Abstract
Aphids are piercing-sucking insect pests and feed on phloem sap. During feeding, aphids inject a battery of salivary proteins into host plant. Some of these proteins function like effectors of microbial pathogens and influence the outcome of plant–aphid interactions. The pea aphid (Acyrthosiphon pisum) is the model aphid and encompasses multiple biotypes each specialized to one or a few legume species, providing an opportunity to investigate the underlying mechanisms of the compatibility between plants and aphid biotypes. We aim to identify the aphid factors that determine the compatibility with host plants, hence involved in the host plant specialization process, and hypothesize that salivary proteins are one of those factors. Agrobacterium-mediated transient gene expression is a powerful tool to perform functional analyses of effector (salivary) proteins in plants. However, the tool was not established for the legume species that A. pisum feeds on. Thus, we decided to optimize the method for legume plants to facilitate the functional analyses of A. pisum salivary proteins. We screened a range of cultivars of pea (Pisum sativum) and alfalfa (Medicago sativa). None of the M. sativa cultivars was suitable for agroinfiltration under the tested conditions; however, we established a protocol for efficient transient gene expression in two cultivars of P. sativum, ZP1109 and ZP1130, using A. tumefaciens AGL-1 strain and the pEAQ-HT-DEST1 vector. We confirmed that the genes are expressed from 3 to 10 days post-infiltration and that aphid lines of the pea adapted biotype fed and reproduced on these two cultivars while lines of alfalfa and clover biotypes did not. Thus, the pea biotype recognizes these two cultivars as typical pea plants. By using a combination of ZP1109 and an A. pisum line, we defined an agroinfiltration procedure to examine the effect of in planta expression of selected salivary proteins on A. pisum fitness and demonstrated that transient expression of one candidate salivary gene increased the fecundity of the aphids. This result confirms that the agroinfiltration can be used to perform functional analyses of salivary proteins in P. sativum and consequently to study the molecular mechanisms underlying host specialization in the pea aphid complex.
Keywords: pea aphid, Acyrthosiphon pisum, Leguminosae, agroinfiltration, salivary proteins, biotypes, host specialization, effector
Introduction
Herbivorous insects present a high level of species diversity and a large majority of them is specialized to feed on certain host plant species. Specialization to different host plants also occurs within single insect species and leads to the existence of distinguishable “host races” or “biotypes” (Dres and Mallet, 2002). The mechanisms of host plant adaptation in herbivorous insects are poorly understood, although these could explain a large part of insect species richness (Simon et al., 2015). Therefore, insect species displaying an array of races or biotypes provide interesting opportunities to study the process of host plant specialization due to the possibility to compare genomes and feeding strategies between closely related races or biotypes.
The pea aphid, Acyrthosiphon pisum Harris, is the first aphid to be genome sequenced and owing to its long history of research, it is the model of aphids and sap-feeding insects (hemipterans; International Aphid Genomics Consortium, 2010). In addition, A. pisum encompasses a range of biotypes each specialized to one or a few closely related legume species but cannot survive or reproduce well on non-host legume plants. So far, 15 biotypes are described (Peccoud et al., 2015), of which alfalfa, clover and pea biotypes are the ones most studied in host specialization (Hawthorne and Via, 2001; Ferrari et al., 2008; Peccoud et al., 2009; Jaquiery et al., 2012; Via et al., 2012). In addition to show strong differences in performances on host and non-host plants, these biotypes are genetically distinct and can be distinguished by using microsatellite markers (Ferrari et al., 2008; Peccoud et al., 2009). Interestingly, all the A. pisum biotypes studied so far feed well on Vicia faba, which is considered as a universal host plant for pea aphids (Ferrari et al., 2008; Peccoud et al., 2009). Many of these A. pisum biotypes can be crossed with other biotypes (Peccoud et al., 2014), and QTL analyses have been used to identify aphid factors that determine the compatibility with the host plants (Hawthorne and Via, 2001; Via et al., 2012; Kanvil et al., 2015).
Aphids feed on plant phloem sap using a specialized mouthpart called stylet. During feeding, aphids may transmit plant pathogenic viruses, inject toxic saliva and remove nutrients from host plants. Hence, aphids are considered among the most serious crop pests. Recent studies gradually revealed that there are intricate molecular interactions between the proteins secreted with aphid saliva and host plant proteins (Elzinga and Jander, 2013; Rodriguez and Bos, 2013; Kaloshian and Walling, 2016). In some cases, salivary proteins trigger plant defense responses (De Vos and Jander, 2009; Chaudhary et al., 2014; Elzinga et al., 2014), in others, they suppress plant defense reactions and promote aphid proliferation (Will et al., 2007; Bos et al., 2010; Atamian et al., 2013; Elzinga et al., 2014; Naessens et al., 2015). Hence, aphid salivary proteins are considered to be analogous to effectors of plant pathogens, and their functions have been examined using similar techniques, such as silencing of salivary genes or in planta expression of salivary proteins (Elzinga and Jander, 2013; Rodriguez and Bos, 2013). The first characterized aphid salivary gene was an A. pisum gene named C002, which is strongly expressed in salivary glands and was detected in plants infested by the aphids. Silencing of A. pisum C002 (ApC002) was achieved by injection of siRNA in aphids. It prevented aphids from feeding on V. faba, while aphid feeding on artificial diet was unaffected (Mutti et al., 2006, 2008). In line with these studies, transient or stable expression of Myzus persicae orthologue of ApC002, MpC002, in Nicotiana benthamiana and Arabidopsis thaliana, respectively, increased the fecundity of M. persicae feeding on these plants, indicating the conserved role of C002 as an effector required for aphid feeding on host plants (Bos et al., 2010; Pitino and Hogenhout, 2013; Elzinga et al., 2014).
Since then, several A. pisum salivary proteins required for aphid full performance have been identified and characterized mostly by using gene silencing induced by siRNA injection to aphids (Guo et al., 2014; Pan et al., 2015; Wang et al., 2015a,b) while several salivary proteins from other aphids, such as M. persicae have been identified using transient or stable in planta expression of salivary genes (Bos et al., 2010; Pitino and Hogenhout, 2013; Elzinga et al., 2014). However, since the A. pisum genome is extensively duplicated and more than 2000 gene families show massive expansion compared to published insect genomes (Rispe et al., 2008; International Aphid Genomics Consortium, 2010; Jaubert-Possamai et al., 2010), it is often difficult to select a siRNA or dsRNA fragment that specifically targets the gene of interest for silencing. In some cases, co-silencing of multiple gene family members need to be examined to determine whether the phenotype observed is due to the silencing of single gene or multiple genes. Furthermore, there is a possibility that gene silencing does not show a strong phenotypic effect on plant–aphid interactions if genes with redundant functions exist or if gene silencing is too transient.
