, chemotherapy and anti-EGFR therapies [16]. Even so, little is known about which ligands and phosphorylation patterns modulate the nuclear translocation process. Here, we show that six different EGFR ligands differ in their effects on receptor phosphorylation sites, nuclear translocation and cell migration behavior.
The current work suggests that EGFR ligands, HB-EGF, TGF-α, β-Cellulin are involved in the EGFR nuclear translocation (Figure 1). Screening of phosphorylation sites showed a strong correlation between ligands promoting nuclear receptor accumulation and certain tyrosine-phosphorylation markers (Figure 2). Specific residues in the EGFR have been implicated in the regulation of its trafficking. For example, mutation of Tyr998 residue rendered receptors defective for endocytosis and interaction with AP-2, a clathrin-associated protein complex [17, 18]. Interestingly, clathrin knockdown is involved in EGFR nuclear translocation [10]. In HEp2 cells, EGFR ligands differentially affect EGFR endocytosis, recycling, and stimulation of the EGFR phosphorylation profile. EGF, β-Cellulin and HB-EGF are the most efficient at stimulating persistent EGFR phosphorylation. The low phosphorylation levels observed after TGF-α, amphiregulin, and epiregulin stimulation was due to limiting ligand concentrations [19].
Corroborating our data, in 32D/EGFR cells, EGF stimulates phosphorylation of EGFR Tyr1045 considerably more than amphiregulin. On the other hand, both ligands lead to similar phosphorylation of Tyr992 [4]. Amphiregulin also stimulated less phosphorylation than EGF, TGF-α or β-Cellulin at many sites on the EGF receptor from CHO cells, and correlate with smaller biological effects [20]. A link between EGFR activation and nuclear localization may be attributed to tyrosine phosphorylation sites; site-specific mutagenesis could identify key phosphorylation residues, and adaptor proteins involved directly in the nuclear translocation of EGF receptor.
Several of the ligands are co-expressed and found in increased concentrations in a large number of breast cancers, where they are involved in tumor progression [21]. Upon EGF stimulation, EGFR translocates to the nucleus where it binds to the proximal region of the cyclin D1 promoter, stimulating its expression, resulting in cell proliferation [22]. EGFR ligands (at concentration of 100ng/ml) did not affect SkHep-1 proliferation, showing no difference in number of cells, DNA synthesis, and mitotic markers compared to the control group (non-stimulated cells) (Figure 3). Specific signaling events in response to high EGF concentrations induce negative effects on proliferation in some cell models [23]. Moreover, in non-small cell lung cancer, EGFR activation by EGF resulted in cell migration stimulation but it did not promote cell proliferation [24]. Our results indicate that single ligand stimulation of serum starved SkHep-1 cells fails to trigger proliferation, while promoting migration. These effects are interesting to evaluate the migration by wound healing assay of these cells after stimulation of different EGFR ligands without the interference of cell proliferation.
In the present study, we have established a role for EGF, HB-EGF, TGF-α and β-Cellulin in promoting migration of SkHep-1, consistent with others cell models [25, 26]. In addiction, epiregulin induced in vitro wound closure, as reported in normal human epidermal keratinocytes and A431 cells [27]. Gefitinib (ZD1839, Iressa) specifically inhibits tyrosine kinase activity and downstream survival signals of EGFR [28]. Interestingly, elevated accumulation of EGFR was observed in the nucleus of gefitinib-resistant cell line and gefitinib treated cells [29, 30]. Furthermore, the EGFR phosphorylation at Ser229 by AKT is critical for EGFR nuclear translocation and gefitinib resistance [30]. In our study, AKT activity (pAKT) was elevated by all four ligands (EGF, HB-EGF, TGF-α and BTC) that mediate EGFR nuclear accumulation (Figure 1, 2). EGF/EGFR-driven cell migration is dependent on AKT activation [31], and the current findings positively correlate EGFR ligands and cell migration effects (Figure 4).
