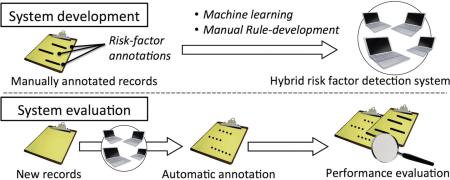
Abstract
In the United States, about 600,000 people die of heart disease every year. The annual cost of care services, medications, and lost productivity reportedly exceeds 108.9 billion dollars. Effective disease risk assessment is critical to prevention, care, and treatment planning. Recent advancements in text analytics have opened up new possibilities of using the rich information in electronic medical records (EMRs) to identify relevant risk factors. The 2014 i2b2/UTHealth Challenge brought together researchers and practitioners of clinical natural language processing (NLP) to tackle the identification of heart disease risk factors reported in EMRs. We participated in this track and developed an NLP system by leveraging existing tools and resources, both public and proprietary. Our system was a hybrid of several machine-learning and rule-based components. The system achieved an overall F1 score of 0.9185, with a recall of 0.9409 and a precision of 0.8972.
Keywords: Medical records, Risk assessment, Natural language processing, Text classification
Graphical abstract
1. Background
In the United States, heart disease is the leading cause of death, accounting for over 600,000 deaths per year [1]. The American Heart Association reports that the annual total cost of care services, medications, and lost productivity exceeds 108.9 billion dollars [2]. The 2014 i2b2/UTHealth Challenge brought together researchers and practitioners of clinical natural language processing (NLP) to tackle problems of common interest, which included the identification of heart disease risk factors reported in electronic medical records (EMRs), a task that will support prevention, care, and treatment planning of the disease. We participated in a track focusing on this task, Track 2 of the 2014 i2b2/UTHealth Challenge.
The goal of the Track-2 task was to annotate diagnoses, risk factors, and associated medications at the record (document) level. The challenge organizer provided a training corpus with gold annotations at the record level, but also made available raw evidence annotations at the phrase level for participants’ system development. The task concerns several clinical NLP topics including disease concept identification, medication detection, and smoking status classification. Each of these topics may require supporting tasks, such as assertion detection, section detection, and temporal information detection, as well as basic NLP tasks, such as part-of-speech tagging. Many of these tasks have been studied over the years [3–14]. Track 2 of the 2014 i2b2/UTHealth Challenge provided a valuable opportunity to determine the generalizability of past research.
Because significant overlap exists between the 2014 Challenge and prior i2b2 Challenges, we reviewed successful prior efforts, and discovered a common technique, which was termed “hot-spot identification” by Cohen [12]. In this technique, a small amount of discriminative words are identified to classify a document. The hot-spot phrases may be identified via hand-coded rules or sequence labeling techniques, such as Conditional Random Field (CRF) [8]. Hot-spot-based techniques have demonstrated repeated success by multiple teams during the 2006 and 2011 i2b2 Challenges.
In the 2006 i2b2 NLP Challenge, hot-spot phrases were leveraged to classify patients’ smoking status [9] (non-smoker, current smoker, past smoker, smoker, or unknown). Among the best performing systems in the challenge were those developed by Aramaki et al. [10], Clark et al. [11], and Cohen [12], which achieved micro-averaged F-measures of 0.88, 0.90, and 0.89, respectively. All of these top performers used hot-spot-based techniques. Aramaki et al. tackled this task in two steps. In the first step, a single sentence reporting the patient's smoking status was selected from a medical record. This selection was based on the occurrence of a handful of keywords: “nicotine”, “smoker”, “smoke”, “smoking”, “tobacco”, and “cigarette.” In the second step, selected sentences were classified using a k-nearest-neighbors method, and then predicted classes were assigned to the corresponding host documents. The approach by Cohen was similar to Aramaki et al. in that they first identified keywords in a medical record which are occurrences of any of the selected stemmed words: “nicotine”, “smok”, “tob”, “tobac”, “cig”, and “packs.” He called these keywords “hot-spots.” Unlike Aramaki et al., however, he did not select a single sentence per document, but used words near the hot-spots as features for Support Vector Machine (SVM) classifiers [13]. Clark et al. used both a two-step approach, similar to Aramaki et al., and a one-step approach, similar to Cohen.
Hot-spot-based techniques were also successful in the 2011 i2b2/VA/Cincinnati Challenge for sentiment analysis of suicide notes [14]. This task was a multi-label/multi-class classification of sentences from suicide notes, where there were 16 target classes (Guilt, Hopefulness, Love, Thankfulness, etc.). In this task, sentences could sometimes be long, but detection of target classes might depend on only a small text segment and often on a very limited vocabulary, e.g., the class Thankfulness was mostly associated with the single word “thank(s)” and the class Love was associated with the word “love.” Among the best performing systems was Hui et al. [15], who used the hot-spot technique through CRF models. They manually annotated “cue phrases” that are indicative of sentence classes in a development data set, and then trained CRF models to automatically detect the same or similar phrases. These “cue phrases” are essentially the same as hot-spot phrases by Cohen, Aramaki et al., and Clark et al. Given a new sentence, trained CRF models were used to identify cue phrases and, if found, associated classes were assigned to that sentence. Leveraging the CRF models, their system achieved the best results in the 2011 Challenge.