On the other hand, in planta expression of saliva gene allows simple characterization of single gene in plant-aphid interactions. While the construction and multiplication of transgenic plants require several months to years of preparation before testing, Agrobacterium mediated transient gene expression (agroinfiltration) can be achieved in a few days; therefore, it is a commonly used technique to identify and characterize effector functions. However, the efficiency of agroinfiltration is highly variable and often depends on the compatibility between the Agrobacterium tumefaciens strain and the plant species or cultivar used (Wroblewski et al., 2005). The technique has been developed in N. benthamiana using a disarmed strain where the virulence factors encoded by the Ti plasmid were deleted (Goodin et al., 2008). Then, the technique was optimized for different plants such as potato (Bhaskar et al., 2009), lettuce (Chen et al., 2016), grapevine (Santos-Rosa et al., 2008), Medicago truncatula (Picard et al., 2013) and recently in soybeans (King et al., 2015). However, the technique is not established in the legume plants, which are hosts for A. pisum.
As mentioned earlier, A. pisum encompasses multiple biotypes which cannot survive on the plants they are not specialized to. We study the commonest and most studied pea aphid biotypes to identify the factors that determine the compatibility between the aphid and legume species as such factors are likely be involved in the host plant specialization process of the aphids. Based on our recent genome analysis of three aphid biotypes respectively specialized on clover, alfalfa and pea, we hypothesized that salivary proteins are one of the factors that are involved in the host plant specialization process in A. pisum (Jaquiery et al., 2012). Hence, we envisaged to identify salivary proteins with biotype specific polymorphisms and to characterize their effects on specific plant–aphid interactions. Some salivary proteins from non-adapted biotypes may induce resistance responses in non-host plants while some salivary proteins from adapted biotypes may suppress specific plant defense reactions and allow non-adapted aphids to feed on non-host plants.
Here, as the first step to reach the objectives and to facilitate identification and functional characterization of A. pisum salivary proteins, we undertook optimization of agroinfiltration in Medicago sativa (alfalfa) and Pisum sativum (pea). We focused on these two plants because (1) significant amount of studies have been done on the aphid biotypes that feed on these plants (Hawthorne and Via, 2001; Jaquiery et al., 2012; Via et al., 2012), (2) these two biotypes show clear-cut performance difference on these two plants (Peccoud et al., 2009), and (3) seeds of various cultivars are easily available in our research center.
Materials and Methods
Aphids, Bacteria Strains, Plasmids and Growth Conditions
Aphid lineages, and bacterial strains and plasmids used in this study are listed in Supplementary Tables S1 and S2, respectively. All aphid lineages were reared in a growth chamber at 18°C with a 16 h day/8 h night photoperiod on the broad bean, Vicia faba (Castel), at low density to avoid the production of winged individuals. Escherichia coli and A. tumefaciens strains were grown on Luria-Bertani medium at 37°C and 30°C, respectively. For solid media, agar was added at a final concentration of 1.5% (w/v). Antibiotics were used at the following concentrations: for all bacteria, 50 μg/ml kanamycin; for A. tumefaciens, 50 μg/ml rifampicin; for E. coli, 10 μg/ml gentamycin.
Plants and Growth Conditions
Pisum sativum (Supplementary Table S3) and Medicago sativa plants were grown in a growth chamber at 18°C with a 16 h day/8 h night photoperiod for 2 and 3 weeks, respectively.
Measurements of Aphid Performances on Pea Cultivars
Life traits of five aphid lineages from pea (Ar_Po_28, Ar_Po_58), alfalfa (L9Ms14) and clover (YR2, T8005) biotypes (Supplementary Table S1) were measured on P. sativum cultivars ZP1130 and ZP1109 (Supplementary Table S3). Adult aphids were installed on both pea cultivars and removed 24 h later, giving them enough time to produce 10 larvae that were left on the plants (day 1). Survival rate of the 10 larvae was measured at day 9 (when they reach adulthood), three surviving adult aphids were then reinstalled on the plants and biomass (the cumulated weight of the three adults and their offspring) of the aphid population was weighted at day 17. The biomass is a good proxy of the number of nymphs produced by adult aphids and reflects well their overall fitness (Peccoud et al., 2009). Five replicates for each aphid lineage on the two tested plants were performed.
Construction of Plasmids
All primers used in this study are listed in Supplementary Table S4. The genes encoding eGFP and the β-glucuronidase with a plant derived intron (GUSi; Vancanneyt et al., 1990) were amplified using GFP-Fw/GFP-Rv primers and GUS-Fw/GUS-Rv primers, respectively, and were added complete attB1 and attB2 sequences by the second PCR with attB1 and attB2 primers. In order to clone aphid salivary genes, cDNAs produced from aphid head total RNA were used to enrich transcripts encoding salivary genes. Adult aphids feeding on V. faba were flash frozen in liquid nitrogen, and decapitated with a scalpel between the first and second pairs of legs. Head RNA was extracted from 10 to 20 individuals using the RNeasy plant mini kit (Qiagen). cDNA synthesis was performed with poly-T primers using the AMV reverse transcriptase system (Promega) according to the manufacturers’ instructions. ACYPI009919 (Ap25) and ACYPI008617 (ApC002) open reading frames encoding mature proteins were amplified from the cDNA of the Ar_Po_58 line (pea biotype) with Phusion DNA polymerase (ThermoFisher Scientific) using AP25-Fw/AP25-Rv and APC002-Fw/APC002-Rv primers (Supplementary Table S4), respectively. attB1 and attB2 sites were added with a second PCR using attB1 and attB2 primers. All amplicons, eGFP, GUSi and two salivary genes, were recombined by BP reaction into pDONR207 (Invitrogen) using BP clonase II (Invitrogen) and produced entry vectors (Table S2). Entry vectors were recombined by LR reaction using LR clonase II (Invitrogen) into pEAQ-HT-DEST1 expression vector (Supplementary Table S2; Sainsbury et al., 2009). Expression vectors were transformed in electro-competent A. tumefaciens cells (Supplementary Table S2).