Nuclear expression of EGFR is correlated with poor prognosis in many cancer types, including breast and ovarian [32, 33]. Moreover, resistance of cancer to radiotherapy is frequently correlated with elevated EGFR expression and nuclear translocation [8, 16]. EGFR ligands are also correlated with clinical-pathological features. High expression of TGF-α and HB-EGF are associated with large tumor and diameter and high histoprognostic grading, while EGF and amphiregulin are associated with small tumor diameter and low histoprognostic grade [21]. Cancer patients with tumors expressing high levels of amphiregulin and epiregulin are more likely to have disease control, with cetuximab showing longer progression-free survival than patients with low expression [34]. Interestingly, EGFR ligands associated with better drug-responses are the ones that do not stimulate EGFR nuclear localization. It is possible that tumors expressing EGFR ligands that do not induce EGFR nuclear translocation will be more sensitive to EGFR targeted treatment. The current work highlights the differential roles of EGFR ligands in mediating nuclear receptor accumulation, corroborating the importance of nuclear signaling in cancer progression.
Highlights.
EGF, HB-EGF, TGF-α, β-Cellulin are involved in the EGFR nuclear translocation
Amphiregulin and epiregulin did not promote nuclear translocation of EGFR
EGF, HB-EGF, TGF-α and β-Cellulin have a role in SkHep-1 cells migration.
EGFR ligands associated with better prognosis don’t stimulate EGFR translocation.
Acknowledgments
The authors would like to thank Dr. Elio Anthony Cino for your advice and comments. This work was supported by NIH grant 1R03TW008709 and grants from CNPq, FAPEMIG and CAPES. The microscopic data shown in this work was obtained using the microscopes of the “Centro de Aquisição e Processamento de Imagens” (CAPI - ICB/UFMG).
Abbreviations
- HB-EGF
heparin-binding EGF-like growth factor
- EGF
epidermal growth factor
- TGF-α
transforming growth factor- α
- EGFR
epidermal growth factor receptor
Footnotes
Author contributions
J.A.Q.A.F. carried out experiments and wrote the manuscript. C.A designed and performed some experiments. A.M.G. provided reagents for this project. D.A.G. AND M.A.R. conceived and directed the project and participated in experiments.
Declarations of interest
We declare that there are no conflicts of interest.
Conflits of interest
All authors declare that there are no conflicts of interests regarding the publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Wilson KJ, Gilmore JL, Foley J, Lemmon MA, et al. Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol Ther. 2009;122:1–8. doi: 10.1016/j.pharmthera.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higashiyama S, Iwabuki H, Morimoto C, Hieda M, et al. Membrane-anchored growth factors, vthe epidermal growth factor family: beyond receptor ligands. Cancer Sci. 2008;99:214–220. doi: 10.1111/j.1349-7006.2007.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson KJ, Mill C, Lambert S, Buchman J, et al. EGFR ligands exhibit functional differences in models of paracrine and autocrine signaling. Growth Factors. 2012;30:107–116. doi: 10.3109/08977194.2011.649918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogiso H, Ishitani R, Nureki O, Fukai SM, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YN, Hung MC. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012;2:13. doi: 10.1186/2045-3701-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dittmann KH, Mayer C, Ohneseit PA, Raju U, et al. Celecoxib induced tumor cell radiosensitization by inhibiting radiation induced nuclear EGFR transport and DNA-repair: a COX-2 independent mechanism. Int J Radiat Oncol Biol Phys. 2008;70:203–212. doi: 10.1016/j.ijrobp.2007.08.065. [DOI] [PubMed] [Google Scholar]
- 9.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 10.De Angelis Campos AC, Rodrigues MA, de Andrade C, de Goes AM, et al. Epidermal growth factor receptors destined for the nucleus are internalized via a clathrin-dependent pathway. Biochem Biophys Res Commun. 2011;412:341–346. doi: 10.1016/j.bbrc.2011.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes DA, Rodrigues MA, Leite MF, Gomez MV, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008;283:4344–4351. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lill NL, Sever NI. Where EGF receptors transmit their signals. Sci Signal. 2012;5:pe41. doi: 10.1126/scisignal.2003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue G, Hemmings BA. PKB/Akt-dependent regulation of cell motility. J Natl Cancer Inst. 2013;105:393–404. doi: 10.1093/jnci/djs648. [DOI] [PubMed] [Google Scholar]
