Abstract
Background
Neurological dysfunction after traumatic brain injury (TBI) poses short-term or long-lasting health issues for family members and health care providers. Presently there are no approved medicines to treat TBI. Epidemiological evidence suggests that TBI may cause neurodegenerative disease later in life. In an effort to illuminate target cellular processes for drug development, we examined the effects of a mild TBI on hippocampal gene expression in mouse.
Methods
mTBI was induced in a closed head, weight drop-system in mice (ICR). Animals were anesthetized and subjected to mTBI (30 g). Fourteen days after injury the ipsilateral hippocampus was utilized for cDNA gene array studies. mTBI animals were compared with sham-operated animals. Genes regulated by TBI were identified to define TBI-induced physiological/pathological processes. mTBI regulated genes were divided into functional groupings to provide gene ontologies. Genes were further divided to identify molecular/cellular pathways regulated by mTBI.
Results
Numerous genes were regulated after a single mTBI event that mapped to many ontologies and molecular pathways related to inflammation and neurological physiology/pathology, including neurodegenerative disease.
Conclusions
These data illustrate diverse transcriptional changes in hippocampal tissues triggered by a single mild injury. The systematic analysis of individual genes that lead to the identification of functional categories, such as gene ontologies and then molecular pathways, illustrate target processes of relevance to TBI pathology. These processes may be further dissected to identify key factors that can be evaluated at the protein level to highlight possible treatments for TBI in human disease and potential biomarkers of neurodegenerative processes.
Keywords: mild traumatic brain injury (mTBI), gene expression, gene ontology, molecular pathway, inflammation, neurological, neurodegeneration, dementia
1. Introduction
Traumatic brain injury (TBI) has become a highly prevalent medical cause for concern, an insult soon to exceed traditional ailments as a main cause of death in the United States (Faul et al., 2010). The induction of TBIs may occur via several mechanisms, such as a concussive event or a high pressure shockwave generated by an explosion (Danshevar et al., 2015). Clinically, TBI can be categorized into several classes such as mild, moderate and severe. These classifications are based upon criteria related to several indices, such as the Glasgow Coma Scale, length of post-traumatic amnesia, results of neuroimaging and whether or not the insult resulted from an open or closed skull injury (Gómez et al., 2014). Clinical cases of TBI in the civilian world are predominantly categorized as ‘mild’ and concussive in nature, requiring a visit to a hospital emergency department followed by discharge (Korley et al., 2015). In addition to the immediate health related effects of TBI on patients, recent epidemiological studies suggest that there is an association between diverse forms of concussive TBI and the subsequent development of neurodegenerative dementia-related illness in later life (Barnes et al., 2014; Danshevar et al., 2015; Gupta and Sen 2015).
Currently, there is no proven effective drug therapy available for the treatment of any form of TBI (Stein et al., 2015; Xiong et al., 2015). This leaves a significant treatment gap for TBI which requires urgent attention from the scientific and medical community. With the multiple types of clinical TBI, no simple animal model best represents clinical injury (Marklund and Hillered 2011). Appropriate models need to be developed to aid in the evaluation of candidate therapies, including the use of novel or repurposed medicines.
Many events triggered by TBI have been described (Choi et al., 1987; Maas et al., 2008; Greve and Zink, 2009; Stoica and Faden, 2010; Barkhoudarian et al., 2011; Cornelius et al., 2013), still further evaluation and description of the molecular events related to the pathology of TBI require study. One powerful approach to address this knowledge gap includes one of a mass screening of central nervous system (CNS) molecular markers to provide insight into the molecular and cellular processes driving the pathology of TBI. The use of large-scale gene array chips and powerful bioinformatics tools to examine an organ or a tissue’s transcriptomic profile has opened up an avenue to perform such studies. The identification of molecular pathways initiated by TBI provides a platform for the systematic evaluation of known and novel therapies that may ameliorate or slow the progression of TBI-induced cognitive or neurological disorders.
As mild TBIs may be the more common form of injury in the clinic, we have opted to study molecular events triggered by a well-characterized mild closed head weight drop model of TBI in the mouse. This model, involving a 30 - 40 g mouse concussed with a 30 g weight, bears face validity to the human condition involving the clash of heads between two similar sized adults in a sports injury. The study described here illustrates the effects of a mild TBI on mouse hippocampal tissue gene expressions 14 days after the initial injury; however, the same methodologies can be applied to other models of TBI, animal species and times post injury. Through the use of sensitive, large-scale gene array chips we identified large numbers of subtle gene regulations driven by a single mTBI event. Additional bioinformatics analysis of the regulated genes indicated that TBI was associated with numerous inflammatory and neurological physiological/pathological processes that provide a basis for targeted drug development programs.
2. Materials and Methods
2.1. Animal Studies - housing and induction of mTBI
ICR mice (Institute for Cancer Research (ICR)) were housed five per cage under a constant 12-h light/dark cycle, at room temperature (23°C). Food (Purina rodent chow) and water were available ad libitum. Each animal was used for one experiment only. The Ethics Committee of the Sackler Faculty of Medicine approved the experimental protocol (M-12-063), in compliance with the guidelines for animal experimentation of the National Institutes of Health (DHEW publication 85–23, revised, 1995). A minimal number of mice were used for the study and all efforts were made to minimize suffering. mTBI was induced as has been described previously (Zohar et al., 2003; Milman et al., 2005; Edut et al., 2011; Baratz et al., 2011), mice (30 - 40 g) were fully anesthetized by exposure to Isoflurane. After full anesthesia was achieved, the animals were placed under the opening of a weight drop device and a weight (30 g) dropped from a height of 80 cm. Immediately thereafter, the animals were placed in a recovery cage and were observed until full recovery from anesthesia occurred and they could be returned to their home cages. For the sham procedure, animals were anesthetized and placed under the weight drop device; however, no weight was dropped. In the present study the group sizes were as follows: sham n=5 and for mTBI n=4.
2.2. Hippocampal cDNA gene array hybridization
Fourteen days after the induction of mTBI, mice were euthanized and the ipsilateral hippocampus extracted for use in cDNA gene array studies. The entire hippocampus was used to prepare RNA and the Qiagen RNeasy Mini Kit used to prepare total RNA using the manufacturer’s specifications (Qiagen, Inc. Valencia CA). The Agilent 2100 Bioanalyzer with RNA 6000 Nano Chips Quantity was used to determine the quality and quantity of the RNA. Biotin-labeled, amplified (aRNA) was created by using the Illumina TotalPrep RNA Amplification Kit (Ambion; Austin, TX, cat # IL1791). A total of 750 ng of aRNA was hybridized at 58°C for 16 hours to Illumina’s SentrixMouse Ref-8, v2 Expression BeadChips (Illumina, San Diego, CA). The arrays were washed and then blocked, after which the biotin-labeled probe was detected by staining with streptavidin-Cy3. Arrays were scanned at a resolution of 0.8 μm using Beadstation 500 X from Illumina, and data intensity extracted from the array image using Illumina BeadStudio software, V3.
2.3.1. Bioinformatic analysis of array data - regulated genes
Hippocampal gene expression profiles were compared between mTBI and sham animal samples, as has been described previously (Tweedie et al., 2013a,b; 2015). Raw array chip hybridization image signals were filtered and processed to generate normalized data that was then transformed to create Z-scores for each gene. Z-score transformed data was then utilized to generate a Z-ratio measurement, which allowed for statistical analysis of the gene expression data sets. Significantly regulated genes were selected by the following criteria: 1) gene expression changes had a z-test value of ≤ 0.05 vs. sham; 2) the absolute value of Z-ratio was calculated to be ≥ 1.5 vs. sham; 3) the False Discovery Rate for the genes was ≤ 0.30; 4) the average Z-score over all sample comparisons were not negative and lastly; 5) an one way independent ANOVA test p-value cut off was ≤ 0.05. Only genes that displayed consistent significant expression changes in all samples from a given treatment group were considered for further statistical analysis. A list of mTBI regulated genes are described in Table 1, with the following information provided: Accession number, gene symbol, fold-change in gene expression compared to sham levels, the statistically significant p-value, and the Z-ratio. Raw and Z-score normalized gene expression data sets are accessible online in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) as SuperSeries GSE71850, with individual accession number GSE44625.
Table 1.
mTBI regulated genes identified in hippocampal tissues 14 days after injury
| Up-regulated genes | ||||
|---|---|---|---|---|
| Accesion number | Symbol | Fold Change | Z-ratio | p value |
| NM_178198.1 | Hist1h2bj | 1.56 | 4.13 | 0.0006 |
| NM_145216.3 | Rasl10a | 1.51 | 4.09 | 0.0262 |
| NM_178200.1 | Hist1h2bm | 1.59 | 4.07 | 0.0023 |
| NM_008218.2 | Hba-a1 | 1.42 | 4.03 | 0.0100 |