Infiltration of Agrobacterium
Agrobacterium tumefaciens-mediated transient expression was performed as described (Rivas et al., 2002). Freshly cultured cells were resuspended in induction buffer [10 mM MgCl2, 10 mM Mes (2-(N-morpholino) ethanesulfonic acid), pH 5.6, and 150 μM acetosyringone] to an O.D.600 (optical density at 600 nm) of 0.5. Cells were syringe infiltrated into leaves of 2 week-old P. sativum (Supplementary Table S1) and 3 week-old M. sativa plants.
GUS Staining
Plant leaves infiltrated with Agrobacterium were detached 3 days post infiltration (dpi) and GUS activity was visualized as described (Jefferson, 1987). Briefly, leaves were vacuum infiltrated with GUS staining solution (61 mM Na2HPO4, 39 mM NaHPO4, 0.1% triton X-100, 10 mM EDTA, 0.3% H2O2 and 1.5 mM 5-bromo, 4-chloro, 3-indolyl glucuronide (X-Glc, Biosynth), pH 7.0) and incubated overnight at 37°C. Then chlorophyll discoloration was performed with successive washes with ethanol at 37°C.
Protein Extraction and Western-Blot Analyses
Three leaf disks per leaf were sampled using a cork borer (area = 0.79 cm2) at 0, 7, and 10 days post-infiltration for GFP protein detection. Leaf disks were flash frozen in liquid nitrogen and stored at -80°C. Proteins were extracted in 120 μl extraction buffer (50 mM tris pH 7.5, 1 μM Dithiothreitol, glycerol 10%, 1 mM PMSF (Phenylmethylsulfonyl fluoride), 0.05% triton X-100). Extracts from pea plants were prepared as described (Canonne et al., 2011) and supernatants were resuspended in 5X loading buffer (0.5 M Tris pH 6.8, SDS 10%, glycerol 50 and 0.001% bromophenol blue). Fifteen microliters of samples were separated by SDS-PAGE (12% polyacrylamide) and transferred on PVDF (Polyvinylidene fluoride) membranes, (Merck Millipore) as described (Witte et al., 2004) with following modifications: PVDF membranes were soaked in methanol before and after transfer, and then washed in water. Methanol in transfer buffer was replaced by ethanol. The rabbit anti-GFP antibody (Biorad) and secondary antibodies (polyclonal goat anti-rabbit antibody peroxidase conjugated; Sigma–Aldrich) were both used at 1:10000. Detection was performed by chemiluminescence using Clarity Western ECL Substrate (Biorad) and CL-XPosureTM Film (Lifetechnologies) according to manufacturer’s instructions. Coomassie stains were performed with 0.2% Coomassie Brilliant Blue R250 (Sigma) in 50:40:10 water, methanol, acetic acid.
Aphid Performance Test on Agroinfiltrated Leaves
One young leaf of the P. sativum ZP1109 cultivar was syringe-infiltrated with A. tumefaciens AGL-1 strain harboring expression vectors. Three days later (at 3 dpi), 6 new-born aphids (1 day-old) born on V. faba were installed on P. sativum agroinfiltrated leaves in custom-built clip cages (area = 2.54 cm2). When aphids were 8 days-old (10 dpi), clip cages were opened and the number of surviving aphids was recorded to estimate the survival rate. From the survivors, one average sized aphid was selected and transferred to a new P. sativum leaf that was infiltrated with the same construct of Agrobacterium 3 days before the transfer. Clip cages were opened when aphids were 12 and 15 days-old to assess the fecundity by counting the number of nymphs produced by each aphid. The nymphs were removed after each counting to avoid overcrowding of the cages. In one experiment, 10 replicates per gene were performed and the same experiment was repeated twice, producing 20 replicates. All the experiments were conducted at 20°C, 16 h day/8 h night photoperiod.
Statistical Analyses
All statistical analyses were conducted in R version 3.1.2 (R Core Team, 2014). Data were checked for approximate normal distribution by graphical visualizing of residuals. The effects of the different factors (pea cultivar, aphid lineage, expressed gene) were tested and the simplest model explaining the data was used. Analyses of survival rates (Figures 2A and 3A) and fecundity counts (Figure 3B) were performed by classical linear regressions using generalized linear models (GLM) with binomial and Poisson distributions, respectively. Both tests were followed by multiple comparisons of means by the Tukey contrast method implemented in the package “multcomp” (Hothorn et al., 2008). The influence of pea cultivars and aphid lineage on aphid biomass (Figure 2B) was analyzed by a two-way ANOVA. Tukey’s post hoc multiple comparisons of means from the R package “agricolae” (De Mendiburu, 2014) were used to reveal differences between groups.
FIGURE 2.

ZP1109 and ZP1130 allow only A. pisum pea biotype reproduction. Survival (A) and biomass (B) of five aphid lineages are measured on the pea cultivars ZP1109 (black bars) and ZP1130 (gray bars). Bars show the average of survival or biomass and standard deviation for five replicates per conditions. Statistical differences between groups are indicated by different letters. a, b, c; indicate groups determined by multiple comparisons tests after GLM and ANOVA analyses for survival and biomass data, respectively.
FIGURE 3.

Transient expression of Ap25 promotes reproduction of A. pisum on ZP1109. Leaves of ZP1109 cultivar of P. sativum were syringe-infiltrated with A. tumefaciens AGL-1 carrying the pEAQ-HT-DEST1 plasmid encoding the genes for ACYPI008617 (ApC002; white bar) and ACYPI009919 (Ap25; gray bar), under the control of the CaMV 35S promoter. A. tumefaciens carrying the pEAQ-HT-DEST1 plasmid encoding the gene for enhanced GFP (eGFP; black bar), under the control of the CaMV 35S promoter was used as control. (A) Survival rate of Ar_Po_58 line is not affected by the transient expression of the salivary genes ApC002 and Ap25. At 3 days post infiltration, six new-born aphids were clip caged on agroinfiltrated leaves and counted when aphids were adults (8 days-old) to check the survival rate. Bars show the average percentage of survivors plus the standard deviation of 20 biological replicates. After the survival test, one aphid per plant was kept and placed on a new 3-day-post-infiltrated leaf to check the fecundity. (B) The transient expression of different genes influenced aphid nymph production. Bars show the average number of nymphs produced plus the standard deviation of 20 biological replicates. Different letters indicate significant differences between groups.