After analyzing the 2014 challenge task, we determined that the task was well suited for a hot-spot-based approach. In designing our system for the 2014 challenge, we leveraged the approaches reported for these past challenge tasks.
2. Materials and Methods
2.1. Annotated corpora
Participants in the Track-2 task were provided with two sets of annotated corpora, the Gold corpus and the Complete corpus. Both corpora contain the same source documents that consisted of 790 de-identified clinical notes.
In the Gold corpus, each medical record is provided as an XML file, and target concepts, if reported anywhere in the record, are annotated with XML tags at the record level (e.g., <CAD time=“during DCT” indicator=“mention” />). The tags and associated attribute-vales are found in Table 1.
Table 1. Distribution of target categories in the training corpus.
Each tag is modified by at most two attribute-value pairs, except for MEDICATION tag that is modified by two mandatory attributes, time and type1, and one optional, rarely used attribute, type2. Each table below shows possible combinations of a particular tag and attribute-value pairs, and the numbers indicate how many medical records are annotated with a tag with particular attribute-value pairs in the Gold training corpus consisting of 790 medical records. For instance, the number at the upper-left corner, 224, in the first table (a) indicates there are 224 medical records annotated with an XML tag <CAD indicator=“event” and time=“before DCT” />. For MEDICATION, since type2 attribute is rarely used, it is omitted in the corresponding table (f).
| (a) Tag: CAD | ||||
|---|---|---|---|---|
| time | ||||
| before DCT | during DCT | after DCT | ||
| indicator | event | 224 | 20 | 2 |
| mention | 260 | 261 | 259 | |
| symptom | 54 | 24 | 3 | |
| (b) Tag: DIABETES | ||||
|---|---|---|---|---|
| time | ||||
| before DCT | during DCT | after DCT | ||
| indicator | A1C | 89 | 21 | 0 |
| glucose | 16 | 9 | 0 | |
| mention | 518 | 524 | 518 | |
| (c) Tag: HYPERLIPIDEMIA | ||||
|---|---|---|---|---|
| time | ||||
| before DCT | during DCT | after DCT | ||
| indicator | high LDL | 23 | 10 | 0 |
| high chol. | 8 | 1 | 0 | |
| mention | 340 | 340 | 340 | |
| (d) Tag: HYPERTENSION | ||||
|---|---|---|---|---|
| time | ||||
| before DCT | during DCT | after DCT | ||
| indicator | high bp | 41 | 322 | 0 |
| mention | 523 | 521 | 519 | |
| (e) Tag: OBESE | ||||
|---|---|---|---|---|
| time | ||||
| before DCT | during DCT | after DCT | ||
| indicator | BMI | 3 | 15 | 2 |
| mention | 133 | 147 | 133 | |
| (f) Tag: MEDICATION | ||||
|---|---|---|---|---|
| time | ||||
| before DCT | during DCT | after DCT | ||
| type (type1) | ACE inhibitor | 326 | 318 | 323 |
| ARB | 98 | 93 | 97 | |
| DPP4 inhibitors | 1 | 0 | 0 | |
| anti diabetes | 1 | 1 | 1 | |
| aspirin | 424 | 435 | 424 | |
| beta blocker | 469 | 472 | 470 | |
| calcium channel blocker | 186 | 178 | 181 | |
| diuretic | 113 | 99 | 106 | |
| ezetimibe | 12 | 12 | 12 | |
| fibrate | 22 | 20 | 22 | |
| insulin | 204 | 218 | 212 | |
| metformin | 187 | 176 | 181 | |
| niacin | 7 | 6 | 7 | |
| nitrate | 117 | 126 | 93 | |
| statin | 436 | 427 | 438 | |
| sulfonylureas | 159 | 155 | 157 | |
| thiazolidinedione | 43 | 41 | 40 | |
| thienopyridine | 97 | 98 | 97 | |
| (g) Tag: SMOKER | ||
|---|---|---|
| status | current | 58 |
| ever | 9 | |
| never | 184 | |
| past | 149 | |
| unknown | 371 | |
| (h) Tag: FAMILY_HIST | ||
|---|---|---|
| indicator | present | 22 |
| not present | 768 | |
In the Complete corpus, segments of text marked up by three clinicians as evidence annotations are also included. In other words, each concept annotated at the document level in this data set has a reference to the text segment providing the evidence in the record (e.g., <CAD start=“3575” end=“3579” text=“CAD” time=“during DCT” indicator=“mention”/>). The corpus is “Complete” in the sense that it includes all the raw annotations by the clinicians, who were requested to record at least the first piece of text that provides supporting evidence in each record in addition to the document level annotation. There could be more than one evidence text segment indicating the same target concept in a record, but they were not exhaustively marked up. Besides, unlike corpora created specifically for training a sequence-labeling model, the annotation boundaries were determined rather arbitrarily.