| NM_053090 | Drctnnb1a | 1.45 | 3.58 | 0.0371 |
| NM_008416.1 | Junb | 1.48 | 3.53 | 0.0001 |
| NM_013603.1 | Mt3 | 1.34 | 3.49 | 0.0294 |
| NM_025396.3 | Pgls | 1.42 | 3.18 | 0.0007 |
| NM_025963.1 | Rps10 | 1.33 | 3.16 | 0.0000 |
| NM_029362.3 | Chmp4b | 1.36 | 3.16 | 0.0187 |
| NM_026938.1 | Tmem160 | 1.35 | 3.12 | 0.0173 |
| NM_173071.2 | Bai2 | 1.43 | 3.06 | 0.0039 |
| NM_026161.3 | C1qtnf4 | 1.34 | 3.05 | 0.0112 |
| NM_183086.1 | Mrps10 | 1.43 | 3.00 | 0.0018 |
| XM_126580.1 | Thra | 1.33 | 2.92 | 0.0158 |
| NM_001003949.3 | ORF61 | 1.38 | 2.91 | 0.0061 |
| NM_020255.2 | Scand1 | 1.35 | 2.90 | 0.0182 |
| NM_009696.2 | Apoe | 1.27 | 2.90 | 0.0044 |
| NM_007457.2 | Ap1s1 | 1.34 | 2.86 | 0.0035 |
| NM_175329.3 | Ndg2 | 1.24 | 2.85 | 0.0060 |
| NM_007590.3 | Calm3 | 1.22 | 2.79 | 0.0294 |
| NM_009942 | Cox5b | 1.22 | 2.76 | 0.0290 |
| NM_026398.2 | Pop5 | 1.29 | 2.70 | 0.0074 |
| NM_031161.2 | Cck | 1.25 | 2.69 | 0.0308 |
| NM_010261.2 | Rabac1 | 1.27 | 2.66 | 0.0238 |
| NM_026938.1 | Tmem160 | 1.31 | 2.59 | 0.0064 |
| NM_029361.2 | Wnk2 | 1.35 | 2.54 | 0.0187 |
| NM_001042487.1 | Dlgap4 | 1.29 | 2.51 | 0.0028 |
| NM_027231.1 | Polr2f | 1.29 | 2.51 | 0.0190 |
| NM_028659.2 | Eif3k | 1.33 | 2.50 | 0.0153 |
| NM_009082.2 | Rpl29 | 1.29 | 2.48 | 0.0000 |
| NM_011218.1 | Ptprs | 1.28 | 2.45 | 0.0004 |
| NM_010261.1 | Rabac1 | 1.22 | 2.45 | 0.0122 |
| NM_009077.2 | Rpl18 | 1.23 | 2.40 | 0.0169 |
| NM_025313.1 | Atp5d | 1.24 | 2.40 | 0.0071 |
| NM_026302.3 | Dctn4 | 1.24 | 2.39 | 0.0164 |
| NM_023172.3 | Ndufb9 | 1.34 | 2.38 | 0.0280 |
| NM_001002267.2 | Tmem158 | 1.32 | 2.38 | 0.0127 |
| NM_009849.1 | Entpd2 | 1.32 | 2.35 | 0.0003 |
| NM_025407.2 | Uqcrc1 | 1.22 | 2.35 | 0.0102 |
| NM_177407.3 | Camk2a | 1.21 | 2.34 | 0.0357 |
| NM_029365.1 | Med25 | 1.24 | 2.33 | 0.0247 |
| NM_026542.1 | Slc25a39 | 1.30 | 2.31 | 0.0004 |
| NM_020024.3 | Taf10 | 1.27 | 2.30 | 0.0117 |
| NM_001017426.1 | Jmjd3 | 1.32 | 2.29 | 0.0001 |
| NM_021604.2 | Agrn | 1.21 | 2.29 | 0.0382 |
| NM_080855.1 | Zcchc14 | 1.29 | 2.28 | 0.0042 |
| NM_009272.2 | Srm | 1.26 | 2.25 | 0.0106 |
| NM_029887.2 | Yif1b | 1.26 | 2.21 | 0.0036 |
| NM_013923.2 | Rnf19a | 1.23 | 2.19 | 0.0014 |
| NM_011722.2 | Dctn6 | 1.24 | 2.17 | 0.0059 |
| NM_025424.1 | Nenf | 1.23 | 2.17 | 0.0322 |
| NM_175177.3 | Bdh1 | 1.27 | 2.16 | 0.0023 |
| NM_001080385.1 | Clta | 1.20 | 2.16 | 0.0260 |
| NM_013842.2 | Xbp1 | 1.22 | 2.16 | 0.0000 |
| NM_007393.3 | Actb | 1.16 | 2.15 | 0.0182 |
| NM_026644.1 | Agpat4 | 1.24 | 2.15 | 0.0000 |
| NM_178600.2 | Vkorc1 | 1.26 | 2.15 | 0.0237 |
| NM_010047.1 | Dgcr6 | 1.26 | 2.10 | 0.0014 |
| NR_003292.1 | Zxda | 1.28 | 2.10 | 0.0089 |
| NM_175665.1 | Hist1h2bk | 1.28 | 2.09 | 0.0297 |
| NM_025592.3 | Rpl35 | 1.15 | 2.08 | 0.0335 |
| NM_016679.3 | Keap1 | 1.25 | 2.07 | 0.0266 |
| NM_001033212.1 | Rprml | 1.20 | 2.05 | 0.0172 |
| NM_007457.2 | Ap1s1 | 1.25 | 2.02 | 0.0050 |
| NM_078479.2 | Mrps21 | 1.22 | 2.02 | 0.0070 |
| NM_025789.4 | Rshl2a | 1.27 | 2.02 | 0.0000 |
| NM_025344.1 | Eif3f | 1.14 | 2.00 | 0.0126 |
| NM_028965.3 | Snx11 | 1.21 | 1.99 | 0.0000 |
| NM_011073.2 | Prf1 | 1.26 | 1.99 | 0.0235 |
| NM_024227.2 | Mrpl28 | 1.20 | 1.98 | 0.0090 |
| NM_007747.2 | Cox5a | 1.15 | 1.96 | 0.0048 |
| NM_029083.1 | Ddit4 | 1.23 | 1.94 | 0.0001 |
| NM_023130.2 | Raly | 1.23 | 1.94 | 0.0117 |
| NM_011846.4 | Mmp17 | 1.17 | 1.93 | 0.0018 |
| NM_011698.1 | Lin7b | 1.19 | 1.92 | 0.0004 |
| NM_023260.1 | Mrps34 | 1.18 | 1.90 | 0.0198 |
| NM_018772.1 | Bri3 | 1.16 | 1.88 | 0.0015 |
| NM_146183.1 | Zfp428 | 1.24 | 1.88 | 0.0020 |
| NM_009976.3 | Cst3 | 1.10 | 1.87 | 0.0307 |
| NM_016707.2 | Bcl11a | 1.17 | 1.87 | 0.0316 |
| NM_009941.1 | Cox4i1 | 1.11 | 1.87 | 0.0345 |
| NM_134002.1 | Csnk1g2 | 1.15 | 1.87 | 0.0261 |
| NM_019568.1 | Cxcl14 | 1.25 | 1.86 | 0.0016 |
| NM_001081030.1 | Sbf1 | 1.18 | 1.86 | 0.0260 |
| NM_016905.2 | Galk1 | 1.26 | 1.85 | 0.0001 |
| NM_011865.3 | Pcbp1 | 1.17 | 1.85 | 0.0158 |
| NM_015807.1 | Nt5c | 1.22 | 1.85 | 0.0002 |
| NM_009272.4 | Srm | 1.20 | 1.83 | 0.0362 |
| NM_133976.1 | Imp3 | 1.18 | 1.83 | 0.0235 |
| NM_144900.1 | Atp1a1 | 1.13 | 1.82 | 0.0264 |
| NM_138596.1 | Med10 | 1.22 | 1.82 | 0.0044 |
| NM_011949.3 | Mapk1 | 1.16 | 1.82 | 0.0317 |
| NM_025587.2 | Rps21 | 1.11 | 1.81 | 0.0296 |
| NM_015816.1 | Lsm4 | 1.17 | 1.80 | 0.0188 |
| NM_026570.1 | Yeats4 | 1.23 | 1.80 | 0.0021 |
| NM_001037741.2 | Gpx4 | 1.19 | 1.79 | 0.0001 |
| NM_019879.1 | Suclg1 | 1.14 | 1.79 | 0.0001 |
| NM_008019.2 | Fkbp1a | 1.14 | 1.78 | 0.0310 |
| NM_007510.2 | Atp6v1e1 | 1.10 | 1.78 | 0.0372 |
| NM_011647.2 | Tsc2 | 1.18 | 1.77 | 0.0233 |
| NM_026552.2 | Arpc4 | 1.19 | 1.76 | 0.0022 |
| NM_029153.1 | Scamp1 | 1.12 | 1.76 | 0.0021 |
| NM_029272.3 | Ndufs7 | 1.14 | 1.74 | 0.0007 |
| NM_023138.3 | Map2k2 | 1.17 | 1.74 | 0.0167 |
| NM_011375.2 | St3gal5 | 1.19 | 1.74 | 0.0250 |
| NM_020569.1 | Park7 | 1.12 | 1.74 | 0.0230 |
| NM_175102.3 | Sf3b5 | 1.18 | 1.72 | 0.0335 |
| NM_010906.2 | Nfix | 1.15 | 1.72 | 0.0349 |
| NM_013892.2 | Pcsk1n | 1.17 | 1.71 | 0.0314 |
| NM_007457.2 | Ap1s1 | 1.19 | 1.70 | 0.0058 |
| NM_026373.3 | Cdk2ap2 | 1.22 | 1.70 | 0.0108 |
| NM_008338.2 | Ifngr2 | 1.22 | 1.70 | 0.0000 |
| NM_028774.2 | Rnf6 | 1.14 | 1.69 | 0.0355 |
| NM_008945.2 | Psmb4 | 1.11 | 1.66 | 0.0285 |
| NM_026729.1 | Ict1 | 1.22 | 1.66 | 0.0279 |
| NM_133224.1 | Atp13a1 | 1.19 | 1.65 | 0.0105 |
| NM_026744.3 | Mrpl53 | 1.15 | 1.65 | 0.0316 |
| NM_010047.3 | Dgcr6 | 1.19 | 1.64 | 0.0269 |
| NM_027215.2 | Tmem147 | 1.13 | 1.63 | 0.0215 |
| NM_007682.2 | Cenpb | 1.15 | 1.63 | 0.0058 |
| NM_008801.2 | Pde6d | 1.18 | 1.63 | 0.0185 |
| NM_013758.2 | Add3 | 1.17 | 1.63 | 0.0007 |
| NM_013680.3 | Syn1 | 1.12 | 1.62 | 0.0263 |
| NM_025516.2 | Ergic3 | 1.16 | 1.60 | 0.0031 |
| NM_025573.3 | Sfrs9 | 1.16 | 1.60 | 0.0092 |
| NM_019722.3 | Arl2 | 1.19 | 1.59 | 0.0078 |
| NM_016894.2 | Ramp1 | 1.16 | 1.58 | 0.0140 |
| NM_145941.2 | Eif4g1 | 1.20 | 1.58 | 0.0272 |
| NM_009672.2 | Anp32a | 1.12 | 1.57 | 0.0071 |
| NM_009221.2 | Snca | 1.07 | 1.56 | 0.0122 |
| NM_144509.1 | Arl6ip4 | 1.13 | 1.56 | 0.0054 |
| NM_024218.2 | Rpl24 | 1.13 | 1.55 | 0.0385 |
| NM_013750.1 | Phlda3 | 1.21 | 1.54 | 0.0007 |
| NM_153529.1 | Nrn1 | 1.10 | 1.53 | 0.0352 |
| NM_025337.2 | Akr7a5 | 1.14 | 1.53 | 0.0353 |
| NM_019390.1 | Lmna | 1.17 | 1.53 | 0.0312 |
| NM_010364.1 | Gtf2h4 | 1.20 | 1.52 | 0.0015 |
| NM_145385.1 | Mlf2 | 1.08 | 1.52 | 0.0061 |
| NM_024177.3 | Mrpl38 | 1.14 | 1.52 | 0.0229 |
| NM_021713.2 | Myg1 | 1.20 | 1.51 | 0.0007 |
| NM_026697.3 | Rab14 | 1.11 | 1.50 | 0.0248 |
| NM_026015.2 | Zmat5 | 1.21 | 1.50 | 0.0177 |
| Down-regulated genes | ||||
| Accesion number | Symbol | Fold Change | Z-ratio | p value |
| NM_010072.2 | Dpm1 | -1.16 | -1.50 | 0.0241 |
| NM_016784.3 | Plrg1 | -1.23 | -1.51 | 0.0239 |
| NM_173374.3 | Sfrs1 | -1.25 | -1.51 | 0.0071 |
| NM_172700.2 | Zmpste24 | -1.15 | -1.51 | 0.0026 |
| NM_011622.2 | Tom1 | -1.15 | -1.52 | 0.0008 |
| NM_026932.3 | Ebna1bp2 | -1.16 | -1.52 | 0.0014 |
| NM_030559.2 | Vps16 | -1.18 | -1.52 | 0.0064 |
| NM_011807.2 | Dlg2 | -1.21 | -1.53 | 0.0153 |
| NM_020618.3 | Smarce1 | -1.17 | -1.53 | 0.0048 |
| NM_019758.2 | Mtch2 | -1.18 | -1.53 | 0.0154 |
| NM_010830.1 | Msh6 | -1.18 | -1.54 | 0.0224 |
| NM_175294.2 | Nucks1 | -1.16 | -1.55 | 0.0300 |
| NM_009963.3 | Cry2 | -1.15 | -1.55 | 0.0143 |
| NM_027678.2 | Zranb3 | -1.16 | -1.56 | 0.0110 |
| NM_173406.2 | Jazf1 | -1.16 | -1.56 | 0.0178 |
| NM_026536.1 | Atp5s | -1.18 | -1.57 | 0.0237 |
| NM_178051.3 | Mterfd2 | -1.15 | -1.57 | 0.0016 |
| NM_001003971.1 | Senp7 | -1.18 | -1.58 | 0.0042 |
| NM_023374.3 | Sdhb | -1.21 | -1.58 | 0.0016 |
| NM_183028.3 | Pcmtd1 | -1.16 | -1.59 | 0.0243 |
| NM_080557.2 | Snx4 | -1.17 | -1.60 | 0.0230 |
| NM_026434.3 | Rbm18 | -1.19 | -1.60 | 0.0021 |
| NM_029738.2 | Cluap1 | -1.15 | -1.60 | 0.0001 |
| NM_031156.2 | Ide | -1.18 | -1.61 | 0.0003 |
| NM_028245.1 | Zfp131 | -1.22 | -1.61 | 0.0110 |
| NM_019734.1 | Asah1 | -1.15 | -1.62 | 0.0172 |
| NM_181070.4 | Rab18 | -1.21 | -1.62 | 0.0163 |
| NM_021510.2 | Hnrph1 | -1.23 | -1.63 | 0.0000 |
| NM_016682.2 | Uba2 | -1.21 | -1.63 | 0.0275 |
| NM_016883.3 | Psmd10 | -1.16 | -1.64 | 0.0003 |
| NM_153091.2 | St7l | -1.18 | -1.65 | 0.0174 |
| NM_025936.1 | Rars | -1.19 | -1.65 | 0.0047 |
| NM_028209.1 | Ttc4 | -1.16 | -1.65 | 0.0006 |
| NM_199448.1 | Fez2 | -1.20 | -1.66 | 0.0228 |
| NM_008917.2 | Ppt1 | -1.18 | -1.66 | 0.0006 |
| NM_001040130.1 | Tmem141 | -1.16 | -1.67 | 0.0002 |
| NM_025918.3 | Ccdc43 | -1.19 | -1.67 | 0.0144 |
| NM_007422.2 | Adss | -1.20 | -1.67 | 0.0002 |
| NM_138314.2 | Nme7 | -1.17 | -1.68 | 0.0063 |
| NM_008974.3 | Ptp4a2 | -1.19 | -1.68 | 0.0103 |
| NM_175548.3 | Lsamp | -1.19 | -1.68 | 0.0182 |
| NM_011871.2 | Prkra | -1.19 | -1.69 | 0.0248 |
| NM_008786.1 | Pcmt1 | -1.22 | -1.69 | 0.0230 |
| NM_016918.2 | Nudt5 | -1.18 | -1.69 | 0.0237 |
| NM_207302.1 | Zranb1 | -1.18 | -1.70 | 0.0221 |
| NM_007958.1 | Smarcad1 | -1.18 | -1.70 | 0.0294 |
| NM_144911.1 | Rpap2 | -1.18 | -1.71 | 0.0304 |
| NM_009206.1 | Slc4a1ap | -1.19 | -1.72 | 0.0014 |
| NM_028521.2 | Phospho2 | -1.21 | -1.72 | 0.0278 |
| NM_016716.4 | Cul3 | -1.26 | -1.73 | 0.0000 |
| NM_011806.2 | Dmtf1 | -1.22 | -1.73 | 0.0062 |
| NM_177128.3 | Iqcb1 | -1.21 | -1.73 | 0.0000 |
| NM_025985.4 | Ube2g1 | -1.21 | -1.74 | 0.0097 |
| NM_010298.5 | Glrb | -1.18 | -1.75 | 0.0385 |
| NM_029949.3 | Snapc3 | -1.19 | -1.77 | 0.0007 |
| NM_145507.1 | Dars | -1.25 | -1.78 | 0.0085 |
| NM_028173.3 | Tram1 | -1.19 | -1.79 | 0.0267 |
| NM_178705.5 | Luzp2 | -1.22 | -1.79 | 0.0131 |
| NM_054102.2 | Ivns1abp | -1.22 | -1.79 | 0.0001 |
| NM_019749.3 | Gabarap | -1.29 | -1.80 | 0.0306 |
| NM_019665.2 | Arl6 | -1.23 | -1.81 | 0.0105 |
| NM_026902.3 | Mcts1 | -1.28 | -1.81 | 0.0305 |
| NM_019445.1 | Fmn2 | -1.20 | -1.81 | 0.0093 |
| NM_011546.2 | Zeb1 | -1.23 | -1.81 | 0.0154 |
| NM_139308.1 | Stard7 | -1.25 | -1.81 | 0.0149 |
| NM_019710.1 | Smc1a | -1.21 | -1.82 | 0.0050 |
| NM_025844.2 | Chordc1 | -1.23 | -1.82 | 0.0217 |
| NM_010303.2 | Gna13 | -1.25 | -1.82 | 0.0066 |
| NM_019697.3 | Kcnd2 | -1.21 | -1.83 | 0.0263 |
| NM_008622.3 | Mpv17 | -1.23 | -1.83 | 0.0156 |