Results
Screening of P. sativum and M. sativa Cultivars for Agroinfiltration
Combinations of A. tumefaciens and various M. sativa and P. sativum cultivars were tested using the β-glucuronidase containing a plant derived intron (GUSi) as a reporter gene (Vancanneyt et al., 1990). Green fluorescence protein (GFP) could not be used as a reporter due to strong auto fluorescence induced in the leaf surface by the infiltration. Initially, we tested two plant expression vectors pGWB402Ω (Nakagawa et al., 2007) and pEAQ-HT-DEST1 (Sainsbury et al., 2009) in some pea cultivars, but the difference in expression levels between the two vectors was not very clear or slightly better when pEAQ-HT-DEST1 was used. Therefore, we used pEAQ-HT-DEST1 for the rest of screening. Also, our initial test showed that a bacterial suspension with an O.D.600 less than 0.3 resulted in a weak transgene expression and more than 0.7 triggered leaf chlorosis a few days after infiltration. Therefore, for the rest of the screening, agroinfiltrations were performed using syringe infiltration method and a bacterial suspension with an O.D.600 = 0.5. Seventeen P. sativum (Supplementary Table S3) and five M. sativa cultivars were selected based on geographic origin and phylogenetic groups in order to screen a large genetic diversity. Each cultivar was infiltrated with three Agrobacterium strains [C58C1, GV3101 and AGL-1 (Supplementary Table S2)] each harboring pEAQ-HT-DEST1-GUSi to identify the combination of plant and bacterium genotypes that produce high amount of GUS proteins. Leaves were analyzed histochemically for GUS activity at 3 dpi. At least three independent experiments were performed for each combination and results are summarized in Table 1. None of the M. sativa cultivars was suitable for Agrobacterium-mediated transient expression in leaves as no GUS staining could be observed in these plants under the tested conditions. High differences between pea cultivars were observed. Most of the pea cultivars had no or weak intensities of GUS staining. Of the three Agrobacterium strains used in this study, AGL-1 induced the highest expression of GUS, and C58C1 was the lowest inducer. Two pea genotypes, ZP1130 and ZP1109, inoculated with AGL-1 showed most intense coloration during GUS staining (Figure 1A). GUS staining could be observed at 3 dpi for both cultivars, ZP1130 and ZP1109. To confirm protein expression in these two cultivars, transient expression of eGFP and detection by western-blot was performed (we could not visualize GFP fluorescence due to autofluorescence induced by wounding). eGFP protein was detected at 7 and 10 dpi for both ZP1109 and ZP1130 (Figure 1B). During this study, yellowing of the leaves starting at 9–10 dpi for ZP1130 and at 12–13 dpi for ZP1109 was observed. This leaf yellowing was probably due to AGL-1 infection as the yellowing was observed in the leaves infiltrated with Agrobacterium with empty vector control, and no yellowing was observed in buffer infiltrated leaves (data not shown). Taken together, we identified two pea cultivars, ZP1130 and ZP1109, and the A. tumefaciens strain AGL-1 as the combinations that are suitable for transient gene expression, and we presumed that 3–8 dpi for ZP1130 and 3–10 dpi for ZP1109 are the timing to examine the effect of transgene expression in the plant or plant–aphid interactions.
Table 1.
Results of screening of P. sativum and M. sativa cultivars for agroinfiltration.
| C58C1b | GV301 | AGL-1 | |
|---|---|---|---|
| Pisum sativuma | |||
| AP3783 | N | I | W |
| AP3830 | N | N | N |
| WP1018 | W | W | W |
| ZP690 | N | N | N |
| ZP748 | N | N | N |
| ZP750 | N | W | W |
| ZP793 | N | W | W |
| ZP747 | N | N | W |
| ZP1109 | N | I | I |
| ZP1124 | N | N | W |
| ZP1130 | N | W | S |
| ZP3495 | N | W | N |
| ZP3508 | N | N | N |
| ZP3514 | N | N | N |
| ZP3535 | N | N | N |
| ZP3570 | N | N | N |
| ZP3664 | N | W | W |
| Medicago sativum | |||
| Comète | nd | nd | N |
| Harpe | nd | nd | N |
| Lux Timbale | nd | nd | N |
| Lux Galaxie | nd | nd | N |
| Cannelle | nd | nd | N |
apea or alfalfa cultivars used in this study. bA. tumefaciens strains used in this study. N, no coloration; W, weak; I, intermediate coloration; S, strong coloration (Figure 1A), nd, not determined.
FIGURE 1.
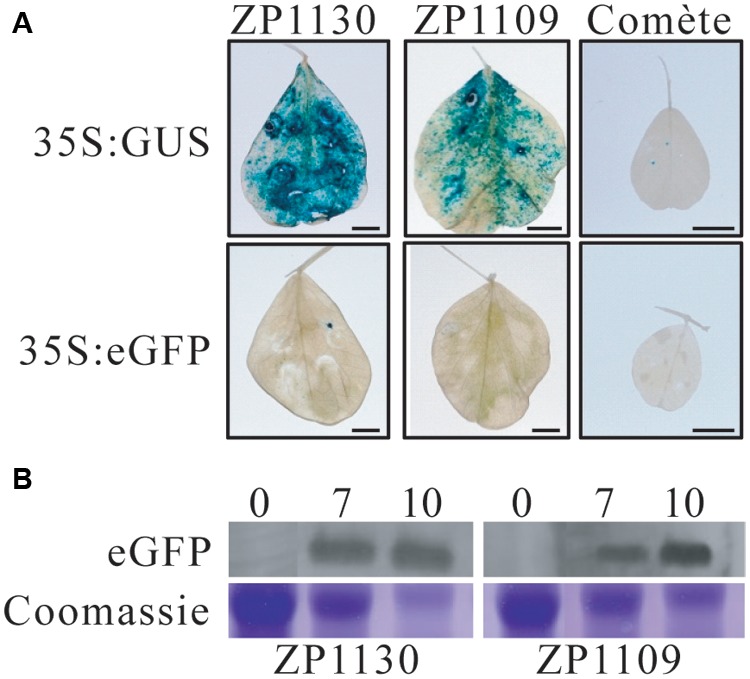
Optimization of agroinfiltration in legume plants. (A) GUS expression in P. sativum and M. sativa. Leaves of ZP1130 and ZP1109 cultivars of P. sativum and comète cultivar of M. sativa were syringe-infiltrated with A. tumefaciens AGL-1 carrying the pEAQ-HT-DEST1 plasmid encoding the gene for β-glucuronidase with an intron (GUSi), under the control of the CaMV 35S promoter (35S:GUSi). GUS staining was performed 3 days post infiltration. Results are representatives of four independent experiments. A. tumefaciens carrying the pEAQ-HT-DEST1 plasmid encoding the gene for enhanced GFP, under the control of the CaMV 35S promoter (35S:eGFP) was used as control. Scale bars represent 0.5 cm. (B) Total protein extracts of leaves from ZP1130 and ZP1109 cultivars expressing eGFP using Agrobacterium-mediated transformation were separated by 12% SDS-PAGE and analyzed by immunoblot using the anti-GFP antibody. Samples were harvested at 0, 7 and 10 days post infiltration. Coomassie stained portions of the gel (Rubisco) are shown to compare sample loading between lanes.