After the system development period, the challenge participants were provided with an evaluation corpus consisting of 514 medical records, which do not include annotations. Participants applied their system to obtain a result, called a run, and submitted up to three different runs to the organizer for evaluation. Further information regarding these data sets can be found in the overview papers of the 2014 i2b2/UTHealth Challenge [16,17].
2.2. Methods
We built a general text classification system to tackle the diverse sub-tasks in Track 2. Given the success of hot-spot features in prior i2b2 Challenges, we focused on this approach. A general text classifier, a smoking status classifier, and a CRF-based classifier were created that leveraged hot-spot features and also features derived from existing NLP systems. Due to the distinctive properties of the smoking status classification sub-task and the potential benefit of having a standalone tool for that sub-task, an independent module was developed for the sub-task. Different ways of integrating the classification components were explored for the submission runs. The UIMA platform [18] and UIMA compliant tools [19,20] were used to ease the integration.
2.2.1. General text classifier
The general classifier was designed to handle diverse classification sub-tasks in Track 2. In this classification system, each combination of a tag and specific attribute-value pairs was regarded as an independent target category. Then, each of such categories was applicable to some medical records (positive instances) but not to the other records (negative instances). For instance, we considered a tag with specific attribute-value pairs, <CAD indicator=“mention” time=“before DCT” />, as an independent target category, and this category was either applicable to a particular record (i.e., the record does contain a mention of CAD as an event that the patient previously had) or not (i.e., the record does not contain such information). Then, this view defined a binary classification task. That is, in Table 1, each cell with a number entry corresponds to one binary classification task. The number represents exactly the quantity of positive instances of the class, and the negative instances are therefore the remaining complement (i.e., the total number of 790 notes in the training set minus the number of positive instances). For example, in Table 1 (a) Tag: CAD, a cell with the number 260 in row 2 (time=“before DCT”) column 1 (indicator=“mention”) corresponds to the binary classification task for the aforementioned category, <CAD indicator=“mention” time=“before DCT” />, where the number of positive and negative instances are 260 and 530 (= 790 - 260) respectively. The Track-2 task was regarded as a collection of many binary classification tasks. For each of these tasks, we trained a supervised machine learning model that consisted of a classification rule set derived by the RIPPER algorithm [21].
2.2.1.1. General text classifier features
Hot-spot features
For each tag (e.g., “CAD”), phrases frequently annotated as evidence text in the Complete corpus (e.g., “coronary artery disease”, “coronary disease”, and “CAD”) were hand-selected and used to identify text segments in a medical record that were immediately relevant to the current classification purpose. Following Cohen [12], we named these selected phrases as hot-spot phrases. Then, selected phrases with similar patterns/concepts were manually grouped together, and a binary feature was defined for the group. For instance, “coronary artery disease”, “coronary disease”, and “coronary heart disease” were grouped together, and if any of these phrases was found in a given medical record, a corresponding feature was set to be 1, or 0 otherwise. The same approach was applied to the MEDICATION tag. For instance, in the Complete corpus, evidence phrases annotated for the ACE inhibitor type (i.e., <MEDICATION type1=“ACE inhibitor” type2=“...” time=“...” />) was predominantly “lisinopril”, followed by “zestril”, “captopril”, etc. These phrases were hand-coded as hot-spot phrases just as those coded for the disease tags.
Hot-spot phrases involving numeric values were also considered with special care for the value range. For instance, a report on high blood pressure was associated with the target category <HYPERTENSION indicator=“high bp” time=“...” />, where the corresponding evidence phrases were often of the form “Blood pressure #/#” or “BP: #/#” (“#” being some numerical value). Any such phrase was considered as a hot-spot if the first value was above 140 (e.g., “Blood pressure 160/58”) and/or the second value was above 90 (“BP: 126/92”), as specified in the annotation guidelines [16]. For each hot-spot feature, n-grams (n from 1 to 5) within ±50 tokens from the hot-spot phrase were also extracted and represented as binary features.