| NM_172476.4 | Tmc7 | -1.22 | -1.83 | 0.0142 |
| NM_019683.3 | Ankrd49 | -1.22 | -1.83 | 0.0112 |
| NM_178726.3 | Ppm1l | -1.24 | -1.84 | 0.0015 |
| NM_028242.1 | Htatsf1 | -1.29 | -1.84 | 0.0075 |
| NM_008825.3 | Pfkfb2 | -1.24 | -1.84 | 0.0275 |
| NM_007840.3 | Ddx5 | -1.19 | -1.85 | 0.0353 |
| NM_009981.3 | Pcyt1a | -1.20 | -1.85 | 0.0068 |
| NM_145600.1 | Zfp330 | -1.20 | -1.85 | 0.0084 |
| NM_026647.2 | Zdhhc21 | -1.23 | -1.85 | 0.0000 |
| NM_146193.2 | Btbd1 | -1.28 | -1.86 | 0.0165 |
| NM_146123.2 | Cacnb4 | -1.22 | -1.87 | 0.0294 |
| NM_022885.2 | Slc30a5 | -1.25 | -1.88 | 0.0284 |
| NM_027901.2 | Gtf3c2 | -1.22 | -1.89 | 0.0102 |
| NM_013758.2 | Add3 | -1.21 | -1.90 | 0.0084 |
| NM_139061.3 | Vps54 | -1.23 | -1.90 | 0.0234 |
| NM_026454.3 | Ube2f | -1.25 | -1.92 | 0.0187 |
| NM_020601.2 | Tbl1x | -1.23 | -1.92 | 0.0282 |
| NM_008994.2 | Pex2 | -1.25 | -1.92 | 0.0366 |
| NM_009136.3 | Scrg1 | -1.26 | -1.94 | 0.0145 |
| NM_178846.1 | Gnl3 | -1.24 | -1.94 | 0.0005 |
| NM_011966.3 | Psma4 | -1.30 | -1.95 | 0.0206 |
| NM_022309.3 | Cbfb | -1.25 | -1.95 | 0.0115 |
| NM_029735.1 | Eprs | -1.20 | -1.95 | 0.0010 |
| NM_021389.3 | Sh3kbp1 | -1.23 | -1.95 | 0.0112 |
| NM_022309.2 | Cbfb | -1.25 | -1.95 | 0.0000 |
| NM_016883.3 | Psmd10 | -1.24 | -1.96 | 0.0016 |
| NM_026176.2 | Pdcl | -1.21 | -1.96 | 0.0057 |
| NM_025647.2 | Cmpk | -1.26 | -1.97 | 0.0110 |
| NM_010472.2 | Hrb | -1.26 | -1.97 | 0.0100 |
| NM_011959.2 | Orc5l | -1.21 | -1.97 | 0.0004 |
| NM_178794.3 | Zrsr2 | -1.25 | -1.99 | 0.0072 |
| NM_021510.2 | Hnrph1 | -1.26 | -2.00 | 0.0331 |
| NM_011278.3 | Rnf4 | -1.22 | -2.00 | 0.0039 |
| NM_175328.2 | Slc6a15 | -1.26 | -2.04 | 0.0056 |
| NM_008133.3 | Glud1 | -1.26 | -2.05 | 0.0074 |
| NM_001077707.1 | Shprh | -1.23 | -2.06 | 0.0252 |
| NM_173374.3 | Sfrs1 | -1.30 | -2.06 | 0.0000 |
| NM_026345.2 | Mansc1 | -1.23 | -2.06 | 0.0029 |
| NM_029826.2 | Hdhd2 | -1.23 | -2.08 | 0.0043 |
| NM_010852.1 | Myef2 | -1.24 | -2.09 | 0.0177 |
| NM_009162.3 | Scg5 | -1.38 | -2.09 | 0.0009 |
| NM_021535.3 | Smu1 | -1.25 | -2.10 | 0.0004 |
| NM_019942.2 | Septin 6 | -1.26 | -2.10 | 0.0170 |
| NM_134255.2 | Elovl5 | -1.31 | -2.10 | 0.0121 |
| NM_133795.1 | Ttc1 | -1.26 | -2.12 | 0.0015 |
| NM_011890.4 | Sgcb | -1.27 | -2.12 | 0.0231 |
| NM_145510.1 | Rabif | -1.23 | -2.12 | 0.0042 |
| NM_033037.3 | Cdo1 | -1.25 | -2.14 | 0.0014 |
| NM_027810.2 | Bbs7 | -1.24 | -2.15 | 0.0086 |
| NM_009261.2 | Strbp | -1.26 | -2.15 | 0.0035 |
| NM_026213.3 | Ttc33 | -1.22 | -2.15 | 0.0050 |
| NM_026081.5 | Gprasp1 | -1.31 | -2.16 | 0.0176 |
| NM_025408.2 | Phca | -1.24 | -2.17 | 0.0002 |
| NM_172871.2 | Klhl9 | -1.25 | -2.18 | 0.0049 |
| NM_016744.3 | Pde1a | -1.29 | -2.18 | 0.0255 |
| NM_012010.3 | Eif2s3x | -1.28 | -2.19 | 0.0243 |
| NM_019656.3 | Tspan6 | -1.26 | -2.19 | 0.0230 |
| NM_029662.1 | Mfsd2 | -1.29 | -2.19 | 0.0044 |
| NM_012001.1 | Cops4 | -1.26 | -2.19 | 0.0183 |
| NM_178041.1 | Eif5 | -1.28 | -2.19 | 0.0028 |
| NM_019428.2 | Rpp30 | -1.26 | -2.23 | 0.0029 |
| NM_027439.3 | Atp6ap2 | -1.29 | -2.24 | 0.0297 |
| NM_026584.2 | Gtf2e2 | -1.25 | -2.24 | 0.0067 |
| NM_009700.1 | Aqp4 | -1.32 | -2.24 | 0.0368 |
| NM_030199.3 | Zfp623 | -1.26 | -2.25 | 0.0006 |
| NM_009460 | Sumo1 | -1.29 | -2.26 | 0.0012 |
| NM_178660.2 | Rbms3 | -1.28 | -2.27 | 0.0333 |
| NM_008098.2 | Mtpn | -1.33 | -2.28 | 0.0106 |
| NM_007960.3 | Etv1 | -1.27 | -2.28 | 0.0006 |
| NM_023565.2 | Cse1l | -1.27 | -2.29 | 0.0019 |
| NM_029478.3 | Tmem49 | -1.28 | -2.30 | 0.0177 |
| NM_178050.3 | Arl6ip2 | -1.26 | -2.31 | 0.0194 |
| NM_144911.1 | Rpap2 | -1.25 | -2.32 | 0.0032 |
| NM_008409.2 | Itm2a | -1.29 | -2.33 | 0.0055 |
| NM_030066.2 | Armcx1 | -1.30 | -2.34 | 0.0271 |
| NM_025681 | Lix1 | -1.25 | -2.35 | 0.0000 |
| NM_001042565.2 | Wsb1 | -1.31 | -2.36 | 0.0041 |
| NM_008722.1 | Npm1 | -1.40 | -2.37 | 0.0038 |
| NM_016746.2 | Ccnc | -1.28 | -2.39 | 0.0274 |
| NM_009584.3 | Dnajc2 | -1.29 | -2.39 | 0.0019 |
| NM_021510.2 | Hnrph1 | -1.34 | -2.40 | 0.0129 |
| NM_016877.3 | Cnot4 | -1.32 | -2.40 | 0.0049 |
| NM_001039202.1 | Hdhd2 | -1.29 | -2.40 | 0.0004 |
| NM_018733.1 | Scn1a | -1.30 | -2.40 | 0.0000 |
| NM_011406.1 | Slc8a1 | -1.32 | -2.41 | 0.0092 |
| NM_145967.1 | Vstm2a | -1.26 | -2.41 | 0.0164 |
| NM_001014288.2 | Ptprd | -1.34 | -2.41 | 0.0000 |
| NM_024196.3 | Tbc1d20 | -1.31 | -2.42 | 0.0063 |
| NM_010497.2 | Idh1 | -1.30 | -2.43 | 0.0130 |
| NM_029068.2 | Snx16 | -1.27 | -2.44 | 0.0194 |
| NM_001001491.1 | Tpm4 | -1.30 | -2.46 | 0.0006 |
| NM_026309.1 | Lsm3 | -1.30 | -2.47 | 0.0038 |
| NM_018886.3 | Lgals8 | -1.28 | -2.47 | 0.0005 |
| NM_177715.4 | Kctd12 | -1.35 | -2.47 | 0.0262 |
| NM_011818.2 | Gmcl1 | -1.30 | -2.47 | 0.0060 |
| NM_019562.1 | Uchl5 | -1.29 | -2.48 | 0.0017 |
| NM_016886.2 | Gria3 | -1.31 | -2.49 | 0.0084 |
| NM_009460.2 | Sumo1 | -1.31 | -2.49 | 0.0040 |
| NM_009065.2 | Rit2 | -1.35 | -2.49 | 0.0136 |
| NM_028817.2 | Acsl3 | -1.37 | -2.53 | 0.0123 |
| NM_029826.2 | Hdhd2 | -1.30 | -2.53 | 0.0000 |
| NM_025942.2 | Ola1 | -1.33 | -2.54 | 0.0032 |
| NM_001039511.1 | Ivns1abp | -1.30 | -2.55 | 0.0000 |
| NM_212433.1 | Fbxo3 | -1.32 | -2.57 | 0.0008 |
| NM_175564.4 | Tmem169 | -1.30 | -2.58 | 0.0000 |
| NM_138668.1 | Ufsp2 | -1.33 | -2.59 | 0.0000 |
| NM_027439.3 | Atp6ap2 | -1.35 | -2.61 | 0.0028 |
| NM_026396.2 | Bxdc2 | -1.33 | -2.64 | 0.0013 |
| NM_010197.3 | Fgf1 | -1.32 | -2.64 | 0.0014 |
| NM_144901.2 | Csde1 | -1.32 | -2.64 | 0.0002 |
| NM_025372.1 | Tipin | -1.32 | -2.64 | 0.0143 |
| NM_134040.1 | Ddx1 | -1.35 | -2.66 | 0.0136 |
| NM_019708.3 | Scoc | -1.42 | -2.67 | 0.0125 |
| NM_138751.1 | Tmem47 | -1.34 | -2.69 | 0.0008 |
| NM_007960.1 | Etv1 | -1.33 | -2.69 | 0.0023 |
| NM_025703.2 | Tceal8 | -1.37 | -2.70 | 0.0037 |
| NM_001005525.1 | Rsrc2 | -1.35 | -2.70 | 0.0065 |
| NM_011818.2 | Gmcl1 | -1.33 | -2.70 | 0.0199 |
| NM_022985.5 | Zfand6 | -1.39 | -2.84 | 0.0125 |
| NM_028264.2 | Tmem55a | -1.40 | -2.86 | 0.0000 |
| NM_008073.2 | Gabrg2 | -1.41 | -2.87 | 0.0366 |
| NM_010169.3 | F2r | -1.38 | -2.87 | 0.0005 |
| NM_153581.2 | Gpm6a | -1.43 | -2.88 | 0.0086 |
| NM_018747.3 | Akap7 | -1.39 | -2.88 | 0.0002 |
| NM_028284.2 | Bbs5 | -1.40 | -2.91 | 0.0155 |
| NM_011252.2 | Rbmx | -1.37 | -2.94 | 0.0006 |
| NM_001098227.1 | Sdcbp | -1.42 | -2.98 | 0.0201 |
| NM_027379.2 | Mlstd2 | -1.36 | -2.99 | 0.0004 |
| NM_007586.1 | Calb2 | -1.40 | -3.03 | 0.0146 |
| NM_020012.1 | Rnf14 | -1.37 | -3.05 | 0.0004 |
| NM_009831.2 | Ccng1 | -1.39 | -3.06 | 0.0012 |
| NM_177408.3 | Gabrg2 | -1.43 | -3.06 | 0.0032 |
| NM_010477.3 | Hspd1 | -1.39 | -3.08 | 0.0041 |
| NM_054089.3 | Tgs1 | -1.39 | -3.09 | 0.0008 |
| NM_207649.1 | Rcan2 | -1.37 | -3.13 | 0.0044 |
| NM_001039389.1 | Wdr37 | -1.37 | -3.13 | 0.0001 |
| NM_025985.4 | Ube2g1 | -1.42 | -3.16 | 0.0173 |
| NM_030245.2 | Tada1l | -1.44 | -3.23 | 0.0002 |
| NM_023503.2 | Ing2 | -1.43 | -3.28 | 0.0000 |
| NM_025822.3 | Rsrc1 | -1.43 | -3.28 | 0.0000 |
| NM_001098231.1 | Ppm2c | -1.43 | -3.36 | 0.0079 |
| NM_027439.2 | Atp6ap2 | -1.45 | -3.42 | 0.0003 |
| NM_007714.4 | Clk4 | -1.47 | -3.42 | 0.0015 |
| NM_016723.1 | Uchl3 | -1.53 | -3.46 | 0.0296 |
| NM_019562.1 | Uchl5 | -1.47 | -3.49 | 0.0282 |
| NM_011666.1 | Ube1c | -1.43 | -3.54 | 0.0137 |
| NM_031396.1 | Cnnm1 | -1.42 | -3.54 | 0.0257 |
| NM_177408.3 | Gabrg2 | -1.48 | -3.57 | 0.0051 |
| NM_172120.3 | Vps41 | -1.51 | -3.76 | 0.0152 |
| NM_028058.1 | Fundc1 | -1.56 | -3.87 | 0.0041 |
| NM_001081377.1 | Pcdh9 | -1.53 | -4.05 | 0.0002 |
| NM_172694.2 | Megf9 | -1.62 | -4.07 | 0.0092 |
| NM_026070.2 | Ccdc53 | -2.42 | -7.47 | 0.0005 |
2.3.2. Bioinformatic analysis of array data - regulated gene ontologies
The data underwent Parametric Analysis of Gene Set Enrichment analysis (PAGE, Kim and Volsky, 2005). This analysis takes the identities of regulated genes observed in the experimental groups and matches them with published databases where sets of genes have been identified to be involved with specific processes or biological themes. mTBI effects on Gene Ontology were subdivided into three classifications: Cellular Component, Molecular Function and Biological Process.
2.3.3. Bioinformatic analysis of array data - regulated molecular pathways
Additional, analysis of significantly regulated genes by use of the Ingenuity Pathway Analysis tool (Ingenuity Systems, Inc., Redwood City, CA, USA) was used to identify significantly regulated molecular pathways induced by mTBI. The molecular pathways were defined by the Broad Institute of MIT and Harvard (https://www.broadinstitute.org/). Consequent to our collaborative group’s interest in neurodegenerative diseases and the inflammatory processes commonly associated with CNS disease, we chose to focus upon GOs and molecular pathways related to neuronal and inflammatory processes.
3. Results
3.1. Hippocampal tissue genes regulated by mTBI
In all, 413 gene probes were observed to be significantly regulated on the Illumina SentrixMouse Ref-8, v2 array chip 14 days after the injury. Of these, 366 of the gene probes had defined gene annotations; 143 genes were significantly up-regulated and 223 genes were significantly down-regulated. The observed fold-changes in gene expressions tended to be relatively small, the maximum positive fold-change was +1.59, and the maximum negative fold-change was -2.42. The full list of mTBI regulated genes is available in Table 1.
3.2. CNS, inflammation and age related gene ontologies derived from mTBI regulated hippocampal tissue genes
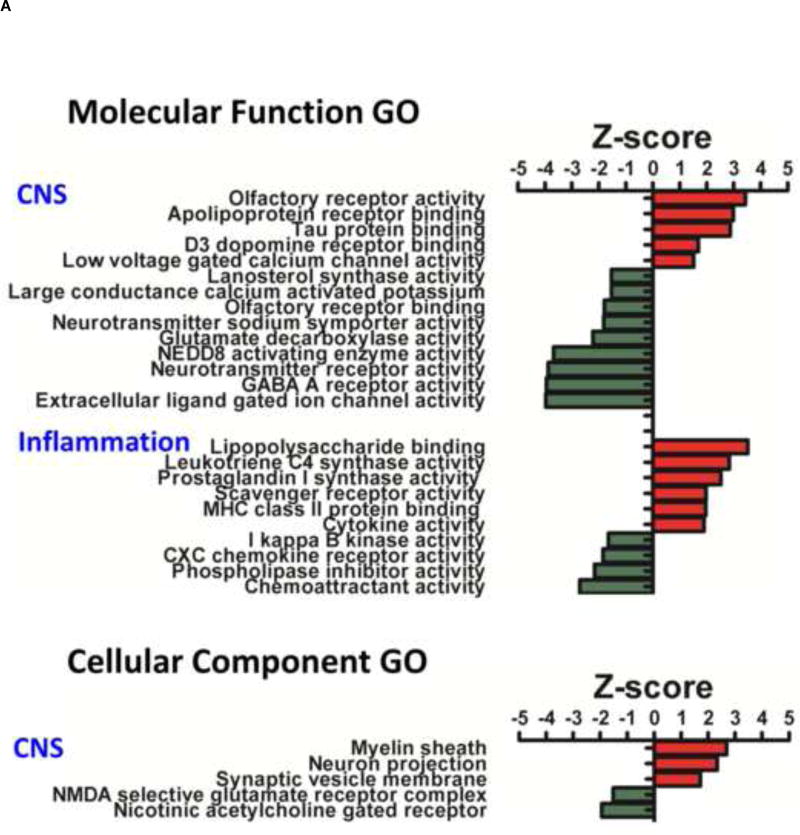
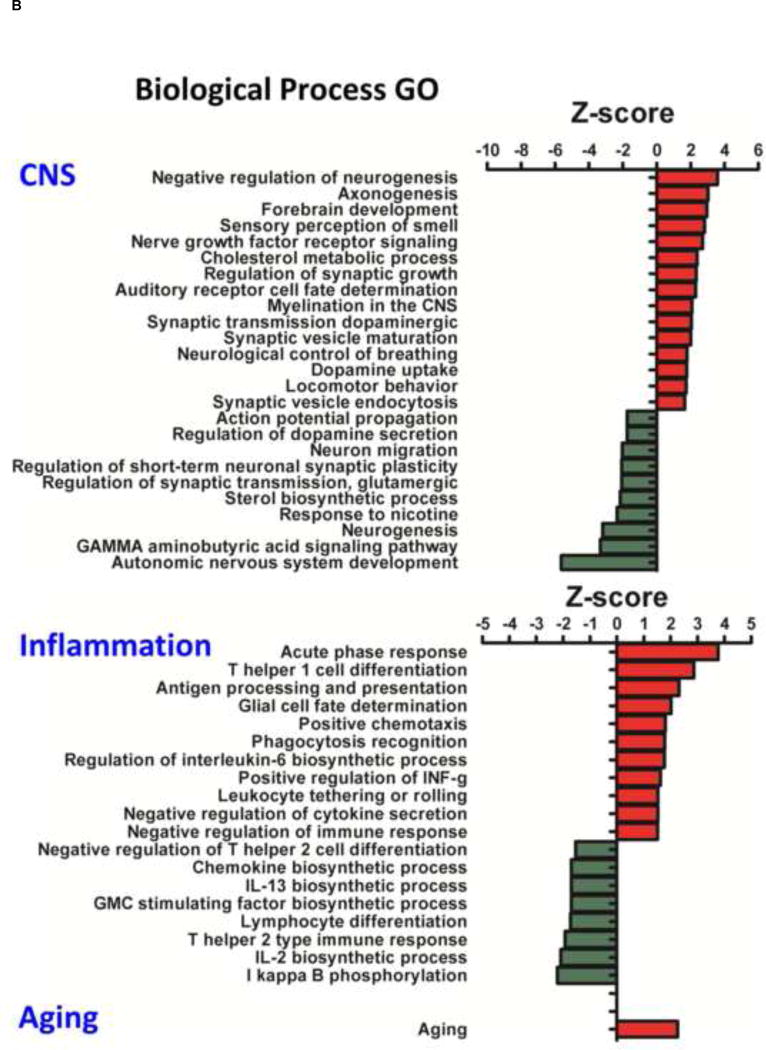
The bioinformatics tool ‘Parametric Analysis of Gene Set Enrichment’ (PAGE) analysis, was used to generate lists of mTBI-regulated functional groups (Gene Ontologies) from the differentially regulated mTBI genes. Gene ontology (GO) data for all GOs including Molecular Function (MFGOs); Cellular Component (CCOGs) and Biological Process (BPGOs), are listed in Table 2, with their GO identifiers and Z-score. The details of the CNS, inflammation or age-related GO categories are illustrated in Figure 1A/B. One age-related BPGO was observed to be regulated by mTBI, which had a positive Z-score (+2.26) Figure 1B.
Table 2.