Pea Cultivars ZP1130 and ZP1109 Are Hosts Only for the A. pisum Pea Biotype
Survival rate and biomass of the five A. pisum lineages belonging to three biotypes (pea, alfalfa and clover; Supplementary Table S1) were assessed on the ZP1130 and ZP1109 pea cultivars we identified as suitable for agroinfiltration (Figure 2). Analysis revealed that the two plant cultivars did not influence the survival rate and produced aphid biomass [χ2= 0.14, P = 0.243; F(5,44) = 129.7, P = 0.261; for survival and biomass, respectively], but pea aphid lineages differed significantly in their survival rates (χ2= 19.04, P < 0.001) and biomass production [F(4,45)= 128.9, P < 0.001]. The pea adapted lineages Ar_Po_28 and Ar_Po_58 showed a higher survival rate on the pea cultivars at day 9 compared to L9Ms14 (alfalfa biotype), YR2 and T8005 (clover biotype). The difference in survival was very pronounced between pea and alfalfa specialized lineages, and intermediate for lineages of the clover biotype (Figure 2A). On both ZP1130 and ZP1109 cultivars, only the lineages of the pea biotype (Ar_Po_28 and Ar_Po_58) produced a substantial biomass. Although Ar_Po_28 had a significantly higher biomass than Ar_Po_58, both lineages performed well on the tested cultivars that they seem to recognize as favorable hosts. By contrast, alfalfa and clover adapted lineages hardly reproduced on the pea cultivars that seem to be non-host plants in these interactions (Figure 2B). Thus, the ZP1130 and ZP1109 cultivars are selective hosts for A. pisum biotypes, allowing to assess host and non-host interactions using agroinfiltration experiments.
Transient Expression of AP25 in ZP1109 Increased A. pisum Fecundity
Next, we expressed two salivary genes in ZP1109 by agroinfiltration using strain AGL-1 and examined their effects on A. pisum feeding on the infiltration site. We chose Ar_Po_58 as a test aphid line as it belongs to the pea biotype and harbors no secondary symbiont, which may interfere with plant–aphid interactions. Mature proteins encoding ACYPI008617 (ApC002) and ACYPI009919, which we named Ap25, were transiently expressed using pEAQ-HT-DEST1 vector. The genes were expressed by CaMV 35S promoter, which is known to be ubiquitously and constitutively activated in various plant tissues including epidermal, mesophyll and phloem tissues (Stockhaus et al., 1989). In the process of establishing phloem feeding, A. pisum punctures various tissues and salivates (Schwarzkopf et al., 2013). When the aphid attempts to feed on non-host legume plant, it punctures epidermal and mesophyll cells but cannot establish phloem feeding: therefore, the factors that determine the compatibility between A. pisum and host plants are present in those tissues (Schwarzkopf et al., 2013). Based on these informations, we thought it is important to express salivary proteins ubiquitously to fully assess their functions in plants and used 35S promoter for transient expression. 35S promoter has been successfully used in other studies on aphid salivary proteins (Bos et al., 2010; Naessens et al., 2015). ApC002 was chosen because it is one of the most studied salivary proteins and is shown to be essential for A. pisum to feed on the universal host plant (V. faba; Ferrari et al., 2008; Peccoud et al., 2009). Ap25 was selected because the gene presents the same features as that of ApC002: the gene was identified in salivary glands by transcriptomic analyses (Carolan et al., 2011), is specifically expressed in salivary glands (Akiko Sugio et al., unpublished data), and encodes a signal peptide and a small (13.9 kDa) mature protein with no predicted function. Although many genes are duplicated in A. pisum genome, Ap25, like ApC002, is single copy in A. pisum and its orthologues exist only in the Aphididae family (Hélène Boulain et al., unpublished data).
In this study, transient protein expression was observed from 3 (detected by GUS activity) to 10 days (detected by western blot) at 20°C after infiltration of Agrobacterium. A. pisum starts to reproduce around 9th day after birth, reaches its peak of reproduction around 5 days later, and slows down but continues to reproduce until its death at an age of approximately 30 days (Tsuchida et al., 2004). By supplying newly infiltrated leaves, we extended the duration of the experiment to characterize the effect of transgene expression on aphid fecundity. Leaves of ZP1109 were infiltrated with AGL-1 harboring expression plasmids of eGFP, ApC002 or Ap25. Three days after the infiltration, six new-born aphids of the pea adapted clone Ar_Po_58 were clip caged on the infiltrated leaves. When the aphids were 8 days-old (at 10 dpi) the cages were opened to count the number of survivors. One aphid was transferred to a new 3-day-post-infiltrated leaf. Production of nymphs of the caged adult was measured when the aphid was 12 and 15-day old corresponding to the peak of reproduction of adults. Survival rate and total number of nymphs of the aphids are shown in Figure 3. There was no difference in the survival rate of the aphids that were fed on the leaves expressing the three tested genes (χ2= 0.01, P = 0.96). Production of nymphs of Ar_Po_58 feeding on ApC002 expressing leaves was same as that of aphids feeding on eGFP expressing leaves, while the aphids produced approximately 12% more offspring on Ap25 expressing leaves than on eGFP expressing leaves (20 biological replicates, χ2= 18.75, P < 0.001). The results indicate that Ap25 plays a role in promoting A. pisum feeding on P. sativum.