Hot spot modifier features
Certain phrases repeatedly observed near the hot-spot phrases were hand-selected as hot-spot modifier features. Similar to hot-spot phrases, identified patterns were grouped together, and aggregated features were derived. These features were:
- Disease-related hot spot modifier features:
- A hot-spot modifier feature, HISTORY_OF, is set when a hot-spot phrase selected for a disease concept (CAD, DIABETES, HYPERLIPIDEMIA, HYPERTENSION, or OBESE) is preceded by selected keywords, such as “history of”, “hx of”, “known”, or “previous”, unless it is preceded by a negation word (e.g., “no”, “denies”, or “denied”). Similarly, the STATUS_POST feature is set when the preceding word is “status post”, “sp”, or “s/p”. These features were frequently observed with time=“before DCT”.
- Medication-related hot spot modifier features:
- The ONGOING feature is set for keywords “ongoing”, “recurrent”, “recurring”, “frequent”, and “progressive”, which commonly precede MEDICATION hot-spot keywords, such as “lisinopril”. Similarly, the START feature is set for the preceding keywords “add”, “prescribe”, “start”, or “try”, and the STOP feature for the preceding keywords “discontinue”, “stop”, or “hold.” When these phrases were looked up in text, inflected forms (e.g., “adds” and “added” in addition to “add”) and words in upper case (e.g., “Stop” as well as “stop”) were also considered.
Disease concept features
An in-house clinical NLP system nQuiry was used to identify disease concepts reported in medical records. Some of its components are described previously in [22,20]. The system detects disease mentions in text, normalizes them to concept IDs (UMLS CUIs), and further determines the assertion classes: if they are reported to be present in the patient (e.g., whether the disease mention is negated, reported about a family member, or stated as hypothetical). Different combinations of the detected CUI, the assertion class, and the phrase in text were considered as features represented as binary values (e.g., CUI alone, CUI with the assertion class, CUI with the assertion class and the phrase). Additionally, from the sentence where a concept was detected, n-grams (n from 1 to 5) were extracted and used as features indicating local contexts of the concept mention.
Medication features
The MedEx system [23] identifies medication information in text and reports normalized drug name, brand name, generic name, and UMLS CUIs, along with the identified phrase. Each of these values was used as a binary classification feature. Similar to the disease concept features described above, n-grams were additionally extracted from the same sentence as medication mentions.
2.2.1.2. Machine learning component
We used the Weka machine learning suite [24] to explore several different choices of supervised machine learning classifiers, including Naïve Bayes, SVM, Decision Tree, Random Forest, and RIPPER. Different features, such as hot-spot phrases, were gradually enriched and revised, while experimenting with different machine learning methods. In an early stage of our development, however, we decided to employ, RIPPER [21], a rule learner producing a set of if-then rules, which performed as good as or better than other algorithms. In the training of a RIPPER classification model, rules were generated and pruned over randomized training data and, hence, the training program had a random seed value as a parameter. By varying this parameter, we built 21 models and used majority vote to form an ensemble classifier.
As described in the previous sections, many feature values were extracted using existing tools and custom rules, e.g., hot-spot phrases, disease concepts, medication information, n-grams near each concept. Extracted feature values were distinguished by the extraction methods (e.g., n-grams are prefixed by the extraction method names), but the information contents might be essentially redundant sometimes. For example, a medication name may be extracted as a hot-spot by a hand-coded rule or as a phrase identified by MedEx. Before applying the RIPPER training program, collected feature values were filtered and reduced to 500 using the information gain measure. Information gain, also known as mutual information, is a widely used measure to quantify the significance of individual features in machine learning classification tasks. For the formula and computation of information gain, we refer to existing literature, such as Forman [25]. Given a training data set in a classification problem, features can be ranked according to assigned information gain values. Only the top ranked features may be used in a classifier to mitigate the training cost and/or to improve the classifier performance. In our task, upfront reduction of features sped up the training time (since we used the ensemble of 21 RIPPER models) and, thus, eased our development efforts, but it did not improve classifier performance. This may be understandable as the RIPPER algorithm uses information gain internally to select features and, hence, prior filtering of features using the same selection method has little impact.
The procedures described above were implemented using Weka. Specifically, for each target category, features were selected using Weka's InfoGainAttributeEval along with Ranker. Selected features were fed into a meta-classifier, Vote, which was configured to apply a majority vote over 21 models of JRip, the Weka implementation of RIPPER.
Once the ensemble classifier was prepared for each target category and given a new medical record, feature extraction was performed in the same way as in the training phase, and the trained ensemble classifier predicted a binary class. The binary prediction (i.e., whether a certain category is applicable or not) was interpreted to assign a corresponding tag with specific attribute-value pairs to the record.
Among the eight target tags, FAMILY_HISTORY and SMOKER are different from the other six tags in that they have a single attribute with exclusive attribute-values (See Table 1). FAMILY_HISTORY has an attribute indicator, and all medical records are annotated with either one of the two attribute-values present or not present. The distribution of these two attribute-values is highly skewed (22 present and 768 not present in the training data set). A trivial classifier that always assigns not present can guarantee good and stable performance, which was found to be difficult to improve on with a machine learning classifier. Therefore, for FAMILY_HISTORY we just applied the majority-guess classifier. As for SMOKER, a dedicated system was developed (see 2.2.2. below).