mTBI regulated Gene Ontology classifications identified in hippocampal tissues 14 days after injury
| Molecular Function GOs regulated 14 days after mTBI | |
|---|---|
| Up-regulated Z-scores | |
| Gene Ontology Term | Z-score |
| GO0008137 NADH DEHYDROGENASE (UBIQUINONE) ACTIVITY | 5.05 |
| GO0005507 COPPER ION BINDING | 4.36 |
| GO0003735 STRUCTURAL CONSTITUENT OF RIBOSOME | 4.04 |
| GO0047391 ALKYLGLYCEROPHOSPHOETHANOLAMINE PHOSPHOD | 3.91 |
| GO0003779 ACTIN BINDING | 3.86 |
| GO0001530 LIPOPOLYSACCHARIDE BINDING | 3.53 |
| GO0004984 OLFACTORY RECEPTOR ACTIVITY | 3.43 |
| GO0005198 STRUCTURAL MOLECULE ACTIVITY | 3.43 |
| GO0003954 NADH DEHYDROGENASE ACTIVITY | 3.39 |
| GO0004129 CYTOCHROME C OXIDASE ACTIVITY | 3.36 |
| GO0003774 MOTOR ACTIVITY | 3.08 |
| GO0050749 APOLIPOPROTEIN E RECEPTOR BINDING | 2.98 |
| GO0008553 HYDROGEN EXPORTING ATPASE ACTIVITY PHOS | 2.90 |
| GO0015662 ATPASE ACTIVITY COUPLED TO TRANSMEMBRAN | 2.88 |
| GO0048156 TAU PROTEIN BINDING | 2.88 |
| GO0004464 LEUKOTRIENE C4 SYNTHASE ACTIVITY | 2.85 |
| GO0019838 GROWTH FACTOR BINDING | 2.80 |
| GO0005520 INSULIN LIKE GROWTH FACTOR BINDING | 2.80 |
| GO0046983 PROTEIN DIMERIZATION ACTIVITY | 2.78 |
| GO0004951 CHOLECYSTOKININ RECEPTOR ACTIVITY | 2.76 |
| GO0004720 PROTEIN LYSINE 6 OXIDASE ACTIVITY | 2.68 |
| GO0004872 RECEPTOR ACTIVITY | 2.67 |
| GO0016820 HYDROLASE ACTIVITY ACTING ON ACID ANHYD | 2.55 |
| GO0008116 PROSTAGLANDIN I SYNTHASE ACTIVITY | 2.53 |
| GO0004727 PRENYLATED PROTEIN TYROSINE PHOSPHATASE | 2.51 |
| GO0046933 HYDROGEN ION TRANSPORTING ATP SYNTHASE A | 2.50 |
| GO0046961 HYDROGEN ION TRANSPORTING ATPASE ACTIVIT | 2.50 |
| GO0050178 PHENYLPYRUVATE TAUTOMERASE ACTIVITY | 2.46 |
| GO0003923 GPI ANCHOR TRANSAMIDASE ACTIVITY | 2.43 |
| GO0016628 OXIDOREDUCTASE ACTIVITY ACTING ON THE C | 2.43 |
| GO0004699 CALCIUM INDEPENDENT PROTEIN KINASE C ACT | 2.40 |
| GO0005179 HORMONE ACTIVITY | 2.38 |
| GO0047196 LONG CHAIN ALCOHOL O FATTY ACYLTRANSFERA | 2.37 |
| GO0005319 LIPID TRANSPORTER ACTIVITY | 2.37 |
| GO0003870 5 AMINOLEVULINATE SYNTHASE ACTIVITY | 2.29 |
| GO0008083 GROWTH FACTOR ACTIVITY | 2.27 |
| GO0030246 CARBOHYDRATE BINDING | 2.23 |
| GO0008242 OMEGA PEPTIDASE ACTIVITY | 2.22 |
| GO0042803 PROTEIN HOMODIMERIZATION ACTIVITY | 2.20 |
| GO0004925 PROLACTIN RECEPTOR ACTIVITY | 2.18 |
| GO0004111 CREATINE KINASE ACTIVITY | 2.12 |
| GO0000171 RIBONUCLEASE MRP ACTIVITY | 2.10 |
| GO0030528 TRANSCRIPTION REGULATOR ACTIVITY | 2.10 |
| GO0008503 BENZODIAZEPINE RECEPTOR ACTIVITY | 2.09 |
| GO0001882 NUCLEOSIDE BINDING | 2.00 |
| GO0005548 PHOSPHOLIPID TRANSPORTER ACTIVITY | 1.99 |
| GO0005201 EXTRACELLULAR MATRIX STRUCTURAL CONSTITU | 1.98 |
| GO0004930 G PROTEIN COUPLED RECEPTOR ACTIVITY | 1.98 |
| GO0005044 SCAVENGER RECEPTOR ACTIVITY | 1.98 |
| GO0005080 PROTEIN KINASE C BINDING | 1.98 |
| GO0042289 MHC CLASS II PROTEIN BINDING | 1.96 |
| GO0015457 AUXILIARY TRANSPORT PROTEIN ACTIVITY | 1.92 |
| GO0004252 SERINE TYPE ENDOPEPTIDASE ACTIVITY | 1.90 |
| GO0019103 PYRIMIDINE NUCLEOTIDE BINDING | 1.90 |
| GO0001691 PSEUDOPHOSPHATASE ACTIVITY | 1.90 |
| GO0005125 CYTOKINE ACTIVITY | 1.90 |
| GO0005534 GALACTOSE BINDING | 1.87 |
| GO0003869 4 NITROPHENYLPHOSPHATASE ACTIVITY | 1.87 |
| GO0047057 VITAMIN K EPOXIDE REDUCTASE (WARFARIN SE | 1.86 |
| GO0003696 SATELLITE DNA BINDING | 1.84 |
| GO0005160 TRANSFORMING GROWTH FACTOR BETA RECEPTOR | 1.83 |
| GO0050517 INOSITOL HEXAKISPHOSPHATE KINASE ACTIVIT | 1.81 |
| GO0042577 LIPID PHOSPHATASE ACTIVITY | 1.75 |
| GO0005055 LAMININ RECEPTOR ACTIVITY | 1.74 |
| GO0015085 CALCIUM ION TRANSMEMBRANE TRANSPORTER AC | 1.73 |
| GO0030731 GUANIDINOACETATE N METHYLTRANSFERASE ACT | 1.71 |
| GO0004243 MITOCHONDRIAL INTERMEDIATE PEPTIDASE ACT | 1.71 |
| GO0031750 D3 DOPAMINE RECEPTOR BINDING | 1.70 |
| GO0046790 VIRION BINDING | 1.70 |
| GO0008095 INOSITOL 1 4 5 TRIPHOSPHATE RECEPTOR ACT | 1.67 |
| GO0008296 3 5 EXODEOXYRIBONUCLEASE ACTIVITY | 1.66 |
| GO0015186 L GLUTAMINE TRANSMEMBRANE TRANSPORTER AC | 1.61 |
| GO0004766 SPERMIDINE SYNTHASE ACTIVITY | 1.61 |
| GO0046870 CADMIUM ION BINDING | 1.59 |
| GO0004630 PHOSPHOLIPASE D ACTIVITY | 1.59 |
| GO0004839 UBIQUITIN ACTIVATING ENZYME ACTIVITY | 1.58 |
| GO0051721 PROTEIN PHOSPHATASE 2A BINDING | 1.57 |
| GO0016641 OXIDOREDUCTASE ACTIVITY ACTING ON THE C | 1.55 |
| GO0004228 GELATINASE A ACTIVITY | 1.54 |
| GO0015056 CORTICOTROPHIN RELEASING FACTOR RECEPTOR | 1.52 |
| GO0008332 LOW VOLTAGE GATED CALCIUM CHANNEL ACTIVI | 1.51 |
| Down-regulated Z-scores | |
| Gene Ontology Term | Z-score |
| GO0004569 GLYCOPROTEIN ENDO ALPHA 1 2 MANNOSIDASE | -1.50 |
| GO0009881 PHOTORECEPTOR ACTIVITY | -1.50 |
| GO0001515 OPIOID PEPTIDE ACTIVITY | -1.50 |
| GO0000250 LANOSTEROL SYNTHASE ACTIVITY | -1.52 |
| GO0060072 LARGE CONDUCTANCE CALCIUM ACTIVATED POTA | -1.52 |
| GO0004394 HEPARAN SULFATE 2 O SULFOTRANSFERASE ACT | -1.52 |
| GO0003878 ATP CITRATE SYNTHASE ACTIVITY | -1.53 |
| GO0004768 STEAROYL COA 9 DESATURASE ACTIVITY | -1.53 |
| GO0004738 PYRUVATE DEHYDROGENASE ACTIVITY | -1.55 |
| GO0004765 SHIKIMATE KINASE ACTIVITY | -1.56 |
| GO0004218 CATHEPSIN S ACTIVITY | -1.57 |
| GO0004001 ADENOSINE KINASE ACTIVITY | -1.60 |
| GO0004022 ALCOHOL DEHYDROGENASE ACTIVITY | -1.64 |
| GO0051903 S (HYDROXYMETHYL)GLUTATHIONE DEHYDROGENA | -1.64 |
| GO0008384 IKAPPAB KINASE ACTIVITY | -1.64 |
| GO0008235 METALLOEXOPEPTIDASE ACTIVITY | -1.65 |
| GO0051061 ADP REDUCTASE ACTIVITY | -1.67 |
| GO0004573 MANNOSYL OLIGOSACCHARIDE GLUCOSIDASE ACT | -1.68 |
| GO0004231 INSULYSIN ACTIVITY | -1.68 |
| GO0005092 GDP DISSOCIATION INHIBITOR ACTIVITY | -1.68 |
| GO0050897 COBALT ION BINDING | -1.69 |
| GO0030060 L MALATE DEHYDROGENASE ACTIVITY | -1.70 |
| GO0008191 METALLOENDOPEPTIDASE INHIBITOR ACTIVITY | -1.71 |
| GO0004814 ARGININE TRNA LIGASE ACTIVITY | -1.71 |
| GO0051879 HSP90 PROTEIN BINDING | -1.72 |
| GO0016790 THIOLESTER HYDROLASE ACTIVITY | -1.73 |
| GO0003725 DOUBLE STRANDED RNA BINDING | -1.73 |
| GO0004742 DIHYDROLIPOYLLYSINE RESIDUE ACETYLTRANSF | -1.74 |
| GO0004217 CATHEPSIN L ACTIVITY | -1.74 |
| GO0004652 POLYNUCLEOTIDE ADENYLYLTRANSFERASE ACTIV | -1.76 |
| GO0005502 11 CIS RETINAL BINDING | -1.78 |
| GO0004239 METHIONYL AMINOPEPTIDASE ACTIVITY | -1.79 |
| GO0008545 JUN KINASE KINASE ACTIVITY | -1.79 |
| GO0008080 N ACETYLTRANSFERASE ACTIVITY | -1.79 |
| GO0031849 OLFACTORY RECEPTOR BINDING | -1.79 |
| GO0015234 THIAMIN TRANSMEMBRANE TRANSPORTER ACTIVI | -1.79 |
| GO0017040 CERAMIDASE ACTIVITY | -1.80 |
| GO0005328 NEUROTRANSMITTER SODIUM SYMPORTER ACTIVI | -1.81 |
| GO0008831 DTDP 4 DEHYDRORHAMNOSE REDUCTASE ACTIVIT | -1.81 |
| GO0048270 METHIONINE ADENOSYLTRANSFERASE REGULATOR | -1.81 |
| GO0047012 STEROL 4 ALPHA CARBOXYLATE 3 DEHYDROGENA | -1.82 |
| GO0035248 ALPHA 1 4 N ACETYLGALACTOSAMINYLTRANSFER | -1.82 |
| GO0030628 PRE MRNA 3 SPLICE SITE BINDING | -1.82 |
| GO0000386 SECOND SPLICEOSOMAL TRANSESTERIFICATION | -1.82 |
| GO0004482 MRNA (GUANINE N7) METHYLTRANSFERASE ACT | -1.84 |
| GO0016494 C X C CHEMOKINE RECEPTOR ACTIVITY | -1.84 |
| GO0004963 FOLLICLE STIMULATING HORMONE RECEPTOR AC | -1.88 |
| GO0004740 PYRUVATE DEHYDROGENASE (LIPOAMIDE) KIN | -1.88 |
| GO0008234 CYSTEINE TYPE PEPTIDASE ACTIVITY | -1.91 |
| GO0016934 EXTRACELLULAR GLYCINE GATED CHLORIDE CHA | -1.91 |
| GO0000900 TRANSLATION REPRESSOR ACTIVITY NUCLEIC | -1.92 |
| GO0003983 UTP GLUCOSE 1 PHOSPHATE URIDYLYLTRANSFER | -1.95 |
| GO0016314 PHOSPHATIDYLINOSITOL 3 4 5 TRISPHOSPHATE | -1.97 |
| GO0051800 PHOSPHATIDYLINOSITOL 3 4 BISPHOSPHATE 3 | -1.97 |
| GO0051717 INOSITOL 1 3 4 5 TETRAKISPHOSPHATE 3 PHO | -1.97 |
| GO0004142 DIACYLGLYCEROL CHOLINEPHOSPHOTRANSFERASE | -2.00 |
| GO0004439 PHOSPHOINOSITIDE 5 PHOSPHATASE ACTIVITY | -2.02 |
| GO0008486 DIPHOSPHOINOSITOL POLYPHOSPHATE DIPHOSPH | -2.02 |
| GO0004127 CYTIDYLATE KINASE ACTIVITY | -2.06 |
| GO0042393 HISTONE BINDING | -2.08 |
| GO0031405 LIPOIC ACID BINDING | -2.11 |
| GO0030675 RAC GTPASE ACTIVATOR ACTIVITY | -2.14 |
| GO0003810 PROTEIN GLUTAMINE GAMMA GLUTAMYLTRANSFER | -2.14 |
| GO0004859 PHOSPHOLIPASE INHIBITOR ACTIVITY | -2.15 |
| GO0016780 PHOSPHOTRANSFERASE ACTIVITY FOR OTHER S | -2.15 |
| GO0030346 PROTEIN PHOSPHATASE 2B BINDING | -2.15 |
| GO0004221 UBIQUITIN THIOLESTERASE ACTIVITY | -2.16 |
| GO0008467 HEPARIN GLUCOSAMINE 3 O SULFOTRANSFERASE | -2.16 |
| GO0004739 PYRUVATE DEHYDROGENASE (ACETYL TRANSFERR | -2.19 |
| GO0043185 VASCULAR ENDOTHELIAL GROWTH FACTOR RECEP | -2.19 |
| GO0003723 RNA BINDING | -2.21 |
| GO0004105 CHOLINE PHOSPHATE CYTIDYLYLTRANSFERASE A | -2.21 |
| GO0004351 GLUTAMATE DECARBOXYLASE ACTIVITY | -2.22 |
| GO0017172 CYSTEINE DIOXYGENASE ACTIVITY | -2.23 |
| GO0008568 MICROTUBULE SEVERING ATPASE ACTIVITY | -2.28 |
| GO0050681 ANDROGEN RECEPTOR BINDING | -2.30 |
| GO0016603 GLUTAMINYL PEPTIDE CYCLOTRANSFERASE ACTI | -2.32 |
| GO0004333 FUMARATE HYDRATASE ACTIVITY | -2.32 |
| GO0003851 2 HYDROXYACYLSPHINGOSINE 1 BETA GALACTOS | -2.35 |
| GO0046872 METAL ION BINDING | -2.42 |
| GO0008120 CERAMIDE GLUCOSYLTRANSFERASE ACTIVITY | -2.47 |
| GO0015189 L LYSINE TRANSMEMBRANE TRANSPORTER ACTIV | -2.56 |
| GO0015326 CATIONIC AMINO ACID TRANSMEMBRANE TRANSP | -2.56 |
| GO0015181 ARGININE TRANSMEMBRANE TRANSPORTER ACTIV | -2.56 |
| GO0030629 U6 SNRNA 3 END BINDING | -2.57 |
| GO0050164 OXOGLUTARATE DEHYDROGENASE (NADP+) ACTIV | -2.65 |
| GO0042056 CHEMOATTRACTANT ACTIVITY | -2.70 |
| GO0004307 ETHANOLAMINEPHOSPHOTRANSFERASE ACTIVITY | -2.80 |
| GO0004062 ARYL SULFOTRANSFERASE ACTIVITY | -2.80 |
| GO0017067 TYROSINE ESTER SULFOTRANSFERASE ACTIVITY | -2.80 |
| GO0004170 DUTP DIPHOSPHATASE ACTIVITY | -3.03 |
| GO0050062 LONG CHAIN FATTY ACYL COA REDUCTASE ACTI | -3.11 |
| GO0008270 ZINC ION BINDING | -3.16 |
| GO0004741 PYRUVATE DEHYDROGENASE (LIPOAMIDE) PHO | -3.49 |
| GO0019781 NEDD8 ACTIVATING ENZYME ACTIVITY | -3.68 |
| GO0030594 NEUROTRANSMITTER RECEPTOR ACTIVITY | -3.87 |
| GO0004890 GABA A RECEPTOR ACTIVITY | -3.94 |
| GO0005230 EXTRACELLULAR LIGAND GATED ION CHANNEL A | -3.96 |
| GO0003676 NUCLEIC ACID BINDING | -4.07 |
| GO0047220 GALACTOSYLXYLOSYLPROTEIN 3 BETA GALACTOS | -6.45 |
| Cellular Component GOs regulated 14 days after mTBI | |
| Up-regulated Z-scores | |
| Gene Ontology Term | Z-score |
| GO0000786 NUCLEOSOME | 6.39 |
| GO0005840 RIBOSOME | 4.10 |
| GO0005694 CHROMOSOME | 4.09 |
| GO0030529 RIBONUCLEOPROTEIN COMPLEX | 4.06 |
| GO0005615 EXTRACELLULAR SPACE | 3.85 |
| GO0005576 EXTRACELLULAR REGION | 3.78 |
| GO0030140 TRANS GOLGI NETWORK TRANSPORT VESICLE | 3.77 |
| GO0005830 CYTOSOLIC RIBOSOME (SENSU EUKARYOTA) | 3.50 |
| GO0005791 ROUGH ENDOPLASMIC RETICULUM | 3.28 |
| GO0035267 NUA4 HISTONE ACETYLTRANSFERASE COMPLEX | 3.24 |
| GO0005605 BASAL LAMINA | 3.15 |
| GO0005843 CYTOSOLIC SMALL RIBOSOMAL SUBUNIT (SENSU | 3.08 |
| GO0035253 CILIARY ROOTLET | 3.07 |
| GO0005856 CYTOSKELETON | 3.04 |
| GO0005624 MEMBRANE FRACTION | 2.84 |
| GO0043209 MYELIN SHEATH | 2.69 |
| GO0042175 NUCLEAR ENVELOPE ENDOPLASMIC RETICULUM N | 2.52 |
| GO0005885 ARP2 OR 3 PROTEIN COMPLEX | 2.50 |
| GO0031143 PSEUDOPODIUM | 2.47 |
| GO0045259 PROTON TRANSPORTING ATP SYNTHASE COMPLEX | 2.46 |
| GO0005739 MITOCHONDRION | 2.41 |
| GO0033185 DOLICHOL PHOSPHATE MANNOSE SYNTHASE COMP | 2.38 |
| GO0043005 NEURON PROJECTION | 2.35 |
| GO0016023 CYTOPLASMIC MEMBRANE BOUND VESICLE | 2.34 |
| GO0005762 MITOCHONDRIAL LARGE RIBOSOMAL SUBUNIT | 2.16 |
| GO0000172 RIBONUCLEASE MRP COMPLEX | 2.10 |
| GO0005771 MULTIVESICULAR BODY | 1.89 |
| GO0005890 SODIUM POTASSIUM EXCHANGING ATPASE COMPL | 1.87 |
| GO0005688 SNRNP U6 | 1.85 |
| GO0043292 CONTRACTILE FIBER | 1.83 |
| GO0043596 NUCLEAR REPLICATION FORK | 1.79 |
| GO0030672 SYNAPTIC VESICLE MEMBRANE | 1.72 |
| GO0044445 CYTOSOLIC PART | 1.63 |
| GO0000125 PCAF COMPLEX | 1.61 |
| GO0031234 EXTRINSIC TO INTERNAL SIDE OF PLASMA MEM | 1.59 |
| GO0030056 HEMIDESMOSOME | 1.52 |
| Down-regulated Z-scores | |
| Gene Ontology Term | Z-score |
| GO0017146 N METHYL D ASPARTATE SELECTIVE GLUTAMATE | -1.51 |
| GO0009346 CITRATE LYASE COMPLEX | -1.53 |
| GO0005583 FIBRILLAR COLLAGEN | -1.55 |
| GO0016591 DNA DIRECTED RNA POLYMERASE II HOLOENZY | -1.59 |
| GO0045254 PYRUVATE DEHYDROGENASE COMPLEX | -1.74 |
| GO0005967 MITOCHONDRIAL PYRUVATE DEHYDROGENASE COM | -1.74 |
| GO0031213 RSF COMPLEX | -1.76 |
| GO0008180 SIGNALOSOME | -1.80 |
| GO0048269 METHIONINE ADENOSYLTRANSFERASE COMPLEX | -1.81 |
| GO0005832 CHAPERONIN CONTAINING T COMPLEX | -1.83 |
| GO0005845 MRNA CAP COMPLEX | -1.84 |
| GO0030127 COPII VESICLE COAT | -1.85 |
| GO0000164 PROTEIN PHOSPHATASE TYPE 1 COMPLEX | -1.92 |
| GO0005892 NICOTINIC ACETYLCHOLINE GATED RECEPTOR C | -1.94 |
| GO0017101 AMINOACYL TRNA SYNTHETASE MULTIENZYME CO | -1.95 |
| GO0005730 NUCLEOLUS | -2.04 |
| GO0000139 GOLGI MEMBRANE | -2.12 |
| GO0005777 PEROXISOME | -2.13 |
| GO0005783 ENDOPLASMIC RETICULUM | -2.27 |
| GO0045239 TRICARBOXYLIC ACID CYCLE ENZYME COMPLEX | -2.32 |
| GO0005775 VACUOLAR LUMEN | -3.18 |
| GO0005797 GOLGI MEDIAL CISTERNA | -6.45 |
| GO0000153 CYTOPLASMIC UBIQUITIN LIGASE COMPLEX | -6.48 |
| Biological Process GOs regulated 14 days after mTBI | |
| Up-regulated Z-score | |
| BPGOs | |
| Gene Ontology Term | Z-score |
| GO0007001 CHROMOSOME ORGANIZATION AND BIOGENESIS | 6.00 |
| GO0006334 NUCLEOSOME ASSEMBLY | 5.48 |
| GO0006953 ACUTE PHASE RESPONSE | 3.77 |
| GO0006875 CELLULAR METAL ION HOMEOSTASIS | 3.67 |
| GO0050768 NEGATIVE REGULATION OF NEUROGENESIS | 3.59 |
| GO0007010 CYTOSKELETON ORGANIZATION AND BIOGENESIS | 3.50 |
| GO0045187 REGULATION OF CIRCADIAN SLEEP OR WAKE CY | 3.26 |
| GO0007409 AXONOGENESIS | 3.02 |
| GO0030900 FOREBRAIN DEVELOPMENT | 2.96 |
| GO0006412 TRANSLATION | 2.94 |