Discussion
Here, we screened cultivars of P. sativum and M. sativa using GUS activity as a reporter and identified two P. sativum cultivars, ZP1130 and 1109, that are amenable to Agrobacterium mediated transient gene expression. We noted that A. tumefaciens strain AGL-1 was the most efficient strain among the three strains tested. This can be explained by the presence of extra virulent factors in this strain (Jin et al., 1987). We also noted that a few days upon infiltration with high concentration of A. tumefaciens (O.D.600> 0.7), chlorosis appeared and was restricted to the agroinfiltrated area. Pruss et al. (2008) also observed that fully virulent and disarmed A. tumefaciens strains also triggered chlorosis restricted to the infiltrated area in tobacco plants. Although the mechanisms underlying this chlorosis have not been well understood, it could be due to a defense response to the A. tumefaciens involving the chloroplasts (Pruss et al., 2008).
Two tested A. pisum lines belonging to the pea biotype reproduced well on these two cultivars, while members of the alfalfa and clover biotypes could not survive and reproduce well on them. This indicates that these two cultivars serve as host plants of the pea biotype only and can be used to characterize candidate aphid salivary genes that may determine the compatibility of A. pisum biotypes with P. sativum. Interestingly, we found differences in aphid performances, as measured by survival and biomass, between the two P. sativum adapted lines on both pea cultivars. In particular, biomass production by Ar_Po_28 was about twice more than that of Ar_Po_58. Since the two lines differ in both genotype and symbiont composition (Ar_Po_28 harbors Rickettsia and Serratia secondary symbionts while Ar_Po_58 is free of any secondary symbiont, Supplementary Table S1), it is difficult to tell which factor (aphid genome or symbiont status), alone or in interaction, accounts for these differences in performances.
Although we optimized agroinfiltration in P. sativum to study the host specialization mechanisms in A. pisum, the system can be used to study the functions of P. sativum genes or effectors of other pea parasites. P. sativum is an important legume crop used in arable rotations for the production of nutritious food for both humans and animals. Various projects to identify genes involved in P. sativum biotic and abiotic stress resistances are ongoing (Lejeune-Henaut et al., 2008; Hamon et al., 2013; Desgroux et al., 2016) and whole-genome sequencing of P. sativum is underway (Alves-Carvalho et al., 2015). Therefore, the P. sativum research community is in need of various tools to analyze the genes of agronomical interest that will be identified in near future. Though P. sativum is reported to be stably transformed (Svabova et al., 2005), it remains to be a time consuming and difficult task. Recent application of virus vectors in P. sativum provides a new tool to express transgene in pea plant relatively quickly, but it is still time consuming (in a few weeks; Meziadi et al., 2016) and the agroinfiltration method described here provides another way to express transgenes in a few days. By using various GatewayTM compatible vectors available for agroinfiltration (Karimi et al., 2002; Nakagawa et al., 2007), fusion proteins or dsRNA will be easily produced in P. sativum leaves. Furthermore, coexpression of a few proteins may be realized by infiltration of A. tumefaciens with different expression constructs.
We transiently expressed ApC002 and Ap25 in P. sativum leaves and examined the survival and fecundity of an A. pisum line of the pea adapted biotype. The aphids grew well in the clip cages fixed on the agroinfiltrated leaves and produced offspring. Since ApC002 is required for A. pisum feeding on V. faba plant, which is a universal plant of all A. pisum biotypes (Ferrari et al., 2008; Peccoud et al., 2009), and in planta (Arabidopsis and N. benthamiana) expression of MpC002 increases the fecundity of M. persicae feeding on the plants, we expected that ApC002 expression in P. sativum leaves would also increase the fecundity of the aphids. However, the survival and fecundity of the aphids fed on ApC002 expressing plants were at the same level as that of the aphids feeding on eGFP expressing plants. As C002 is one of the abundantly expressed salivary genes in A. pisum (Mutti et al., 2006), the aphids may produce enough of this protein and may not benefit significantly from extra production of ApC002 in P. sativum leaves. On the other hand, expression of Ap25 in P. sativum leaves increased the fecundity of the aphids. Ap25 is an Aphididae specific gene which encodes a small protein with a signal peptide. As the protein does not show homology with known proteins, the function of Ap25 is unknown. It is possible that the protein interferes with plant defense reactions triggered by aphid feeding and facilitates nutrient acquisition from the pea plant. Carolan et al. (2011) identified more than 300 salivary genes in A. pisum and more than half of the identified genes encode proteins with unknown function (Carolan et al., 2011). The agroinfiltration method described here provides a mean to examine the functions of those salivary proteins in relatively short time and also allows us to investigate whether those genes are determinants of compatibility between P. sativum and A. pisum biotypes. As the second step of this study, we envisage to express salivary proteins with biotype specific sequences in the pea leaves and examine how they affect the performance of different pea biotypes installed on the leaves. Further, the agroinfiltration technique can be combined with aphid gene silencing to investigate whether a gene expressed in leaves can complement the silenced gene function (Naessens et al., 2015). Studies on plant–insect interactions at a molecular level are less advanced compared to plant-microbe interaction studies partly because it is not yet possible to transform insect herbivores. The tools to manipulate host plants, like the method described here, can provide alternative ways to examine plant–insect interactions at a molecular level and will be able to contribute to advance the field.