2.2.2. Smoking status classifier
The SMOKER tag has mutually exclusive attribute-value pairs, namely status={current, past, ever, never, unknown}. Additionally, categorization features were assumed to be highly unique for smoker. Therefore, it was not straightforward to use the general text classifier on this sub-task. For instance, a collection of independent binary classifiers could yield conflicting prediction results. We developed a dedicated module for the smoking status sub-task.
2.2.2.1. Smoking status classifier features
While the smoker status system was developed separately from the general classifier, a similar approach based on hot-spot phrases was found to be effective. Hot-spot candidates were collected from evidence phrases annotated in the Complete corpus, and the list of phrases was manually reviewed. Hot-spot phrases selected include “tobacco”, “smoking”, and “quit”. Given each document, features were collected at and around each hot-spot occurrence. Specifically, we used (a) the hot-spot keyword itself, (b) three tokens left of the hot-spot keyword, and (c) three tokens right of the hot-spot keyword. The window size of three was chosen based on our review of the hot-spot context patterns in the training data set.
2.2.2.2. Smoking status classification rules
We used a hybrid of machine learning and rule-based methods to tackle this sub-task. Provided with the Gold corpus, the target category and the features described above were fed into LibSVM [26] to train a linear SVM classifier. The trained classifier achieved accuracy of around 80% in 5-fold cross-validation tests on the training corpus. Errors seen in the cross validation test were manually reviewed, and rules based on regular expression patterns were created accordingly to overwrite the SVM decision at run time.
Given a new medical record, feature extraction was performed in the same way through the hot-spot keywords as in training. If any feature was available for an input record, the SVM classifier was applied. If no feature was extracted, the value ‘unknown’ was assigned. The rules learned in the training phase were applied to overwrite SVM decisions.
2.2.3. Alternative sequence-labeling-based classifier
The evidence text annotations in the Complete corpus were not exhaustive (see Section 2.1, Annotated corpora). They, however, could be sufficient to train sequence-labeling models to replicate the clinicians’ phrase annotations. With such phrase annotation models, a new medical record could be automatically marked-up with evidence phrases, and then the record containing those phrases could be assigned with the corresponding categories. This is essentially the same approach as Hui et al., employed for sentiment classification (see the Background section). We used the Stanford NER tool [27] through the ClearTK toolkit [19] to train phrase annotation models for selected categories, for which there were sufficient training examples, namely tags of CAD, DIABETES, HYPERLIPIDEMIA, HYPERTENSION, and OBESE with the attribute-value of indicator=“mention”. Our selected target categories usually appeared with all three “time” attribute-values, and evidence text segments we wished to identify in records were nearly the same for those three “time” attribute-values. For instance, in the training corpus, there were 261 records assigned with the label <CAD indicator=“mention” time=“during DCT” />, 260 assigned with nearly the same label but time=“before DCT”, and 259 assigned with time=“after DCT” (See Table 1). In case of the label < HYPERLIPIDEMIA indicator=“mention” time=“...” />, the same 340 records were assigned with time=“before DCT”, time=“during DCT”, and time=“after DCT”. Observing that, we assumed that CRF models trained for different “time” attribute-values would perform the same or nearly the same. As a heuristic practice, for each tag we aggregated labels with different “time” attribute-values into a single class during model training. When applying the trained model on test data, a medical record assigned with the aggregated tag was re-assigned with the three original labels.
This approach based on sequence labeling was applicable only to the aforementioned subset of categories with sufficient training data. We used it to supplement the results of the general classifiers through post-combinations, as described in the next section.
3. Results
Three runs were compiled for the final evaluation on the test corpus. The first run was derived using the general classifier (for all of the targets except for smoking status) and the smoking status classifier (“General + Smoking status classifier” in Table 2). The second and third runs were obtained by additionally integrating the results of the alternative sequence-labeling-based classifier. Two integration strategies were considered. In the first approach (“Merged, with precedence” in Table 2), prediction results of Stanford NER were adopted for selected categories where they performed better than the general classifier in our tests on the training data set (the tag is CAD or OBESE and the attribute-value pair is indicator=“mention”). In the second approach (“Merged, with set-union” in Table 2), the set-union of predicted categories was computed. Namely, given a medical record, if any of the prediction systems predicts that a particular category was applicable to the record, then that was adopted.
Table 2.