| GO0006120 MITOCHONDRIAL ELECTRON TRANSPORT NADH T | 2.89 |
| GO0042989 SEQUESTERING OF ACTIN MONOMERS | 2.88 |
| GO0008064 REGULATION OF ACTIN POLYMERIZATION AND O | 2.88 |
| GO0045063 T HELPER 1 CELL DIFFERENTIATION | 2.87 |
| GO0007275 MULTICELLULAR ORGANISMAL DEVELOPMENT | 2.85 |
| GO0007608 SENSORY PERCEPTION OF SMELL | 2.82 |
| GO0007166 CELL SURFACE RECEPTOR LINKED SIGNAL TRAN | 2.81 |
| GO0048011 NERVE GROWTH FACTOR RECEPTOR SIGNALING P | 2.71 |
| GO0032114 REGULATION OF GLUCOSE 6 PHOSPHATASE ACTI | 2.71 |
| GO0007566 EMBRYO IMPLANTATION | 2.69 |
| GO0009887 ORGAN MORPHOGENESIS | 2.69 |
| GO0001837 EPITHELIAL TO MESENCHYMAL TRANSITION | 2.64 |
| GO0008286 INSULIN RECEPTOR SIGNALING PATHWAY | 2.61 |
| GO0040011 LOCOMOTION | 2.58 |
| GO0007155 CELL ADHESION | 2.51 |
| GO0001886 ENDOTHELIAL CELL MORPHOGENESIS | 2.48 |
| GO0006878 CELLULAR COPPER ION HOMEOSTASIS | 2.46 |
| GO0006936 MUSCLE CONTRACTION | 2.45 |
| GO0016255 ATTACHMENT OF GPI ANCHOR TO PROTEIN | 2.43 |
| GO0031639 PLASMINOGEN ACTIVATION | 2.43 |
| GO0006098 PENTOSE PHOSPHATE SHUNT | 2.43 |
| GO0009181 PURINE RIBONUCLEOSIDE DIPHOSPHATE CATABO | 2.41 |
| GO0045822 NEGATIVE REGULATION OF HEART CONTRACTION | 2.41 |
| GO0008203 CHOLESTEROL METABOLIC PROCESS | 2.40 |
| GO0006754 ATP BIOSYNTHETIC PROCESS | 2.37 |
| GO0045823 POSITIVE REGULATION OF HEART CONTRACTION | 2.37 |
| GO0007186 G PROTEIN COUPLED RECEPTOR PROTEIN SIGNA | 2.37 |
| GO0008582 REGULATION OF SYNAPTIC GROWTH AT NEUROMU | 2.35 |
| GO0007009 PLASMA MEMBRANE ORGANIZATION AND BIOGENE | 2.35 |
| GO0043388 POSITIVE REGULATION OF DNA BINDING | 2.33 |
| GO0019882 ANTIGEN PROCESSING AND PRESENTATION | 2.32 |
| GO0042668 AUDITORY RECEPTOR CELL FATE DETERMINATIO | 2.31 |
| GO0030198 EXTRACELLULAR MATRIX ORGANIZATION AND BI | 2.30 |
| GO0006346 METHYLATION DEPENDENT CHROMATIN SILENCIN | 2.30 |
| GO0007568 AGING | 2.26 |
| GO0007603 PHOTOTRANSDUCTION VISIBLE LIGHT | 2.26 |
| GO0001501 SKELETAL DEVELOPMENT | 2.26 |
| GO0045165 CELL FATE COMMITMENT | 2.26 |
| GO0042977 TYROSINE PHOSPHORYLATION OF JAK2 PROTEIN | 2.18 |
| GO0001676 LONG CHAIN FATTY ACID METABOLIC PROCESS | 2.16 |
| GO0031638 ZYMOGEN ACTIVATION | 2.10 |
| GO0022010 MYELINATION IN THE CENTRAL NERVOUS SYSTE | 2.09 |
| GO0001963 SYNAPTIC TRANSMISSION DOPAMINERGIC | 2.06 |
| GO0009888 TISSUE DEVELOPMENT | 2.06 |
| GO0006469 NEGATIVE REGULATION OF PROTEIN KINASE AC | 2.04 |
| GO0045446 ENDOTHELIAL CELL DIFFERENTIATION | 2.03 |
| GO0007403 GLIAL CELL FATE DETERMINATION | 2.02 |
| GO0007595 LACTATION | 2.02 |
| GO0040007 GROWTH | 2.01 |
| GO0009605 RESPONSE TO EXTERNAL STIMULUS | 2.01 |
| GO0016188 SYNAPTIC VESICLE MATURATION | 2.00 |
| GO0032417 POSITIVE REGULATION OF SODIUM HYDROGEN A | 2.00 |
| GO0006649 PHOSPHOLIPID TRANSFER TO MEMBRANE | 1.99 |
| GO0009399 NITROGEN FIXATION | 1.94 |
| GO0007017 MICROTUBULE BASED PROCESS | 1.94 |
| GO0007016 CYTOSKELETAL ANCHORING | 1.93 |
| GO0009060 AEROBIC RESPIRATION | 1.90 |
| GO0030201 HEPARAN SULFATE PROTEOGLYCAN METABOLIC P | 1.90 |
| GO0006020 INOSITOL METABOLIC PROCESS | 1.90 |
| GO0010003 GASTRULATION (SENSU MAMMALIA) | 1.89 |
| GO0030218 ERYTHROCYTE DIFFERENTIATION | 1.88 |
| GO0030641 CELLULAR HYDROGEN ION HOMEOSTASIS | 1.87 |
| GO0031947 NEGATIVE REGULATION OF GLUCOCORTICOID BI | 1.87 |
| GO0045989 POSITIVE REGULATION OF STRIATED MUSCLE C | 1.87 |
| GO0006729 TETRAHYDROBIOPTERIN BIOSYNTHETIC PROCESS | 1.86 |
| GO0050820 POSITIVE REGULATION OF COAGULATION | 1.86 |
| GO0042371 VITAMIN K BIOSYNTHETIC PROCESS | 1.86 |
| GO0001835 BLASTOCYST HATCHING | 1.86 |
| GO0051481 REDUCTION OF CYTOSOLIC CALCIUM ION CONCE | 1.80 |
| GO0002087 NEUROLOGICAL CONTROL OF BREATHING | 1.80 |
| GO0006942 REGULATION OF STRIATED MUSCLE CONTRACTIO | 1.80 |
| GO0045988 NEGATIVE REGULATION OF STRIATED MUSCLE C | 1.80 |
| GO0046626 REGULATION OF INSULIN RECEPTOR SIGNALING | 1.80 |
| GO0014067 NEGATIVE REGULATION OF PHOSPHOINOSITIDE | 1.80 |
| GO0050918 POSITIVE CHEMOTAXIS | 1.80 |
| GO0009209 PYRIMIDINE RIBONUCLEOSIDE TRIPHOSPHATE B | 1.78 |
| GO0051583 DOPAMINE UPTAKE | 1.78 |
| GO0006910 PHAGOCYTOSIS RECOGNITION | 1.78 |
| GO0045408 REGULATION OF INTERLEUKIN 6 BIOSYNTHETIC | 1.77 |
| GO0007626 LOCOMOTORY BEHAVIOR | 1.76 |
| GO0045634 REGULATION OF MELANOCYTE DIFFERENTIATION | 1.74 |
| GO0046655 FOLIC ACID METABOLIC PROCESS | 1.72 |
| GO0006620 POSTTRANSLATIONAL PROTEIN TARGETING TO M | 1.72 |
| GO0015884 FOLIC ACID TRANSPORT | 1.72 |
| GO0019538 PROTEIN METABOLIC PROCESS | 1.71 |
| GO0006601 CREATINE BIOSYNTHETIC PROCESS | 1.71 |
| GO0051225 SPINDLE ASSEMBLY | 1.70 |
| GO0001541 OVARIAN FOLLICLE DEVELOPMENT | 1.69 |
| GO0006790 SULFUR METABOLIC PROCESS | 1.67 |
| GO0048488 SYNAPTIC VESICLE ENDOCYTOSIS | 1.66 |
| GO0045597 POSITIVE REGULATION OF CELL DIFFERENTIAT | 1.66 |
| GO0001708 CELL FATE SPECIFICATION | 1.65 |
| GO0000184 MRNA CATABOLIC PROCESS NONSENSE MEDIATE | 1.65 |
| GO0051289 PROTEIN HOMOTETRAMERIZATION | 1.64 |
| GO0032729 POSITIVE REGULATION OF INTERFERON GAMMA | 1.62 |
| GO0006868 GLUTAMINE TRANSPORT | 1.61 |
| GO0000320 RE ENTRY INTO MITOTIC CELL CYCLE | 1.60 |
| GO0019048 VIRUS HOST INTERACTION | 1.59 |
| GO0006644 PHOSPHOLIPID METABOLIC PROCESS | 1.59 |
| GO0006818 HYDROGEN TRANSPORT | 1.59 |
| GO0007231 OSMOSENSORY SIGNALING PATHWAY | 1.58 |
| GO0042538 HYPEROSMOTIC SALINITY RESPONSE | 1.58 |
| GO0030103 VASOPRESSIN SECRETION | 1.58 |
| GO0047484 REGULATION OF RESPONSE TO OSMOTIC STRESS | 1.58 |
| GO0009948 ANTERIOR OR POSTERIOR AXIS SPECIFICATION | 1.56 |
| GO0018277 PROTEIN AMINO ACID DEAMINATION | 1.55 |
| GO0007179 TRANSFORMING GROWTH FACTOR BETA RECEPTOR | 1.55 |
| GO0007223 WNT RECEPTOR SIGNALING PATHWAY CALCIUM | 1.54 |
| GO0042773 ATP SYNTHESIS COUPLED ELECTRON TRANSPORT | 1.54 |
| GO0045672 POSITIVE REGULATION OF OSTEOCLAST DIFFER | 1.54 |
| GO0000003 REPRODUCTION | 1.54 |
| GO0005980 GLYCOGEN CATABOLIC PROCESS | 1.54 |
| GO0050901 LEUKOCYTE TETHERING OR ROLLING | 1.53 |
| GO0050710 NEGATIVE REGULATION OF CYTOKINE SECRETIO | 1.53 |
| GO0030178 NEGATIVE REGULATION OF WNT RECEPTOR SIGN | 1.52 |
| GO0031110 REGULATION OF MICROTUBULE POLYMERIZATION | 1.52 |
| GO0051258 PROTEIN POLYMERIZATION | 1.52 |
| GO0035303 REGULATION OF DEPHOSPHORYLATION | 1.51 |
| GO0000038 VERY LONG CHAIN FATTY ACID METABOLIC PRO | 1.51 |
| GO0050777 NEGATIVE REGULATION OF IMMUNE RESPONSE | 1.51 |
| GO0009612 RESPONSE TO MECHANICAL STIMULUS | 1.51 |
| Down-regulated Z-score | |
| BPGOs | |
| Gene Ontology Term | Z-score |
| GO0043089 POSITIVE REGULATION OF CDC42 GTPASE ACTI | -1.50 |
| GO0006541 GLUTAMINE METABOLIC PROCESS | -1.51 |
| GO0000055 RIBOSOMAL LARGE SUBUNIT EXPORT FROM NUCL | -1.51 |
| GO0048661 POSITIVE REGULATION OF SMOOTH MUSCLE CEL | -1.52 |
| GO0060082 EYE BLINK REFLEX | -1.52 |
| GO0045794 NEGATIVE REGULATION OF CELL VOLUME | -1.52 |
| GO0032344 REGULATION OF ALDOSTERONE METABOLIC PROC | -1.52 |
| GO0046541 SALIVA SECRETION | -1.52 |
| GO0060073 MICTURITION | -1.52 |
| GO0019228 GENERATION OF ACTION POTENTIAL | -1.52 |
| GO0030202 HEPARIN METABOLIC PROCESS | -1.52 |
| GO0009566 FERTILIZATION | -1.53 |
| GO0051146 STRIATED MUSCLE CELL DIFFERENTIATION | -1.53 |
| GO0015936 COENZYME A METABOLIC PROCESS | -1.53 |
| GO0006200 ATP CATABOLIC PROCESS | -1.53 |
| GO0045629 NEGATIVE REGULATION OF T HELPER 2 CELL D | -1.53 |
| GO0060048 CARDIAC MUSCLE CONTRACTION | -1.54 |
| GO0008299 ISOPRENOID BIOSYNTHETIC PROCESS | -1.54 |
| GO0042462 EYE PHOTORECEPTOR CELL DEVELOPMENT | -1.58 |
| GO0006991 RESPONSE TO STEROL DEPLETION | -1.59 |
| GO0045815 POSITIVE REGULATION OF GENE EXPRESSION | -1.60 |
| GO0006613 COTRANSLATIONAL PROTEIN TARGETING TO MEM | -1.61 |
| GO0016584 NUCLEOSOME POSITIONING | -1.62 |
| GO0048732 GLAND DEVELOPMENT | -1.64 |
| GO0006405 RNA EXPORT FROM NUCLEUS | -1.64 |
| GO0006398 HISTONE MRNA 3 END PROCESSING | -1.66 |
| GO0035095 BEHAVIORAL RESPONSE TO NICOTINE | -1.67 |
| GO0060080 REGULATION OF INHIBITORY POSTSYNAPTIC ME | -1.67 |
| GO0019303 D RIBOSE CATABOLIC PROCESS | -1.67 |
| GO0043631 RNA POLYADENYLATION | -1.69 |
| GO0030431 SLEEP | -1.69 |
| GO0042033 CHEMOKINE BIOSYNTHETIC PROCESS | -1.69 |
| GO0042231 INTERLEUKIN 13 BIOSYNTHETIC PROCESS | -1.69 |
| GO0042253 GRANULOCYTE MACROPHAGE COLONY STIMULATIN | -1.69 |
| GO0016572 HISTONE PHOSPHORYLATION | -1.70 |
| GO0015853 ADENINE TRANSPORT | -1.71 |
| GO0006420 ARGINYL TRNA AMINOACYLATION | -1.71 |
| GO0030098 LYMPHOCYTE DIFFERENTIATION | -1.72 |
| GO0051186 COFACTOR METABOLIC PROCESS | -1.73 |
| GO0031579 LIPID RAFT ORGANIZATION AND BIOGENESIS | -1.73 |
| GO0002084 PROTEIN DEPALMITOYLATION | -1.73 |
| GO0032429 REGULATION OF PHOSPHOLIPASE A2 ACTIVITY | -1.73 |
| GO0051181 COFACTOR TRANSPORT | -1.73 |
| GO0019227 ACTION POTENTIAL PROPAGATION | -1.73 |
| GO0043486 HISTONE EXCHANGE | -1.74 |
| GO0014059 REGULATION OF DOPAMINE SECRETION | -1.74 |
| GO0048536 SPLEEN DEVELOPMENT | -1.75 |
| GO0042473 OUTER EAR MORPHOGENESIS | -1.77 |
| GO0045918 NEGATIVE REGULATION OF CYTOLYSIS | -1.79 |
| GO0001971 NEGATIVE REGULATION OF ACTIVATION OF MEM | -1.79 |
| GO0015888 THIAMIN TRANSPORT | -1.79 |
| GO0042147 RETROGRADE TRANSPORT ENDOSOME TO GOLGI | -1.80 |
| GO0007368 DETERMINATION OF LEFT OR RIGHT SYMMETRY | -1.81 |
| GO0045226 EXTRACELLULAR POLYSACCHARIDE BIOSYNTHETI | -1.81 |
| GO0000380 ALTERNATIVE NUCLEAR MRNA SPLICING VIA S | -1.82 |
| GO0051154 NEGATIVE REGULATION OF STRIATED MUSCLE C | -1.83 |
| GO0000173 INACTIVATION OF MAPK ACTIVITY DURING OSM | -1.84 |
| GO0001919 REGULATION OF RECEPTOR RECYCLING | -1.85 |
| GO0051252 REGULATION OF RNA METABOLIC PROCESS | -1.85 |
| GO0006657 CDP CHOLINE PATHWAY | -1.85 |
| GO0006663 PLATELET ACTIVATING FACTOR BIOSYNTHETIC | -1.85 |
| GO0050929 INDUCTION OF NEGATIVE CHEMOTAXIS | -1.88 |
| GO0007509 MESODERM MIGRATION | -1.88 |
| GO0009156 RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHETI | -1.89 |
| GO0043069 NEGATIVE REGULATION OF PROGRAMMED CELL D | -1.91 |
| GO0001953 NEGATIVE REGULATION OF CELL MATRIX ADHES | -1.92 |
| GO0045768 POSITIVE REGULATION OF ANTI APOPTOSIS | -1.93 |
| GO0042092 T HELPER 2 TYPE IMMUNE RESPONSE | -1.93 |
| GO0031647 REGULATION OF PROTEIN STABILITY | -1.97 |
| GO0046856 PHOSPHOINOSITIDE DEPHOSPHORYLATION | -1.97 |
| GO0051895 NEGATIVE REGULATION OF FOCAL ADHESION FO | -1.97 |
| GO0006099 TRICARBOXYLIC ACID CYCLE | -1.97 |
| GO0006672 CERAMIDE METABOLIC PROCESS | -1.97 |
| GO0040016 EMBRYONIC CLEAVAGE | -2.01 |
| GO0006694 STEROID BIOSYNTHETIC PROCESS | -2.02 |
| GO0001764 NEURON MIGRATION | -2.04 |
| GO0009220 PYRIMIDINE RIBONUCLEOTIDE BIOSYNTHETIC P | -2.06 |
| GO0048172 REGULATION OF SHORT TERM NEURONAL SYNAPT | -2.07 |
| GO0051966 REGULATION OF SYNAPTIC TRANSMISSION GLU | -2.07 |
| GO0006907 PINOCYTOSIS | -2.08 |
| GO0001834 TROPHECTODERMAL CELL PROLIFERATION | -2.08 |
| GO0042094 INTERLEUKIN 2 BIOSYNTHETIC PROCESS | -2.09 |
| GO0051205 PROTEIN INSERTION INTO MEMBRANE | -2.11 |
| GO0035108 LIMB MORPHOGENESIS | -2.12 |
| GO0043517 POSITIVE REGULATION OF DNA DAMAGE RESPON | -2.12 |
| GO0006556 S ADENOSYLMETHIONINE BIOSYNTHETIC PROCES | -2.13 |
| GO0050760 NEGATIVE REGULATION OF THYMIDYLATE SYNTH | -2.13 |
| GO0006397 MRNA PROCESSING | -2.14 |
| GO0016126 STEROL BIOSYNTHETIC PROCESS | -2.15 |
| GO0001947 HEART LOOPING | -2.17 |
| GO0046883 REGULATION OF HORMONE SECRETION | -2.18 |
| GO0030521 ANDROGEN RECEPTOR SIGNALING PATHWAY | -2.18 |
| GO0007178 TRANSMEMBRANE RECEPTOR PROTEIN SERINE OR | -2.18 |
| GO0006370 MRNA CAPPING | -2.19 |
| GO0007252 I KAPPAB PHOSPHORYLATION | -2.21 |
| GO0048566 EMBRYONIC GUT DEVELOPMENT | -2.21 |
| GO0042412 TAURINE BIOSYNTHETIC PROCESS | -2.23 |
| GO0000097 SULFUR AMINO ACID BIOSYNTHETIC PROCESS | -2.23 |
| GO0046439 L CYSTEINE METABOLIC PROCESS | -2.23 |
| GO0044267 CELLULAR PROTEIN METABOLIC PROCESS | -2.29 |
| GO0006281 DNA REPAIR | -2.30 |
| GO0001937 NEGATIVE REGULATION OF ENDOTHELIAL CELL | -2.31 |
| GO0048873 HOMEOSTASIS OF NUMBER OF CELLS WITHIN A | -2.32 |
| GO0006106 FUMARATE METABOLIC PROCESS | -2.32 |
| GO0042471 EAR MORPHOGENESIS | -2.34 |
| GO0035094 RESPONSE TO NICOTINE | -2.34 |
| GO0016998 CELL WALL CATABOLIC PROCESS | -2.41 |
| GO0045110 INTERMEDIATE FILAMENT BUNDLE ASSEMBLY | -2.42 |
| GO0000910 CYTOKINESIS | -2.42 |
| GO0009968 NEGATIVE REGULATION OF SIGNAL TRANSDUCTI | -2.43 |
| GO0002063 CHONDROCYTE DEVELOPMENT | -2.44 |
| GO0045749 NEGATIVE REGULATION OF S PHASE OF MITOTI | -2.45 |
| GO0043516 REGULATION OF DNA DAMAGE RESPONSE SIGNA | -2.48 |
| GO0002072 OPTIC CUP MORPHOGENESIS INVOLVED IN CAME | -2.48 |
| GO0050674 UROTHELIAL CELL PROLIFERATION | -2.49 |
| GO0050677 POSITIVE REGULATION OF UROTHELIAL CELL P | -2.49 |
| GO0030177 POSITIVE REGULATION OF WNT RECEPTOR SIGN | -2.49 |
| GO0007067 MITOSIS | -2.55 |
| GO0015819 LYSINE TRANSPORT | -2.56 |
| GO0006396 RNA PROCESSING | -2.56 |
| GO0007217 TACHYKININ SIGNALING PATHWAY | -2.57 |
| GO0006374 NUCLEAR MRNA SPLICING VIA U2 TYPE SPLICE | -2.57 |
| GO0030538 EMBRYONIC GENITALIA MORPHOGENESIS | -2.59 |
| GO0048619 EMBRYONIC HINDGUT MORPHOGENESIS | -2.59 |
| GO0015031 PROTEIN TRANSPORT | -2.63 |
| GO0000070 MITOTIC SISTER CHROMATID SEGREGATION | -2.68 |
| GO0030949 POSITIVE REGULATION OF VASCULAR ENDOTHEL | -2.77 |
| GO0048260 POSITIVE REGULATION OF RECEPTOR MEDIATED | -2.86 |
| GO0006086 ACETYL COA BIOSYNTHETIC PROCESS FROM PYR | -2.88 |
| GO0006399 TRNA METABOLIC PROCESS | -3.03 |
| GO0006512 UBIQUITIN CYCLE | -3.05 |
| GO0022008 NEUROGENESIS | -3.18 |
| GO0051301 CELL DIVISION | -3.26 |
| GO0007214 GAMMA AMINOBUTYRIC ACID SIGNALING PATHWA | -3.33 |
| GO0048557 EMBRYONIC DIGESTIVE TRACT MORPHOGENESIS | -3.37 |
| GO0007113 ENDOMITOTIC CELL CYCLE | -3.68 |
| GO0048483 AUTONOMIC NERVOUS SYSTEM DEVELOPMENT | -5.63 |
Figure 1.