Author Contributions
EG, HB, YA, CLP, KC, SM, J-CS, and AS designed the experiments. EG, HB, YA, CLP, KC, SM, KS, J-CS and AS conducted the experiments. EG, HB, GK, J-CS and AS edited the manuscript. GK, J-CS and AS provided funding for the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors acknowledge to Gaëtan Denis and Jean-François Le Gallic for experimental support, Drs Marie-Laure Pilet-Nayel and Alain Baranger and their team for providing pea seeds, Anne Moussart for providing alfalfa seeds, Dr. George P. Lomonossoff for providing pEAQ vectors, Prof. Tsuyoshi Nakagawa for providing pGWB vectors and Mathew Smoker for Agrobacterium strains. This project was supported by the French Agence Nationale de la Recherche grant (13-JSV7-0012-01-Bugspit) to AS and by Procope German-French collaboration program funded by the German Academic Exchange Service (DAAD) and Campus France to GK (57049694) and J-CS (30753SE). Ph.D. grant to HB is funded by INRA SPE and Région Bretagne.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01171
References
- Alves-Carvalho S., Aubert G., Carrere S., Cruaud C., Brochot A. L., Jacquin F., et al. (2015). Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 84 1–19. 10.1111/tpj.12967 [DOI] [PubMed] [Google Scholar]
- Atamian H. S., Chaudhary R., Dal Cin V., Bao E., Girke T., Kaloshian I. (2013). In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol. Plant Microbe Interact. 26 67–74. 10.1094/mpmi-06-12-0144-fi [DOI] [PubMed] [Google Scholar]
- Bhaskar P. B., Venkateshwaran M., Wu L., Ane J. M., Jiang J. M. (2009). Agrobacterium-mediated transient gene expression and silencing: a rapid tool for functional gene assay in potato. PLoS ONE 4:e5812 10.1371/journal.pone.0005812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. I., Prince D., Pitino M., Maffei M. E., Win J., Hogenhout S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6:e1001216 10.1371/journal.pgen.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canonne J., Marino D., Jauneau A., Pouzet C., Briere C., Roby D., et al. (2011). The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell 23 3498–3511. 10.1105/tpc.111.088815 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carolan J. C., Caragea D., Reardon K. T., Mutti N. S., Dittmer N., Pappan K., et al. (2011). Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): a dual transcriptomic/proteomic approach. J. Proteome Res. 10 1505–1518. 10.1021/pr100881q [DOI] [PubMed] [Google Scholar]
- Chaudhary R., Atamian H. S., Shen Z. X., Brigg S. P., Kaloshian I. (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. U.S.A. 111 8919–8924. 10.1073/pnas.1407687111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Dent M., Hurtado J., Stahnke J., McNulty A., Leuzinger K., et al. (2016). Transient protein expression by agroinfiltration in lettuce. Methods Mol. Biol. 1385 55–67. 10.1007/978-1-4939-3289-4_4 [DOI] [PubMed] [Google Scholar]
- De Mendiburu F. (2014). Agricolae: Statistical Procedures for Agricultural Research. R Package Version 1.2-1. Available at: http://CRAN.R-project.org/package=agricolae [Google Scholar]
- De Vos M., Jander G. (2009). Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana. Plant Cell Environ. 32 1548–1560. 10.1111/j.1365-3040.2009.02019.x [DOI] [PubMed] [Google Scholar]
- Desgroux A., L’Anthoene V., Roux-Duparque M., Riviere J. P., Aubert G., Tayeh N., et al. (2016). Genome-wide association mapping of partial resistance to Aphanomyces euteiches in pea. BMC Genomics 17:124 10.1186/s12864-016-2429-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dres M., Mallet J. (2002). Host races in plant-feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. B Biol. Sci. 357 471–492. 10.1098/rstb.2002.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga D. A., De Vos M., Jander G. (2014). Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact. 27 747–756. 10.1094/mpmi-01-14-0018-r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga D. A., Jander G. (2013). The role of protein effectors in plant-aphid interactions. Curr. Opin. Plant Biol. 16 451–456. 10.1016/j.pbi.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Ferrari J., Via S., Godfray H. C. (2008). Population differentiation and genetic variation in performance on eight hosts in the pea aphid complex. Evolution 62 2508–2524. 10.1111/j.1558-5646.2008.00468.x [DOI] [PubMed] [Google Scholar]
- Goodin M. M., Zaitlin D., Naidu R. A., Lommel S. A. (2008). Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21 1015–1026. 10.1094/mpmi-21-8-1015 [DOI] [PubMed] [Google Scholar]
- Guo K., Wang W., Luo L., Chen J., Guo Y., Cui F. (2014). Characterization of an aphid-specific, cysteine-rich protein enriched in salivary glands. Biophys. Chem. 189 25–32. 10.1016/j.bpc.2014.03.006 [DOI] [PubMed] [Google Scholar]
- Hamon C., Coyne C. J., McGee R. J., Lesne A., Esnault R., Mangin P., et al. (2013). QTL meta-analysis provides a comprehensive view of loci controlling partial resistance to Aphanomyces euteiches in four sources of resistance in pea. BMC Plant Biol. 13:45 10.1186/1471-2229-13-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. J., Via S. (2001). Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412 904–907. 10.1038/35091062 [DOI] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P. (2008). Simultaneous inference in general parametric models. Biom. J. 50 346–363. 10.1002/bimj.200810425 [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313 10.1371/journal.pbio.1000313.g001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiery J., Stoeckel S., Nouhaud P., Mieuzet L., Maheo F., Legeai F., et al. (2012). Genome scans reveal candidate regions involved in the adaptation to host plant in the pea aphid complex. Mol. Ecol. 21 5251–5264. 10.1111/mec.12048 [DOI] [PubMed] [Google Scholar]
- Jaubert-Possamai S., Rispe C., Tanguy S., Gordon K., Walsh T., Edwards O., et al. (2010). Expansion of the miRNA pathway in the Hemipteran Insect Acyrthosiphon pisum. Mol. Biol. Evol. 27 979–987. 10.1093/molbev/msp256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A. (1987). Assaying chimeric genes in plants: the GUS Gene fusion system. Plant Mol. Biol. Rep. 5 387–405. 10.1007/BF02667740 [DOI] [Google Scholar]
- Jin S., Komari T., Gordon M. P., Nester E. W. (1987). Genes responsible for the supervirulence phenotype of Agrobacterium tumefaciens A281. J. Bacteriol. 169 4417–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloshian I., Walling L. L. (2016). Hemipteran and dipteran pests: effectors and plant host immune regulators. J. Integr. Plant Biol. 58 350–361. 10.1111/jipb.12438 [DOI] [PubMed] [Google Scholar]
- Kanvil S., Collins C. M., Powell G., Turnbull C. G. N. (2015). Cryptic virulence and avirulence alleles revealed by controlled sexual recombination in pea aphids. Genetics 199 581–593. 10.1534/genetics.114.173088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. 10.1016/s1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- King J. L., Finer J. J., McHale L. K. (2015). Development and optimization of agroinfiltration for soybean. Plant Cell Rep. 34 133–140. 10.1007/s00299-014-1694-4 [DOI] [PubMed] [Google Scholar]