The performance measures of the systems on the test corpus (precision/recall/F1).
| Run | System/Configuration | Precision | Recall | F1 |
|---|---|---|---|---|
| 1 | General + Smoking status classifier | 0.9010 | 0.9356 | 0.9180 |
| - | Sequence-labeling-based (not submitted) | 0.9171 | 0.3799 | 0.5373 |
| 2 | Merged, with precedence | 0.9027 | 0.9290 | 0.9156 |
| 3 | Merged, set-union | 0.8972 | 0.9409 | 0.9185 |
The performance measures of our systems are summarized in Table 2 and 3, which were calculated using an evaluation script provided by the challenge organizer (the script ver. 2.1.0)a. The highest F1 score among our submitted runs, 0.9185, was achieved by set-union merging of different approaches as in Table 2. The effect of different merging approaches (refer to the run definitions above) for each target category is detailed in Table 3. After merging, the F1 score was improved for HYPERLIPIDEMIA, HYPERTENSION, DIABETES, and OBESE but was degraded on CAD. Since the sequence-labeling models were applied only for those five tags, the performance for the rest of the tags was not affected.
Table 3.
Per-tag results on the test corpus.
| Tag | Records | Run | Precision | Recall | F1 |
|---|---|---|---|---|---|
| CAD | 784 | 1 | 0.8480 | 0.8253 | 0.8365 |
| 2 | 0.8480 | 0.7398 | 0.7902 | ||
| 3 | 0.8226 | 0.8342 | 0.8284 | ||
| DIABETES | 1,180 | 1, 2 | 0.9403 | 0.9339 | 0.9371 |
| 3 | 0.9349 | 0.9492 | 0.9420 | ||
| HYPERLIPIDEMIA | 751 | 1, 2 | 0.9607 | 0.9108 | 0.9351 |
| 3 | 0.9570 | 0.9188 | 0.9375 | ||
| HYPERTENSION | 1,293 | 1, 2 | 0.9472 | 0.9706 | 0.9587 |
| 3 | 0.9399 | 0.9791 | 0.9591 | ||
| OBESE | 262 | 1 | 0.8536 | 0.9122 | 0.8819 |
| 2 | 0.9066 | 0.8893 | 0.8979 | ||
| 3 | 0.8388 | 0.9733 | 0.9011 | ||
| MEDICATION | 5,674 | 1, 2, 3 | 0.8799 | 0.9478 | 0.9126 |
| SMOKER | 512 | 1, 2, 3 | 0.9027 | 0.9062 | 0.9045 |
| FAMILY_HIST | 514 | 1, 2, 3 | 0.9630 | 0.9630 | 0.9630 |
4. Discussion
We found the tasks posed were challenging, but they were realistic in that those challenges faced could potentially be encountered in real life:
The task appeared to be text classification (i.e., annotations in the Gold corpus are provided at the record level), but the underlying task that we really needed to address was identification of specific evidence text (i.e., the Gold corpus annotations were attributed to short text segments as in the Complete corpus). It would be ideal to have a corpus annotated with adjudicated evidence text segments, but that would be very expensive to achieve.
The Track-2 task consists of many diverse sub-tasks. There are eight target tags, and each of which is modified by a set of attribute-value pairs. Moreover, the distribution of these annotations was highly skewed and there might be very small numbers of instances (see Table 1). Accordingly, we had to examine each of these categories thoroughly and develop a framework that exploited both the unique and shared properties. There were, however, too many categories to consider.
Regarding the lack of comprehensive evidence annotations, we found the pre-adjudication corpus was very helpful to build NLP systems in the current task. With that corpus, we could identify informative key phrases for classification and exploit them through hot-spot techniques [10,11,21]. For some categories, we could also train sequence labeling models that automatically annotate evidence phrases [28].
Regarding the large number of target categories, we mitigated this challenge by designing a general text classification system to cover diverse target categories in the same manner. Use of existing resources to extract features, such as disease concepts and medication information, helped tackle many different categories without building extensive customized programs. When applying an alternative approach based on sequence labeling, we merged frequently co-occurring categories so that we did not need to train or apply models for a nearly redundant set of instances.
One interesting observation for the general classifier approach was that only a handful of feature-values were actually selected for use in each of the RIPPER classifiers. For instance, in a RIPPER classifier for the target category of “<CAD indicator=“mention” time=“during DCT” />”, a list of four rules was learned from the entire training set (790 notes), and it involved a mere four feature-values (Table 4). Note that, for each target category, we derived 21 constituent RIPPER classifiers for an ensemble classifier. For example, the first rule of another rule list derived for the same target was “IF nQuiry detects two or more phrases with C0010054, THEN Positive; [Applicable to 125 of 790 notes, of which 125 are correct].” As seen in these example rules, an accurate rule supported by many training instances was learned first, followed by less accurate rules supported by fewer instances. As also exemplified by these rules, RIPPER might be viewed as a rule selector, where rules were selected from a large number of knowledge-rich features derived with the elaborated NLP systems (nQuiry and MedEx) or hand-coded patterns (hot-spots). This, in turn, suggests the performance of our systems largely owed to, and was limited by, the accuracy of the underlying NLP systems and hand-coded rules. For instance, the general classifier failed to identify the phrase “Chlropropamiden” [sic] as MEDICATION (Sulfonylurea type), which could not be readily detected by NLP systems or custom hand-coded patterns due to the typographical error as well as its rare occurrence in the training data (a rare mention of chlorpropamide as well as the rare typographical error pattern in the training data).