- 14.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues MA, Gomes DA, Andrade VA, Leite MF, et al. Insulin induces calcium signals in the nucleus of rat hepatocytes. Hepatology. 2008;48:1621–1631. doi: 10.1002/hep.22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brand TM, Iida M, Luthar N, Starr MM, et al. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108:370–377. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 17.Sorkin A, Mazzotti M, Sorkina T, Scotto L, et al. Epidermal growth factor receptor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J Biol Chem. 1996;271:13377–13384. doi: 10.1074/jbc.271.23.13377. [DOI] [PubMed] [Google Scholar]
- 18.Tong J, Taylor P, Peterman SM, Prakash A, et al. Epidermal growth factor receptor phosphorylation sites Ser991 and Tyr998 are implicated in the regulation of receptor endocytosis and phosphorylations at Ser1039 and Thr1041. Mol Cell Proteomics. 2009;8:2131–2144. doi: 10.1074/mcp.M900148-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, et al. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 2009;10:1115–1127. doi: 10.1111/j.1600-0854.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macdonald-Obermann JL, Pike LJ. Different epidermal growth factor (EGF) receptor ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. J Biol Chem. 2014;289:26178–26188. doi: 10.1074/jbc.M114.586826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revillion F, Lhotellier V, Hornez L, Bonneterre J, et al. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Ann Oncol. 2008;19:73–80. doi: 10.1093/annonc/mdm431. [DOI] [PubMed] [Google Scholar]
- 22.Lin SY, Makino K, Xia W, Matin A, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 23.Krall JA, Beyer EM, MacBeath G. High- and low-affinity epidermal growth factor receptor-ligand interactions activate distinct signaling pathways. PLoS One. 2011;6:e15945. doi: 10.1371/journal.pone.0015945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauand C, Rezende-Teixeira P, Cortez BA, Niero EL, et al. Independent of ErbB1 gene copy number, EGF stimulates migration but is not associated with cell proliferation in non-small cell lung cancer. Cancer Cell Int. 2013;13:38. doi: 10.1186/1475-2867-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson JL, Phelps ED, Doll MA, Schaal S, et al. The role of endogenous epidermal growth factor receptor ligands in mediating corneal epithelial homeostasis. Invest Ophthalmol Vis Sci. 2014;55:2870–2880. doi: 10.1167/iovs.13-12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrientos S, Stojadinovic O, Golinko MS, Brem H, et al. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 27.Draper BK, Komurasaki T, Davidson MK, Nanney LB. Epiregulin is more potent than EGF or TGFalpha in promoting in vitro wound closure due to enhanced ERK/MAPK activation. J Cell Biochem. 2003;89:1126–1137. doi: 10.1002/jcb.10584. [DOI] [PubMed] [Google Scholar]
- 28.Penne K, Bohlin C, Schneider S, Allen D. Gefitinib (Iressa, ZD1839) and tyrosine kinase inhibitors: the wave of the future in cancer therapy. Cancer Nurs. 2005;28:481–486. doi: 10.1097/00002820-200511000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura Y, Yoshioka K, Bereczky B, Itoh K. Evidence for efficient phosphorylation of EGFR and rapid endocytosis of phosphorylated EGFR via the early/late endocytic pathway in a gefitinib-sensitive non-small cell lung cancer cell line. Mol Cancer. 2008;7:42. doi: 10.1186/1476-4598-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan Y, Shi C, Inge L, Hibner M, et al. Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene. 2010;29:4947–4958. doi: 10.1038/onc.2010.240. [DOI] [PubMed] [Google Scholar]
- 32.Lo HW, Xia W, Wei Y, Ali-Seyed M, et al. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 33.Xia W, Wei Y, Du Y, Liu J, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]