Effects of mTBI on CNS, Inflammation and Age related Molecular Function, Cellular Component and Biological Process Gene Ontology (GOs) Z-scores are presented. Down regulated (green) and up regulated (red) GOs are illustrated. A: CNS MFGOs of note include ‘apolipoprotein receptor binding’; ‘tau protein binding’ and ‘GABA A receptor activity’. Inflammation MFGOs of note include ‘leukotriene C4 synthase activity’; ‘prostaglandin I synthase activity’ and ‘phospholipase inhibitor activity’. CNS CCGOs of note include ‘myelin sheath’ and ‘neuron projection’. B: CNS BPGOs of note include ‘negative regulation of neurogenesis’; ‘axonogenesis’; ‘myelination in the CNS’; ‘neuron migration’; ‘neurogenesis’ and ‘autonomic nervous system development’. Inflammation BPGOs of note include ‘acute phase response’; ‘glial cell fate determination’ and ‘I kappa B phosphorylation’. One additional BPGO of interest was ‘aging’, which was also observed to be upregulated by mTBI.
3.2.1. CNS related gene ontologies
The largest majority of regulated MFGOs were associated with neurotransmitter receptor mediated processes related to neuronal physiology and function, most had negative Z-scores yet some had positive Z-scores (Figure 1A). For example ‘glutamate decarboxylase activity’, ‘GABA A receptor activity’, ‘neurotransmitter sodium symporter activity’, ‘neurotransmitter receptor activity’, ‘D3 dopamine receptor binding’. Changes in 2 MFGOs involved in the sense of smell were also observed, they were ‘olfactory receptor binding’, ‘olfactory receptor activity’. A number of MFGOs were linked with neurodegenerative processes, namely Alzheimer’s disease. These were ‘tau protein binding’ and ‘apolipoprotein receptor binding’, which both had positive Z-scores, and ‘NEDD8 activating enzyme activity’ that had a negative Z-score. The full list of mTBI regulated MFGOs with Z-scores are indicated in Figure 1A. Several mTBI regulated CCGOs were related to neuron physiology and function as well as neuronal plasticity. These were ‘myelin sheath’, ‘neuron projection’ and ‘synaptic vesicle membrane’, which had positive Z-scores, and ‘nicotinic acetylcholine gated receptor’ and ‘NMDA selective glutamate receptor complex’ that had negative Z-scores. These Z-scores are indicated in Figure 1A. Interestingly, the ‘NMDA selective glutamate receptor complex’ may be of relevance to Alzheimer’s disease. Numerous BPGOs were observed to be regulated. A series of BPGOs of particular interest to neurodegeneration were associated with mTBI-induced inhibition of neurogenesis, these BPGOs were: ‘neuron migration’ and ‘neurogenesis’, which had negative Z-scores, and ‘negative regulation of neurogenesis’ that had a positive Z-score. Many others were associated with neurotransmitters and neuronal physiology, function and plasticity, for example ‘regulation of synaptic growth’, ‘myelination in the CNS’, ‘synaptic transmission dopaminergic’, ‘synaptic vesicle maturation’ and, ‘dopamine uptake’, all of which had positive Z-scores. Others that had negative Z-scores were ‘regulation of dopamine secretion’, ‘regulation of short-term neuronal synaptic plasticity’ and ‘regulation of synaptic transmission, glutamergic’. A complete list of BPGOs and Z-scores are presented in Figure 1B.
3.2.2. Inflammation related gene ontologies
Numerous inflammation related MFGOs were observed. Several of the MFGOs were linked with arachidonic acid metabolism and included ‘leukotriene C4 synthase activity’ and ‘prostaglandin I synthase activity’, which had positive Z-scores. MFGOs linked with cell mediated endocytosis were ‘scavenger receptor activity’ and ‘MHC class II protein binding’ that were ontologies with positive Z-scores Cytokine signaling event MFGOs included ‘cytokine activity’, which had a positive Z-score, and ‘I kappa B kinase activity’ and ‘CXC chemokine receptor activity’ both of which had negative Z-scores. A complete list of MFGOs with appropriate Z-scores is provided in Figure 1A. mTBI-regulated BPGOs were linked with metabolic processes that generate interleukins and involved ‘regulation of interleukin-6 biosynthetic process’, which had a positive Z-score, as well as ‘IL-13 biosynthetic process’ and ‘IL-2 biosynthetic process’ that both had negative Z-scores. Several BPGOs were related to the regulation of T helper cells and included ‘T helper 1 cell differentiation’, which had a positive Z-score, as well as ‘negative regulation of T helper 2 cell differentiation’ and ‘T helper 2 type immune response’ that both had negative Z-scores. Cytokine signaling process BPGOs were also regulated and comprised ‘acute phase response’, ‘positive chemotaxis’ and ‘positive regulation of INF-γ production’, which all had positive Z-scores, in addition to ‘I kappa B phosphorylation’ that had a negative Z-score. The complete list of inflammation related BPGOs with appropriate Z-scores is presented in Figure 1B.
3.3. Molecular pathways derived from mTBI regulated hippocampal tissue genes
Regulated genes underwent additional analysis by the Ingenuity Pathway Analysis tool to identify molecular pathways. At two weeks post-mTBI many molecular pathways were regulated. The pathways are summarized in Table 3, with the pathway name and Z-scores provided. Several of the observed pathways were associated with the CNS or inflammation, and are illustrated in Figure 2.
Table 3.
mTBI regulated molecular pathways identified in hippocampal tissues 14 days after injury
| Up-regulated Z-score | |
|---|---|
| Pathway | Z-score |
| RIBOSOMAL PROTEINS | 6.01 |
| ELECTRON TRANSPORT CHAIN | 5.45 |
| TARTE MATURE PC | 5.27 |
| MOOTHA VOXPHOS | 4.92 |
| INNEREAR UP | 4.26 |
| ZUCCHI EPITHELIAL DN | 4.14 |
| UVB NHEK1 UP | 3.84 |
| IFN BETA GLIOMA DN | 3.83 |
| FALT BCLL UP | 3.78 |
| REOVIRUS HEK293 DN | 3.62 |
| DAC PANC UP | 3.50 |
| WALLACE JAK2 DIFF | 3.43 |
| OXIDATIVE PHOSPHORYLATION | 3.41 |
| SANSOM APC 5 DN | 3.39 |
| HIPPOCAMPUS DEVELOPMENT POSTNATAL | 3.30 |
| MOREAUX TACI HI VS LOW UP | 3.29 |
| REFRACTORY GASTRIC UP | 3.28 |
| CIS RESIST LUNG DN | 3.28 |
| PROSTAGLANDIN AND LEUKOTRIENE METABOLISM | 3.27 |
| UVB NHEK2 UP | 3.09 |
| BCNU GLIOMA MGMT 48HRS DN | 3.05 |
| UVB NHEK1 C4 | 2.92 |
| SHEPARD GENES COMMON BW CB MO | 2.91 |
| STRESS IONIZING SPECIFIC UP | 2.91 |
| UCALPAINPATHWAY | 2.91 |
| ALZHEIMERS DISEASE UP | 2.90 |
| NOS1 PATHWAY | 2.88 |
| NADLER OBESITY UP | 2.85 |
| ATRIA UP | 2.84 |
| INOS ALL DN | 2.84 |
| ICHIBA GVHD | 2.83 |
| BREAST CANCER ESTROGEN SIGNALING | 2.76 |
| BCNU GLIOMA NOMGMT 48HRS DN | 2.74 |
| GUO HEX DN | 2.73 |
| AKAP CENTROSOME PATHWAY | 2.72 |
| AGUIRRE PANCREAS CHR19 | 2.68 |
| HPV31 DN | 2.62 |
| IL6 FIBRO UP | 2.54 |
| FETAL LIVER VS ADULT LIVER GNF2 | 2.50 |
| TGF BETA C4 UP | 2.46 |
| KLEIN PEL DN | 2.44 |
| UBIQUINONE BIOSYNTHESIS | 2.40 |
| MPR PATHWAY | 2.37 |
| INFLAMMATION PATHWAY | 2.36 |
| ST GAQ PATHWAY | 2.34 |
| CHESLER BRAIN NEURAL HIGH GENES | 2.34 |
| LIAN MYELOID DIFF TF | 2.32 |
| ST MYOCYTE AD PATHWAY | 2.29 |
| UVC TTD 4HR UP | 2.28 |
| ABRAHAM MM VS AL DN | 2.27 |
| BCRABL HL60 AFFY DN | 2.26 |
| FERRANDO TAL1 NEIGHBORS | 2.24 |
| PASSERINI OXIDATION | 2.22 |
| SIG BCR SIGNALING PATHWAY | 2.18 |
| HEART FAILURE ATRIA UP | 2.17 |
| BLOOD CLOTTING CASCADE | 2.16 |
| MOREAUX TACI XG 13 DN | 2.15 |
| HANSON NFKAPPB IND | 2.14 |
| ABRAHAM AL VS MM UP | 2.13 |
| PGC1A PATHWAY | 2.13 |
| WNT PATHWAY | 2.10 |
| SARCOMAS SCHWANNOMA UP | 2.09 |
| POD1 KO UP | 2.08 |
| MITOCHONDRIA | 2.06 |
| LAL KO 6MO UP | 2.06 |
| CSK PATHWAY | 2.05 |
| AMI PATHWAY | 2.05 |
| XU CBP DN | 2.05 |
| MTA3 PATHWAY | 2.01 |
| BRENTANI HORMONAL FUNCTION | 2.01 |
| ROS MOUSE AORTA UP | 1.98 |
| GALE FLT3ANDAPL DN | 1.94 |
| CK1 PATHWAY | 1.90 |
| UVB NHEK4 24HRS DN | 1.84 |
| YANG OSTECLASTS SIG | 1.79 |
| BRENTANI IMMUNE FUNCTION | 1.72 |
| AKAP13 PATHWAY | 1.70 |
| ADIPOCYTE PPARG UP | 1.68 |
| YAO P4 KO VS WT DN | 1.64 |
| CROONQUIST IL6 RAS DN | 1.64 |
| TPA SKIN DN | 1.63 |
| Down-regulated Z-score | |
| Pathway | Z-score |
| CHEOK HDMTX DN | -1.62 |
| SARS PATHWAY | -1.72 |
| CELL CYCLE CHECKPOINT | -1.80 |
| MMS HUMAN LYMPH LOW 4HRS DN | -1.84 |
| HDACI COLON BUT30MIN DN | -2.01 |
| AGEING LYMPH DN | -2.05 |
| MANALO HYPOXIA DN | -2.06 |
| ROME INSULIN 2F UP | -2.07 |
| ALZHEIMERS INCIPIENT DN | -2.10 |
| HUMAN TISSUE TESTIS | -2.18 |
| OLD WERNER FIBRO UP | -2.22 |
| BOQUEST CD31PLUS VS CD31MINUS UP | -2.26 |
| NI2 MOUSE UP | -2.28 |
| UVB SCC UP | -2.30 |
| STOSSI ER UP | -2.37 |
| UVC HIGH D3 DN | -2.46 |
| UVC TTD-XPCS COMMON DN | -2.56 |
| IGF VS PDGF DN | -2.59 |
| CROMER HYPOPHARYNGEAL MET VS NON UP | -2.60 |
| UVB NHEK3 ALL | -2.64 |
| GH GHRHR KO 24HRS UP | -2.66 |
| UVC TTD ALL DN | -2.79 |
| UVB NHEK3 C5 | -2.81 |
| VALINE LEUCINE AND ISOLEUCINE BIOSYNTHESIS | -2.82 |
| CHEN HOXA5 TARGETS UP | -2.87 |
| DIAB NEPH DN | -2.88 |
| BLEO MOUSE LYMPH LOW 24HRS DN | -2.92 |
| UVB NHEK2 DN | -3.01 |
| YAGI AML PROG FAB | -3.06 |
| BRCA BRCA1 POS | -3.08 |
| ET743 SARCOMA 24HRS DN | -3.11 |
| VHL NORMAL UP | -3.11 |
| SANA TNFA ENDOTHELIAL DN | -3.12 |
| CHOLESTEROL BIOSYNTHESIS | -3.16 |
| IDX TSA DN CLUSTER6 | -3.31 |
| LEE T CELLS1 UP | -3.34 |
| LEE T CELLS10 UP | -3.34 |
| LEE T CELLS8 UP | -3.34 |
| ZHAN MM CD1 VS CD2 UP | -3.34 |
| CMV HCMV TIMECOURSE 24HRS DN | -3.35 |
| UVC TTD 4HR DN | -3.35 |
| BRCA1KO MEF DN | -3.42 |
| REOVIRUS HEK293 UP | -3.45 |
| TARTE PLASMA BLASTIC | -3.49 |
| HSC LATEPROGENITORS SHARED | -3.63 |
| HSC LATEPROGENITORS ADULT | -3.64 |
| HSC LATEPROGENITORS FETAL | -3.72 |
| BRCA PROGNOSIS NEG | -3.73 |
| UVC HIGH ALL DN | -3.81 |
| RCC NL UP | -3.82 |
| ET743 SARCOMA 72HRS DN | -3.85 |
| STEMCELL COMMON UP | -3.89 |
| LEE T CELLS2 UP | -3.95 |
| GH GHRHR KO 24HRS DN | -4.03 |
| KREBS TCA CYCLE | -4.13 |
| UVC XPCS 4HR DN | -4.16 |
| FLECHNER KIDNEY TRANSPLANT REJECTION DN | -4.16 |
| ET743 SARCOMA 48HRS DN | -4.16 |
| UVC XPCS 8HR DN | -4.48 |
| ET743 SARCOMA DN | -4.97 |
| ALZHEIMERS DISEASE DN | -5.25 |
| UVC XPCS ALL DN | -5.40 |
| POD1 KO DN | -5.90 |
| FLECHNER KIDNEY TRANSPLANT WELL UP | -6.68 |
| STEMCELL EMBRYONIC UP | -10.99 |
| STEMCELL NEURAL UP | -11.37 |
Figure 2.