Table 4.
An example of a list of if-then rules derived by RIPPER. The target category here is “<CAD indicator=“mention” and time=“during DCT” />”, i.e., the rule list predicts if this category label should be assigned to a given note (Positive) or not (Negative).
| Rule order | Rule |
|---|---|
| 1 |
IF nQuiry detects a phrase assigned with the CUI “C0010054: Coronary Arteriosclerosis” whose assertion type is “certain” (i.e., not negated, not hypothetical, etc.), THEN Positive. [Applicable to 267 of 790 notes, of which 244 are correct] |
| 2 |
IF a hot-spot rule detects a phrase matching the regular expression “^coronary\W* ((artery|heart|vascull?ar)\W*)?(diseases?|arteriosclerosis)$” (case insensitive; e.g., “Coronary artery disease”, “Coronary Arteriosclerosis”, ...) AND nQuiry does not detect the CUI “C0375113: Diabetes mellitus without mention of complication, type II or unspecified type, not stated as uncontrolled”, THEN Positive. [Applicable to 14 of 523 remaining notes, of which 12 are correct] |
| 3 |
IF a hot-spot rule detects a phrase matching with the regular expression “^lad$” (case insensitive; e.g., “LAD”) along with the 1-gram (unigram) “CAD” nearby, THEN Positive. [Applicable to 2 of 509 remaining notes, of which 2 are correct] |
| 4 |
OTHERWISE Negative. [Applicable to 507 remaining notes, of which 504 are correct] |
Another consideration pertaining to the hand-coded rules was their coverage. We looked at the coverage of hot-spot phrases for different tags, and found that at least one hot-spot phrase could be found in almost all positive training records (e.g., in over 99% of the positive records for CAD, DIABETES, HYPERLIPIDEMIA, HYPERTENSION, and OBESE, where the attribute-values are indicator=“mention” and time=“during DCT”). While the coverage of positive records was good, we also noticed some hot-spot phrases occurred in many negative records. For example, at least one hot-spot phrase could be found in 73% of negative records for the aforementioned CAD label, indicating limited discriminative power of this feature type (e.g., the phrase “MI”, selected as one of the hot-spot phrases for CAD, was found in 71 positive and 60 negative records). The corresponding percentages for DIABETES, HYPERLIPIDEMIA, HYPERTENSION, and OBESE, however, were much lower: 24%, 22%, 41%, and 44%, respectively. This analysis suggests difficulty of handling the CAD tag, for which our classifier indeed underperformed in comparison to the other tags (see Table 3). To further improve the system performance, it would be helpful to refine the selection of hot-spot phrases based on their occurrence patterns in negative records as well as in positive records.
Despite the overall strong performance we achieved among other participants in the Track-2 task, there are several limitations in our system and in the current study. During our system development, we reviewed the entire training data to derive many of the classification rules. Therefore, it was not feasible to obtain objective evaluation metrics on the training set, and the current evaluation results were solely based on the one test set. For example, our smoker status classifier achieved F1 of 0.9689 (knowingly over-tuned) on the full training set, compared to the 0.9045 on the final independent test set. With the large number of ensemble and hybrid classifiers involved in our approach, it was prohibitive to conduct extensive experiments for all the target categories and analyze every factor that contributed to the final system performance. In real life, there can be even more target categories that need to be addressed by a clinical NLP system. An effective approach to analyzing system components needs to be continuously considered.
5. Conclusions
To participate in Track 2 of the 2014 i2b2/UTHealth Challenge, we leveraged existing techniques and resources. We were able to demonstrate that the hot-spot technique used in prior i2b2 Challenges could be successfully adapted to this challenge. Combining hot-spot features with nQuiry [20,22], MedEx [23], Weka [24] (RIPPER [21]), LibSVM [26], and Stanford NER [27] to develop three type of classifiers: the general classifier, the dedicated smoking status classifier, and the sequence-labeling-based classifier. The general classifier with the smoker status classifier achieved an F-score of 0.9180 with a precision of 0.9010 and a recall of 0.9356. After supplementing results from the sequence-labeling-based classifier, the performance measures were improved for some of the target categories with different degrees. Over all, our final system merging different approaches achieved an F-score of 0.9185 with a precision of 0.8972 and a recall of 0.9409.
Highlights.