- Lejeune-Henaut I., Hanocq E., Bethencourt L., Fontaine V., Delbreil B., Morin J., et al. (2008). The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor. Appl. Genet. 116 1105–1116. 10.1007/s00122-008-0739-x [DOI] [PubMed] [Google Scholar]
- Meziadi C., Blanchet S., Richard M. M. S., Pilet-Nayel M. L., Geffroy V., Pflieger S. (2016). Bean pod mottle virus: a new powerful tool for functional genomics studies in Pisum sativum. Plant Biotechnol. J. 14 1777–1787. 10.1111/pbi.12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti N. S., Louis J., Pappan L. K., Pappan K., Begum K., Chen M. S., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. U.S.A. 105 9965–9969. 10.1073/pnas.0708958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti N. S., Park Y., Reese J. C., Reeck G. R. (2006). RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J. Insect Sci. 6 1–7. 10.1673/031.006.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naessens E., Dubreuil G., Giordanengo P., Baron O. L., Minet-Kebdani N., Keller H., et al. (2015). A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr. Biol. 25 1898–1903. 10.1016/j.cub.2015.05.047 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., et al. (2007). Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71 2095–2100. 10.1271/bbb.70216 [DOI] [PubMed] [Google Scholar]
- Pan Y. M., Zhu J. J., Luo L., Kang L., Cui F. (2015). High expression of a unique aphid protein in the salivary glands of Acyrthosiphon pisum. Physiol. Mol.Plant Pathol. 92 175–180. 10.1016/j.pmpp.2015.04.006 [DOI] [Google Scholar]
- Peccoud J., de la Huerta M., Bonhomme J., Laurence C., Outreman Y., Smadja C. M., et al. (2014). Widespread host-dependent hybrid unfitness in the pea aphid species complex. Evolution 68 2983–2995. 10.1111/evo.12478 [DOI] [PubMed] [Google Scholar]
- Peccoud J., Maheo F., De La Huerta M., Laurence C., Simon J. C. (2015). Genetic characterisation of new host-specialised biotypes and novel associations with bacterial symbionts in the pea aphid complex. Insect Conserv. Divers. 8 484–492. 10.1111/icad.12131 [DOI] [Google Scholar]
- Peccoud J., Ollivier A., Plantegenest M., Simon J. C. (2009). A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl. Acad. Sci. U.S.A. 106 7495–7500. 10.1073/pnas.0811117106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard K., Lee R., Hellens R., Macknight R. (2013). Transient gene expression in Medicago truncatula leaves via Agroinfiltration. Methods Mol. Biol. 1069 215–226. 10.1007/978-1-62703-613-9_15 [DOI] [PubMed] [Google Scholar]
- Pitino M., Hogenhout S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe Interact. 26 130–139. 10.1094/MPMI-07-12-0172-FI [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Nester E. W., Vance V. (2008). Infiltration with Agrobacterium tumefaciens induces host defense and development-dependent responses in the infiltrated zone. Mol. Plant Microbe Interact. 21 1528–1538. 10.1094/MPMI-21-12-1528 [DOI] [PubMed] [Google Scholar]
- R Core Team (2014). A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rispe C., Kutsukake M., Doublet V., Hudaverdian S., Legeai F., Simon J. C., et al. (2008). Large gene family expansion and variable selective pressures for cathepsin B in aphids. Mol. Biol. Evol. 25 5–17. 10.1093/molbev/msm222 [DOI] [PubMed] [Google Scholar]
- Rivas S., Romeis T., Jones J. D. G. (2002). The Cf-9 disease resistance protein is present in an approximately 420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14 689–702. 10.1105/tpc.010357 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rodriguez P. A., Bos J. I. B. (2013). Toward understanding the role of aphid effectors in plant infestation. Mol. Plant Microbe Interact. 26 25–30. 10.1094/mpmi-05-12-0119-fi [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E. C., Lomonossoff G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7 682–693. 10.1111/j.1467-7652.2009.00434.x [DOI] [PubMed] [Google Scholar]
- Santos-Rosa M., Poutaraud A., Merdinoglu D., Mestre P. (2008). Development of a transient expression system in grapevine via agro-infiltration. Plant Cell Rep. 27 1053–1063. 10.1007/s00299-008-0531-z [DOI] [PubMed] [Google Scholar]
- Schwarzkopf A., Rosenberg D., Niebergall M., Gershenzon J., Kunert G. (2013). To Feed or not to feed: plant factors located in the epidermis, mesophyll, and sieve elements influence pea aphid’s ability to feed on legume species. PLoS ONE 8:e75298 10.1371/journal.pone.0075298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. C., d’Alencon E., Guy E., Jacquin-Joly E., Jaquiery J., Nouhaud P., et al. (2015). Genomics of adaptation to host-plants in herbivorous insects. Brief. Funct. Genomics. 14 413–423. 10.1093/bfgp/elv015 [DOI] [PubMed] [Google Scholar]
- Stockhaus J., Schell J., Willmitzer L. (1989). Correlation of the expression of the nuclear photosynthetic gene ST-LS1 with the presence of chloroplasts. EMBO J. 8 2445–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svabova L., Smykal P., Griga M., Ondrej V. (2005). Agrobacterium-mediated transformation of Pisum sativum in vitro and in vivo. Biol. Plant. 49 361–370. 10.1007/s10535-005-0009-6 [DOI] [Google Scholar]
- Tsuchida T., Koga R., Fukatsu T. (2004). Host plant specialization governed by facultative symbiont. Science 303:1989 10.1126/science.1094611 [DOI] [PubMed] [Google Scholar]
- Vancanneyt G., Schmidt R., O’Connor-Sanchez A., Willmitzer L., Rocha-Sosa M. (1990). Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. 220 245–250. 10.1007/BF00260489 [DOI] [PubMed] [Google Scholar]
- Via S., Conte G., Mason-Foley C., Mills K. (2012). Localizing F(ST) outliers on a QTL map reveals evidence for large genomic regions of reduced gene exchange during speciation-with-gene-flow. Mol. Ecol. 21 5546–5560. 10.1111/mec.12021 [DOI] [PubMed] [Google Scholar]
- Wang W., Dai H. E., Zhang Y., Chandrasekar R., Luo L., Hiromasa Y., et al. (2015a). Armet is an effector protein mediating aphid-plant interactions. Faseb J. 29 2032–2045. 10.1096/fj.14-266023 [DOI] [PubMed] [Google Scholar]
- Wang W., Luo L., Lu H., Chen S. L., Kang L., Cui F. (2015b). Angiotensin-converting enzymes modulate aphid-plant interactions. Sci. Rep. 5:8885 10.1038/srep08885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T., Tjallingii W. F., Thonnessen A., van Bel A. J. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. U.S.A. 104 10536–10541. 10.1073/pnas.0703535104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C. P., Noël L. D., Gielbert J., Parker J. E., Romeis T. (2004). Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55 135–147. 10.1007/s11103-004-0501-y [DOI] [PubMed] [Google Scholar]
- Wroblewski T., Tomczak A., Michelmore R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3 259–273. 10.1111/j.1467-7652.2005.00123.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