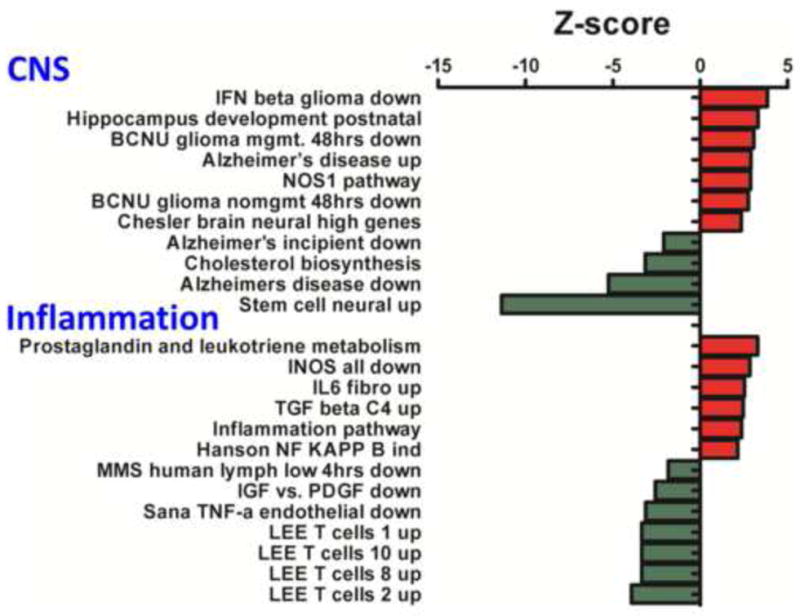
Effects of mTBI on CNS and Inflammation related Molecular Pathways Z-scores are presented. Down regulated (green) and up regulated (red) GOs are illustrated. CNS pathways of note include ‘Alzheimer’s disease up and down’ and ‘Alzheimer’s incipient down’. Inflammation pathways of note include ‘prostaglandin and leukotriene metabolism’; ‘iNOS all down’ and ‘inflammation pathway’ and ‘Sana TNF-α endothelial down’.
3.3.1. CNS and inflammation related molecular pathways
Key CNS molecular pathways were related to neurodegenerative disease, namely Alzheimer’s disease. Those pathways were ‘Alzheimer’s disease up’, which had a positive Z-score, and ‘Alzheimer’s incipient down’ and ‘Alzheimer’s disease down’ that both had negative Z-scores. An additional pathway of high relevance to neurogenesis observed to be regulated was ‘stem cell neural up’ with a negative Z-score. Inflammation related pathways regulated by mTBI included one related to arachidonic acid metabolism, ‘prostaglandin and leukotriene metabolism’, which had a positive Z-score, and several had relevance to T cell function, and comprised ‘Lee T cells 1 up’, ‘Lee T cells 10 up’, ‘Lee T cells 8 up’ and ‘Lee T cells 2 up’ that all had negative Z-scores. Finally, several pathways associated with cytokine related signaling events were evident; specifically, ‘IL-6 fibro up’, ‘Hanson NF kappa B ind’ and ‘inflammation pathway’ that all had positive Z-scores. Each of the CNS and inflammation pathways observed to be regulated by mTBI are indicated Figure 2, and appropriate Z-scores are shown.
4. Discussion
A thorough discussion of all TBI-induced gene regulations and their associated functions is beyond the limits of this article; we have therefore focused on key physiological/pathological areas of interest to us, namely processes involving neuronal function and inflammation. Using the same methodologies other areas of interest of equal scientific importance could readily be evaluated. Prior studies using the same mouse weight drop model of mTBI have illustrated long-lasting cognitive deficits (Zohar et al., 2003; Milman et al., 2005; Edut et al., 2011; Baratz et al., 2011). Parallel studies have illustrated that these cognitive deficits were associated with a diffuse form of injury showing an apoptotic and neurodegenerative pathology in several brain regions associated with learning and memory (Tashlykov et al., 2007; Tweedie et al., 2007; Tashlykov et al., 2009; Rachmany et al., 2013).
The use of cDNA microarray methods has proven to be a valuable tool for identifying subtle changes in cellular gene expressions, but their interpretation requires careful consideration of possible confounding factors that relate to the original source of the RNA sample. Typically, these studies are performed on intact brain tissues and not, exclusively neuronal cells. Herein, total RNA was generated from hippocampal tissue and hence it is very probable that numerous changes in array probes identified in these studies reflect changes in non-neuron cell types in addition to hippocampal neuron cells. Significant sources of non-neuronal transcripts will include those from microglia and astrocytes, the brain resident immune cells. Using methodologies of the current study, it is not possible to distinguish between changes in neuronal and/or glial cell transcripts due to the abundance and co-localization of glial and astrocytes with neuronal cells and the intricate relationship of these cells with neurons and neuron physiology (Azevedo et al., 2009; Lent et al., 2012; Papa et al., 2014).
The present study, importantly, highlights physiological and pathological events downstream of mTBI that were determined by the use of a stepwise analysis of gene transcripts from the level of a single regulated gene or a set of genes; to a large number of functional gene ontologies derived from observations in our samples and known, published gene function databases (PAGE analysis, Table 2). More specifically we have been able to identify many diverse mTBI-induced functional changes in inflammatory and neurological systems in the hippocampal tissue (Figure 1A/B). Furthermore, analysis of the genes with the Ingenuity Pathway Analysis tool (Ingenuity Systems, Inc., Redwood City, CA, USA) identified a host of TBI regulated molecular pathways (Table 3), several of which had relevance to inflammation and neuronal physiological and pathological processes (Figure 2). These GOs and molecular pathways likely play a key role in the observed apoptotic/neurodegenerative processes that underpin the cognitive deficits in this model, and provides a platform to help to define candidate cellular processes in TBI to aid in the development of therapeutic agents.
A prior study using this same animal model likewise documented altered brain tissue gene regulations (Israelsson et al., 2009), evaluated by microarray (neocortex assessed on day 3 after injury) and quantitative RT-PCR (q-RT-PCR) analysis methods (4 hours up to 7 days after injury). Data indicated numerous changes in inflammatory genes at several time points earlier than the one examined in the present study (Israelsson et al., 2009). Israelsson and co-workers described up-regulated genes associated with inflammation at 3 days after injury in ipsilateral cortical tissues. These results are in accord with and extended their prior studies of mice challenged with controlled cortical impact TBI in which time-dependent gene expression changes in proinflammatory chemokines and cytokines were similarly evident (Israelsson et al., 2008). This indicates, that across different forms of TBI, secondary injury chiefly involves inflammatory processes and chemokine signaling that can then drive both regenerative and/or neurodegenerative processes. Findings from our study corroborate those of Israelsson, in the identification of numerous inflammation related GO classifications and molecular pathways regulated by a single mTBI event (Figure 1A/B and 2). The present study in accord with others (Israelsson et al., 2008, 2009) highlights inflammation as a promising target for therapeutic intervention, which has been validated by Baratz and co-workers. Baratz used a small molecule drug protein synthesis inhibitor of TNF-alpha, the pro-inflammatory early phase cytokine, as a pre-injury and post-injury treatment and observed benefits to TBI-induced behavioral deficits, accompanied by mitigation of neuronal loss and neuroinflammation, with a drug therapeutic window of up to 12 hours after the injury (Baratz et al., 2011; 2015). Interestingly numerous models of diverse forms of TBI implicate inflammation as a favorable target for drug development (Kovesdi et al., 2012; Naseem and Parvez, 2014; Tuttolomondo et al., 2014; Bergold 2015); however, the key question has related to the choice of the specific element within the process to best inhibit.
The identification of various Alzheimer’s disease (AD) GOs (GO0050749 apolipoprotein receptor binding, GO0048156 tau protein binding and GO0019781 NEDD8 activating enzyme activity) and molecular pathways (Alzheimer’s disease up and down and Alzheimer’s incipient down) regulated by TBI in our study, may suggest that agents that show efficacy in AD models may possess benefits in the setting of TBI. Thus far, as for TBI, therapeutic approaches focused to treat AD progression have proved unsuccessful (Becker et al., 2010, 2014, 2015), albeit symptomatic agents are of some value. This has, in part, been shown to be the case in TBI too. Donepezil a U.S. Food and Drug Administration (FDA) approved anticholinesterase for the treatment of AD has been found to be moderately beneficial in improving memory deficits in several clinical studies and in some, but not all, experimental models of TBI (Taverni et al., 1998; Whelan et al., 2000; Masanic et al., 2001; Balesteros et al., 2008; Fujiki et al., 2008; Shaw et al., 2013; Yu et al., 2015). The N-methyl-d-aspartate antagonist memantine, a further FDA-approved drug for the symptomatic treatment of AD, has similarly been shown to be neuroprotective and to possess anti-lipid peroxidation properties in several preclinical studies involving experimentally induced TBI in rodents (Rao et al., 2001; Ozsuer et al., 2005). In contrast, other animal studies have reported injury exacerbation (Ikonomidou et al., 2000), and hence proposed caution in its use. Clinical trials of memantine in TBI are ongoing (ClinicalTrials.gov Identifier: NCT02240589), and thus evaluation of its efficacy will have to be awaited. A concern with such symptomatic agents is whether potential improvements in neuropsychological measures are long lasting, impacting neurodegenerative processes, or are merely acute and restricted to the period of drug dosing (e.g., symptomatic). Consequent to a number of shared mechanisms that underpin the progressive degenerative disorders of type 2 diabetes mellitus (T2DM) and AD, several approved drugs effective and well tolerated in the former are being evaluated in the latter, and more recently in TBI. As an example, the incretin mimetic exendin-4 that functions as a long-acting glucagon-like peptide-1 agonist (Salcedo et al., 2012; Greig et al., 2014; Holscher 2014) has neurotrophic and neuroprotective actions in animal models of AD and Parkinson’s disease (Perry et al., 2002; Perry 2003; Li et al., 2009; Li et al., 2010; Wang et al., 2015; Xu et al., 2015), and has been shown to be beneficial across multiple concussive and blast models of TBI (Rachmany et al., 2013a; Eakin et al., 2013; Tweedie et al., 2013b; 2015). These studies have been cross-validated by use of the clinically approved incretin mimetic liraglutide (Li et al., 2015), further promoting interest in evaluation of this drug class in TBI. More recently Kondo and co-workers (Kondo et al., 2015) identified an early marker of neurodegeneration (cis phosporylated-tau) that is induced by TBI and has a direct role in the formation of neurofibrillary tangles, a hallmark feature of AD (Nakamura et al., 2012) as well as TBI associated chronic traumatic encephalopathy. More importantly, they illustrated the utility of an antibody treatment directed against cis P-tau in preventing TBI-induced pathology (Kondo et al., 2015). The occurrence of these GOs and pathways and the work of Kondo, in large part support the claims of Barnes, Danshevar, Gupta and Sen and co-workers of a TBI-induced increased risk of developing neurodegenerative diseases later in life (Barnes et al., 2014; Danshevar et al., 2015; Gupta and Sen 2015).
As the direct relationship between mRNA levels and protein levels in biological systems is complex (Gry et al., 2009; Maier eta al., 2009), the next logical step to adopt in studying data derived from cDNA microarray platforms, as described herein, involves a proteomic approach. Parallel proteomic assessments of tissues should help to advance our understanding of the consequences of TBI on brain physiology and pathology at the protein level. An analysis of protein transcript probes identified by microarray methods can be undertaken on several levels. Typically the following methods have been applied in an attempt to identify ‘bio-markers’ of TBI in plasma and cerebral spinal fluid, however, the same techniques can be used to identify TBI-induced physiological/pathological protein alterations. Individual proteins can be assessed in tissues by Western Blot where likely protein(s) of interest identified from clinical observations can be assessed in rodent models (Harris et al., 2009; Siman et al., 2009; Kabadi et al., 2012). Additional forms of analysis could employ a multiple protein target analysis using a multiplex ELISA method or protein array chips (Buttram et al., 2007; Mukherjee et al., 2011; Gyorgy et al., 2010; Kovesdi et al., 2012). For a yet larger scale detection of proteins changed by TBI, one could employ a mass spectrometry analysis approach (Ding et al., 2015; Sheth et al., 2015; Walls et al., 2015).
4.1. Conclusions
Numerous cellular processes were identified to be regulated by a single mild TBI event 14 days after injury. These processes create a platform for drug development for application to and evaluation in human disease. Based upon this and other studies, neurological and, perhaps more so, inflammatory processes likely mediate the cognitive deficits and secondary neuronal loss associated with models of TBI. Recent studies using anti-apoptotic agents suggest that secondary neuronal cell death can be halted (Rachmany et al., 2013b; Yang et al., 2015) and cognitive impairments thereby mitigated. These observations point towards a focused investigation of anti-inflammatory as well as type 2 diabetes mellitus and AD medicines for clinical utility in TBI. Future studies addressing TBI altered protein levels in brain will further aid in the validation of these therapeutic approaches for the treatment of human TBI.
Highlights.
A single mild concussive traumatic brain injury in mice is associated with many changes in hippocampal gene expressions.
Changes in hippocampal genes were associated with inflammation and neurological functional gene ontology and molecular pathways.
Several of the altered gene ontology and molecular pathways were associated with neurodegenerative processes, namely Alzheimers Disease.
The identification of altered inflammation and neurological processes following mild traumatic brain injury will aid in the development of drugs to treat clinical brain injury.
Acknowledgments
This research was supported in part by (i) the Intramural Research Program of the National Institute on Aging, National Institutes of Health, (ii) the Sackler School of Medicine, Tel-Aviv University, (iii) the Ari and Regine Aprijaskis Fund at Tel-Aviv University, (iv) a grant from the Israel Science Foundation, grant number 108/09, (v) and the Joseph Sagol fellowships and (vi) the National Science Council of Taiwan. This work was performed in partial fulfillment of the requirements for a Ph.D. degree of Lital Rachmany, Sackler Faculty of Medicine, Tel Aviv University, Israel.
Abbreviations
- TBI
traumatic brain injury
- mTBI
mild traumatic brain injury
- ICR
Institute for Cancer Research mouse
- AD
Alzheimer’s disease
- CNS
central nervous system
- cDNA
(cDNA) complementary DNA
- cRNA
complementary RNA
- PAGE
Parametric Analysis of Gene Set Enrichment
- GO
gene ontology term
- q-RT-PCR
quantitative reverse-polymerase chain reaction
- FDA
U.S. Food and Drug Administration
Footnotes
None of the authors have competing interests in the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- Ballesteros J, Güemes I, Ibarra N, Quemada JI. The effectiveness of donepezil for cognitive rehabilitation after traumatic brain injury: a systematic review. J Head Trauma Rehabil. 2008;23:171–180. doi: 10.1097/01.HTR.0000319935.99837.96. [DOI] [PubMed] [Google Scholar]
- Barkhoudarian G, Hovda DA, Giza CC. The molecular pathophysiology of concussive brain injury. Clini Sports Med. 2011;30:33–48. doi: 10.1016/j.csm.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Kaup A, Kirby KA, Byers AL, Diaz-Arrastia R, Yaffe K. Traumatic brain injury and risk of dementia in older veterans. Neurology. 2014;83:312–319. doi: 10.1212/WNL.0000000000000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Greig NH. Lost in translation: neuropsychiatric drug development. Sci Transl Med. 2010;2:61rv6. doi: 10.1126/scitranslmed.3000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Greig NH, Giacobini E, Schneider LS, Ferrucci L. A new roadmap for drug development for Alzheimer’s disease. Nat Rev Drug Discov. 2014;13:156. doi: 10.1038/nrd3842-c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Seeman MV, Greig NH, Lahiri DK. What can triumphs and tribulations from drug research in Alzheimer’s disease tell us about the development of psychotropic drugs in general? Lancet Psychiatry. 2015;2:756–764. doi: 10.1016/S2215-0366(15)00214-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergold PJ. Treatment of traumatic brain injury with anti-inflammatory drugs. Exp Neurol. 2015;23 doi: 10.1016/j.expneurol.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttram SD, Wisniewski SR, Jackson EK, Adelson PD, Feldman K, Bayir H, Berger RP, Clark RS, Kochanek PM. Multiplex assessment of cytokine and chemokine levels in cerebrospinal fluid following severe pediatric traumatic brain injury: effects of moderate hypothermia. J Neurotrauma. 2007;24:1707–1717. doi: 10.1089/neu.2007.0349. [DOI] [PubMed] [Google Scholar]
- Choi DW, Mauluccigedde M, Kriegstein AR. Glutamate neurotoxicity in cortical cell-culture. J Neurosci. 1987;7:357–368. doi: 10.1523/JNEUROSCI.07-02-00357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memantine for neuroprotection and cognitive enhancement following traumatic brain injury. ClinicalTrials.gov Identifier: NCT02240589. Initiated 2014 < https://clinicaltrials.gov/ct2/show/NCT02240589>.