Risk factor detection in electronic medical records (EMR) was automated;
Existing tools and techniques were leveraged to build detection systems;
A general binary classification system was used to extract various risk factors;
Additional classifiers were built for subsets of target risk factors;
The hybrid approach combining our systems achieved F-score of 0.92.
Acknowledgement
We thank the organizers of the 2014 i2b2/UTHealth Challenge. We acknowledge Navdeep Sood in the Medical Informatics group at Kaiser Permanente Southern California for the design and implementation of a supporting tool used in our pipeline and Michael J. Cairelli for his help in editing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy S, Xu J, Kochanek K. Deaths: final data for 2010. National vital statistics reports. 2013;61(4):1–118. [PubMed] [Google Scholar]
- 2.Heidenreich PA, Trogdon JG, Khavjou OA, et al. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933–44. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 3.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inf. 2008:128–44. [PubMed] [Google Scholar]
- 4.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42:760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny JC. Mining Electronic Health Records in the Genomics Era. PLoS Comput Biol. 2012;8:e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Rindflesch TC, Corn M. Natural language processing: state of the art and prospects for significant progress, a workshop sponsored by the National Library of Medicine. J Biomed Inform. 2013;46:765–73. doi: 10.1016/j.jbi.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Doan S, Conway M, Phuong TM, et al. Natural language processing in biomedicine: a unified system architecture overview. Methods Mol Biol Clifton NJ. 2014;1168:275–94. doi: 10.1007/978-1-4939-0847-9_16. [DOI] [PubMed] [Google Scholar]
- 8.Lafferty J, McCallum A, Pereira F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of the 18th International Conference on Machine Learning. 2001;2001:282–9. [Google Scholar]
- 9.Uzuner O, Goldstein I, Luo Y, et al. Identifying Patient Smoking Status from Medical Discharge Records. J Am Med Inform Assoc. 2008;15:14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aramaki E, Imai T, Miyo K, et al. Patient Status Classification by Using Rule based Sentence Extraction and BM25 kNN-based Classifier. Proc. of the i2b2 workshop. 2006 [Google Scholar]
- 11.Clark C, Good K, Jezierny L, et al. Identifying Smokers with a Medical Extraction System. J Am Med Inform Assoc. 2008;15:36–9. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen AM. Five-way smoking status classification using text hot-spot identification and error-correcting output codes. J Am Med Inform Assoc JAMIA. 2008;15:32–5. doi: 10.1197/jamia.M2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. [Google Scholar]
- 14.Pestian J, Pestian J, Matykiewicz Pawel, et al. Sentiment Analysis of Suicide Notes: A Shared Task. Biomed Inform Insights. 2012;3 doi: 10.4137/BII.S9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Yang, Willis Alistair, et al. A Hybrid Model for Automatic Emotion Recognition in Suicide Notes. Biomed Inform Insights. 2012;17 doi: 10.4137/BII.S8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stubbs A, Uzuner O. Annotating risk factors for heart disease in clinical narratives for diabetic patients. J Biomed Inform Published Online First. 2015 May 21; doi: 10.1016/j.jbi.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stubbs A, Kotfila C, Xu H, et al. Identifying risk factors for heart disease over time: Overview of 2014 i2b2/UTHealth shared task Track 2. J Biomed Inform Published Online First. 2015 Jul 22; doi: 10.1016/j.jbi.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10:327–48. [Google Scholar]
- 19.Bethard S, Ogren P, Becker L. ClearTK 2.0: Design patterns for machine learning in UIMA. Proceedings of the Ninth International Conference on Language Resource and Evaluation (LREC’14) 2014:3289–93. [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J, Sood N, Huang Y. Disorder Concept Identification from Clinical Notes: an Experience with the ShARe/CLEF 2013 Challenge.. Online Working Notes of the CLEF 2013 Evaluation Labs and Workshop; Valencia, Spain. 2013. [Google Scholar]
- 21.Cohen WW. Fast Effective Rule Induction. In: Proceedings of the Twelfth International Conference on Machine Learning. Morgan Kaufmann. 1995:115–23. [Google Scholar]
- 22.Fan J, Prasad R, Yabut RM, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2011;2011:382–91. [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc JAMIA. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall M, Frank E, Holmes G, et al. The WEKA data mining software: an update. ACM SIGKDD Explor Newsl. 2009;11:10. [Google Scholar]
- 25.Forman G. An extensive empirical study of feature selection metrics for text classification. J Mach Learn Res. 2003;3:1289–305. [Google Scholar]
- 26.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- 27.Manning CD, Surdeanu M, Bauer J, et al. The Stanford CoreNLP Natural Language Processing Toolkit. Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations. 2014:55–60. [Google Scholar]
- 28.Yang H, Nenadic G, Keane JA. Identification of transcription factor contexts in literature using machine learning approaches. BMC Bioinformatics. 2008:9. doi: 10.1186/1471-2105-9-S3-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]