- Cornelius C, Crupi R, Calabrese V, Graziano A, Milone P, Pennisi G. Traumatic brain injury: oxidative stress and neuroprotection. Antioxid Redox Signal. 2013;19:836–853. doi: 10.1089/ars.2012.4981. [DOI] [PubMed] [Google Scholar]
- Daneshvar DH, Goldstein LE, Kiernan PT, Stein TD, McKee AC. Post-traumatic neurodegeneration and chronic traumatic encephalopathy. Mol Cell Neurosci. 2015;66:81–90. doi: 10.1016/j.mcn.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Ding J, Ding Z, Yuan F, Guo J, Chen H, Gao W, Wang R, Gu Y, Chen J, Guo Y, Tian H. Proteomics analysis after traumatic brain injury in rats: the search for potential biomarkers. Arq Neuropsiquiatr. 2015;73:342–349. doi: 10.1590/0004-282X20150006. [DOI] [PubMed] [Google Scholar]
- Eakin K, Li Y, Chiang YH, Hoffer BJ, Rosenheim H, Greig NH, Miller JP. Exendin-4 ameliorates traumatic brain injury-induced cognitive impairment in rats. PLoS One. 2013;8:e82016. doi: 10.1371/journal.pone.0082016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edut S, Rubovitch V, Schreiber S, Pick CG. The intriguing effects of ecstasy (MDMA) on cognitive function in mice subjected to a minimal traumatic brain injury (mTBI) Psychopharmacology (Berl) 2011;214:877–889. doi: 10.1007/s00213-010-2098-y. [DOI] [PubMed] [Google Scholar]
- Faul M, Xu L, Wald MM, Coronado VG. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations and Deaths 2002–2006. Atlanta (GA): Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; 2010. [Google Scholar]
- Fujiki M, Kubo T, Kamida T, Sugita K, Hikawa T, Abe T, Ishii K, Kobayashi H. Neuroprotective and antiamnesic effect of donepezil, a nicotinic acetylcholine-receptor activator, on rats with concussive mild traumatic brain injury. J Clin Neurosci. 2008;15:791–796. doi: 10.1016/j.jocn.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Gómez PA, de-la-Cruz J, Lora D, Jiménez-Roldán L, Rodríguez-Boto G, Sarabia R, Sahuquillo J, Lastra R, Morera J, Lazo E, Dominguez J, Ibañez J, Brell M, de-la-Lama A, Lobato RD, Lagares A. Validation of a prognostic score for early mortality in severe head injury cases. J Neurosurg. 2014;121:1314–1322. doi: 10.3171/2014.7.JNS131874. [DOI] [PubMed] [Google Scholar]
- Greig NH, Tweedie D, Rachmany L, Li Y, Rubovitch V, Schreiber S, Chiang YH, Hoffer BJ, Miller J, Lahiri DK, Sambamurti K, Becker RE, Pick CG. Incretin mimetics as pharmacologic tools to elucidate and as a new drug strategy to treat traumatic brain injury. Alzheimers Dement. 2014;10(1 Suppl):S62–75. doi: 10.1016/j.jalz.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve MW, Zink BJ. Pathophysiology of traumatic brain injury. Mt Sinai J Med. 2009;76:97–104. doi: 10.1002/msj.20104. [DOI] [PubMed] [Google Scholar]
- Gry M, Rimini R, Strömberg S, Asplund A, Pontén F, Uhlén M, Nilsson P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Sen N. Traumatic brain injury: a risk factor for neurodegenerative diseases. Rev Neurosci. 2015 doi: 10.1515/revneuro-2015-0017. [DOI] [PubMed] [Google Scholar]
- Gyorgy AB, Walker J, Wingo D, Eidelman O, Pollard HB, Molnar A, Agoston DV. Reverse phase protein microarray technology in traumatic brain injury. J Neurosci Methods. 2010;192:96–101. doi: 10.1016/j.jneumeth.2010.07.029. [DOI] [PubMed] [Google Scholar]
- Hölscher C. Insulin, incretins and other growth factors as potential novel treatments for Alzheimer’s and Parkinson’s diseases. Biochem Soc Trans. 2014;42:593–599. doi: 10.1042/BST20140016. [DOI] [PubMed] [Google Scholar]
- Ikonomidou C, Stefovska V, Turski L. Neuronal death enhanced by N-methyl-D-aspartate antagonists. Proc Natl Acad Sci U S A. 2000;97:12885–12890. doi: 10.1073/pnas.220412197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson C, Bengtsson H, Kylberg A, Kullander K, Lewén A, Hillered L, Ebendal T. Distinct cellular patterns of upregulated chemokine expression supporting a prominent inflammatory role in traumatic brain injury. J Neurotrauma. 2008;25:959–974. doi: 10.1089/neu.2008.0562. [DOI] [PubMed] [Google Scholar]
- Israelsson C, Wang Y, Kylberg A, Pick CG, Hoffer BJ, Ebendal T. Closed head injury in a mouse model results in molecular changes indicating inflammatory responses. J Neurotrauma. 2009;26:1307–1314. doi: 10.1089/neu.2008.0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabadi SV, Stoica BA, Byrnes KR, Hanscom M, Loane DJ, Faden AI. Selective CDK inhibitor limits neuroinflammation and progressive neurodegeneration after brain trauma. J Cereb Blood Flow Metab. 2012;32:137–149. doi: 10.1038/jcbfm.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo A, Shahpasand K, Mannix R, Qiu J, Moncaster J, Chen CH, Yao Y, Lin YM, Driver JA, Sun Y, Wei S, Luo ML, Albayram O, Huang P, Rotenberg A, Ryo A, Goldstein LE, Pascual-Leone A, McKee AC, Meehan W, Zhou XZ, Lu KP. Antibody against early driver of neurodegeneration cis P-tau blocks brain injury and tauopathy. Nature. 2015;523:431–436. doi: 10.1038/nature14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korley FK, Kelen GD, Jones CM, Diaz-Arrastia R. Emergency Department Evaluation of Traumatic Brain Injury in the United States, 2009-2010. J Head Trauma Rehabil. 2015 doi: 10.1097/HTR.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovesdi E, Kamnaksh A, Wingo D, Ahmed F, Grunberg NE, Long JB, Kasper CE, Agoston DV. Acute minocycline treatment mitigates the symptoms of mild blast-induced traumatic brain injury. Front Neurol. 2012;3:111. doi: 10.3389/fneur.2012.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lent R, Azevedo FA, Andrade-Moraes CH, Pinto AV. How many neurons do you have? Some dogmas of quantitative neuroscience under revision. Eur J Neurosci. 2012;35:1–9. doi: 10.1111/j.1460-9568.2011.07923.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Perry T, Kindy MS, Harvey BK, Tweedie D, Holloway HW, Powers K, Shen H, Egan JM, Sambamurti K, Brossi A, Lahiri DK, Mattson MP, Hoffer BJ, Wang Y, Greig NH. GLP-1 receptor stimulation preserves primary cortical and dopaminergic neurons in cellular and rodent models of stroke and Parkinsonism. Proc Natl Acad Sci U S A. 2009;106:1285–1290. doi: 10.1073/pnas.0806720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Duffy KB, Ottinger MA, Ray B, Bailey JA, Holloway HW, Tweedie D, Perry T, Mattson MP, Kapogiannis D, Sambamurti K, Lahiri DK, Greig NH. GLP-1 receptor stimulation reduces amyloid-beta peptide accumulation and cytotoxicity in cellular and animal models of Alzheimer’s disease. J Alzheimers Dis. 2010;19:1205–1219. doi: 10.3233/JAD-2010-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bader M, Tamargo I, Rubovitch V, Tweedie D, Pick CG, Greig NH. Liraglutide is neurotrophic and neuroprotective in neuronal cultures and mitigates mild traumatic brain injury in mice. J Neurochem. 2015 May 15; doi: 10.1111/jnc.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas AIR, Stocchetti N, Bullock R. Moderate and severe traumatic brain injury in adults. Lancet Neurol. 2008;7:728–741. doi: 10.1016/S1474-4422(08)70164-9. [DOI] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Masanic CA, Bayley MT, VanReekum R, Simard M. Open-label study of donepezil in traumatic brain injury. Arch Phys Med Rehabil. 2001;82:896–901. doi: 10.1053/apmr.2001.23833. [DOI] [PubMed] [Google Scholar]
- Marklund N, Hillered L. Animal modeling of traumatic brain injury in preclinical drug development: where do we go from here? Br J Pharmacol. 2011;164:1207–1229. doi: 10.1111/j.1476-5381.2010.01163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman A, Rosenberg A, Weizman R, Pick CG. Mild traumatic brain injury induces persistent cognitive deficits and behavioral disturbances in mice. J Neurotrauma. 2005;9:1003–1010. doi: 10.1089/neu.2005.22.1003. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Katki K, Arisi GM, Foresti ML, Shapiro LA. Early TBI-Induced Cytokine Alterations are Similarly Detected by Two Distinct Methods of Multiplex Assay. Front Mol Neurosci. 2011;4:21. doi: 10.3389/fnmol.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer’s disease. Cell. 2012;149:232–244. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem M, Parvez S. Role of melatonin in traumatic brain injury and spinal cord injury. Scientific World Journal. 2014:586270. doi: 10.1155/2014/586270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsüer H, Görgülü A, Kiriş T, Cobanoğlu S. The effects of memantine on lipid peroxidation following closed-head trauma in rats. Neurosurg Rev. 2005;28:143–147. doi: 10.1007/s10143-004-0374-1. [DOI] [PubMed] [Google Scholar]
- Papa M, De Luca C, Petta F, Alberghina L, Cirillo G. Astrocyte-neuron interplay in maladaptive plasticity. Neurosci Biobehav Rev. 2014;42:35–54. doi: 10.1016/j.neubiorev.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Perry T, Haughey NJ, Mattson MP, Egan JM, Greig NH. Protection and reversal of excitotoxic neuronal damage by glucagon-like peptide-1 and exendin-4. J Pharmacol Exp Ther. 2002;302:881–888. doi: 10.1124/jpet.102.037481. [DOI] [PubMed] [Google Scholar]
- Perry T, Lahiri DK, Sambamurti K, Chen D, Mattson MP, Egan JM, Greig NH. Glucagon-like peptide-1 decreases endogenous amyloid-beta peptide (Abeta) levels and protects hippocampal neurons from death induced by Abeta and iron. J Neurosci Res. 2003;72:603–612. doi: 10.1002/jnr.10611. [DOI] [PubMed] [Google Scholar]
- Rachmany L, Tweedie D, Li Y, Rubovitch V, Holloway HW, Miller J, Hoffer BJ, Greig NH, Pick CG. Exendin-4 induced glucagon-like peptide-1 receptor activation reverses behavioral impairments of mild traumatic brain injury in mice. Age (Dordr) 2013a;35:1621–1636. doi: 10.1007/s11357-012-9464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmany L, Tweedie D, Rubovitch V, Yu QS, Li Y, Wang JY, Pick CG, Greig NH. Cognitive impairments accompanying rodent mild traumatic brain injury involve p53-dependent neuronal cell death and are ameliorated by the tetrahydrobenzothiazole PFT-α. PLoS One. 2013b;8:e79837. doi: 10.1371/journal.pone.0079837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VL, Dogan A, Todd KG, Bowen KK, Dempsey RJ. Neuroprotection by memantine, a non-competitive NMDA receptor antagonist after traumatic brain injury in rats. Brain Res. 2001;911:96–100. doi: 10.1016/s0006-8993(01)02617-8. [DOI] [PubMed] [Google Scholar]
- Salcedo I, Tweedie D, Li Y, Greig NH. Neuroprotective and neurotrophic actions of glucagon-like peptide-1: an emerging opportunity to treat neurodegenerative and cerebrovascular disorders. Br J Pharmacol. 2012;166:1586–1599. doi: 10.1111/j.1476-5381.2012.01971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KE, Bondi CO, Light SH, Massimino LA, McAloon RL, Monaco CM, Kline AE. Donepezil is ineffective in promoting motor and cognitive benefits after controlled cortical impact injury in male rats. J Neurotrauma. 2013;30:557–564. doi: 10.1089/neu.2012.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth SA, Iavarone AT, Liebeskind DS, Won SJ, Swanson RA. Targeted Lipid Profiling Discovers Plasma Biomarkers of Acute Brain Injury. PLoS One. 2015;10(6):e0129735. doi: 10.1371/journal.pone.0129735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siman R, Toraskar N, Dang A, McNeil E, McGarvey M, Plaum J, Maloney E, Grady MS. A panel of neuron-enriched proteins as markers for traumatic brain injury in humans. J Neurotrauma. 2009;26:1867–1877. doi: 10.1089/neu.2009.0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DG, Geddes RI, Sribnick EA. Recent developments in clinical trials for the treatment of traumatic brain injury. Handb Clin Neurol. 2015;127:433–451. doi: 10.1016/B978-0-444-52892-6.00028-3. [DOI] [PubMed] [Google Scholar]
- Stoica BA, Faden AI. Cell death mechanisms and modulation in traumatic brain injury. Neurotherapeutics. 2010;7:3–12. doi: 10.1016/j.nurt.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverni JP, Seliger G, Lichtman SW. Donepezil medicated memory improvement in traumatic brain injury during post-acute rehabilitation. Brain Inj. 1998;12:77–80. doi: 10.1080/026990598122881. [DOI] [PubMed] [Google Scholar]
- Tashlykov V, Katz Y, Volkov A, Gazit V, Schreiber S, Zohar O, Pick CG. Minimal traumatic brain injury induce apoptotic cell death in mice. J Mol Neurosci. 2009;37:16–24. doi: 10.1007/s12031-008-9094-2. [DOI] [PubMed] [Google Scholar]
- Tuttolomondo A, Pecoraro R, Pinto A. Studies of selective TNF inhibitors in the treatment of brain injury from stroke and trauma: a review of the evidence to date. Drug Des Devel Ther. 2014;8:2221–2238. doi: 10.2147/DDDT.S67655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie D, Milman A, Holloway HW, Li Y, Harvey BK, Shen H, Pistell PJ, Lahiri DK, Hoffer BJ, Wang Y, Pick CG, Greig NH. Apoptotic and behavioral sequelae of mild brain trauma in mice. J Neurosci Res. 2007;85:805–815. doi: 10.1002/jnr.21160. [DOI] [PubMed] [Google Scholar]
- Tweedie D, Rachmany L, Rubovitch V, Zhang Y, Becker KG, Perez E, Hoffer BJ, Pick CG, Greig NH. Changes in mouse cognition and hippocampal gene expression observed in a mild physical- and blast-traumatic brain injury. Neurobiol Dis. 2013a;54:1–11. doi: 10.1016/j.nbd.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie D, Rachmany L, Rubovitch V, Lehrmann E, Zhang Y, Becker KG, Perez E, Miller J, Hoffer BJ, Greig NH, Pick CG. Exendin-4, a glucagon-like peptide-1 receptor agonist prevents mTBI-induced changes in hippocampus gene expression and memory deficits in mice. Exp Neurol. 2013b;239:170–182. doi: 10.1016/j.expneurol.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie D, Rachmany L, Rubovitch V, Li Y, Holloway HW, Lehrmann E, Zhang Y, Becker KG, Perez E, Hoffer BJ, Pick CG, Greig NH. Blast traumatic brain injury-induced cognitive deficits are attenuated by preinjury or postinjury treatment with the glucagon-like peptide-1 receptor agonist, exendin-4. Alzheimers Dement. 2015 doi: 10.1016/j.jalz.2015.07.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls MK, Race N, Zheng L, Vega-Alvarez SM, Acosta G, Park J, Shi R. Structural and biochemical abnormalities in the absence of acute deficits in mild primary blast-induced head trauma. J Neurosurg. 2015:1–12. doi: 10.3171/2015.1.JNS141571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang L, Jiang R, Yuan Y, Yu Q, Li Y. Exendin-4 antagonizes Aβ1-42-induced suppression of long-term potentiation by regulating intracellular calcium homeostasis in rat hippocampal neurons. Brain Res. 2015 doi: 10.1016/j.brainres.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Whelan FJ, Walker MS, Schultz SK. Donepezil in the treatment of cognitive dysfunction associated with traumatic brain injury. Ann Clin Psychiatry. 2000;12:131–135. doi: 10.1023/a:1009008816928. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Zhang Y, Mahmood A, Chopp M. Investigational agents for treatment of traumatic brain injury. Expert Opin Investig Drugs. 2015;24:743–760. doi: 10.1517/13543784.2015.1021919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang Y, Yuan G, Zhu W, Ma D, Hu S. Exendin-4, a glucagon-like peptide-1 receptor agonist, reduces Alzheimer disease-associated tau hyperphosphorylation in the hippocampus of rats with type 2 diabetes. J Investig Med. 2015;63:267–272. doi: 10.1097/JIM.0000000000000129. [DOI] [PubMed] [Google Scholar]
- Yang LY, Chu YH, Tweedie D, Yu QS, Pick CG, Hoffer BJ, Greig NH, Wang JY. Post-trauma administration of the pifithrin-α oxygen analog improves histological and functional outcomes after experimental traumatic brain injury. Exp Neurol. 2015;269:56–66. doi: 10.1016/j.expneurol.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kim A, Kernie SG. Donepezil rescues spatial learning and memory deficits following traumatic brain injury independent of its effects on neurogenesis. PLoS One. 2015;10:e0118793. doi: 10.1371/journal.pone.0118793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar O, Schreiber S, Getslev V, Schwartz JP, Mullins PG, Pick CG. Closed-head minimal traumatic brain injury produces long-term cognitive deficits in mice. Neuroscience. 2003;118:949–955. doi: 10.1016/s0306-4522(03)00048-4. [DOI] [PubMed] [Google Scholar]