Abstract
Cancer-stromal interaction is a critical process in tumorigenesis. Conventional dish-based co-culture assays simply mix two cell types in the same dish; thus, they are deficient in controlling cell locations and precisely tracking single cell behavior from heterogeneous cell populations. Microfluidic technology can provide a good spatial temporal control of microenvironments, but the control has been typically realized by using external pumps, making long-term cultures cumbersome and bulky. In this work, we present a cell-cell interaction microfluidic platform that can accurately control co-culture microenvironment by using a novel electrolytic cell isolation scheme without using any valves or pneumatic pumps. The proposed microfluidic platform can also precisely control the number of interacting cells and pairing ratios to emulate cancer niches. More than 80% of the chambers captured the desired number of cells. The duration of cell isolation can be adjusted by electrolytic bubble generation and removal. We verified that electrolytic process has a negligible effect on cell viability and proliferation in our platform. To the best of our knowledge, this work is the first attempt to incorporate electrolytic bubble generation as a cell isolation method in microfluidics. For proof of feasibility, we performed cell-cell interaction assays between prostate cancer (PC3) cells and myoblast (C2C12) cells. The preliminary results demonstrated the potential of using electrolysis for micro-environmental control during cell culture. Also, the ratio controlled cell-cell interaction assays was successfully performed showing that the cell pairing ratios of PC3 to C2C12 affected the proliferation rate of myoblast cells due to increased secretion of growth factors from prostate cancer cells.
Introduction
The cancer cell niche is a complex microenvironment, consisting of cancer cells, endothelial cells (EC), macrophages and mesenchymal stem cells (MSC); and tumor-stromal interaction is one of critical factors effecting the development of tumors.1-3 It is believed that tumor cells can exploit nearby normal cells to enhance tumor growth, metastasis and drug resistance. Without establishing or accessing a proper micro-environment, the cancer cells may die or stay in senescence forever.4-5 Recent papers revealed the interaction feedback loops between breast cancer and mesenchymal stem cells.6 For example, SUM159 (breast cancer) cells form a positive feedback interaction with mesenchymal stem cells via IL-6 and CXCL7 cytokines. As a result, the existence of mesenchymal stem cells in the cancer niche can accelerate tumor development. It has been also reported that immune cells play a critical role in cancer metastasis by triggering inflammatory response in the tumor microenvironment.7-8. Tumor associated macrophages (TAM) can enhance angiogenesis, and thus metastasis, by secreting a wide range of growth factors and cytokines. Endothelial cells also contribute to the invasion and metastasis of cancer by promoting cancer stem cell phenotypes and enhancing cancer metastasis.9-10 Compared to the late stage tumor cells, these tumor associated cells are less drug resistant; thus killing these tumor associated normal cells can be used to deter the cancer development.11 Inhibiting the interaction between tumor cells and tumor associated normal cells can be an alternative therapy. As a result understanding cancer-niche interactions is of great importance for developing cancer therapeutics.
Conventionally, cell interactions can be studied by co-culturing two cell types in the same petri dish.12 However, dish-based co-culture methods are limited in several key aspects. Metastatic cancer cells are typically transported as a single CTC, and tumorigenesis from a single cell is quite different from co-culturing many cells.13 As cancer metastases account for more than 90% of cancer-related mortality, modelling the tumorigenesis process in an appropriate microenvironment from a single cell is essential for metastasis study.3,14-15 As the cell behaviour can be affected by neighbouring cells, the conventional dish culture cannot ideally model the tumorigenesis process.16 Another limitation of conventional co-culture assays is its poor spatial control. In conventional interaction experiments, two cell populations are simply mixed in a dish, so the spatial distribution of two cell types can vary from one place to another. Some cells may be surrounded by a large number of different types of cells, while others may form aggregation of the same type of cells. Hence, the precise ratio controlled co-culture cannot be achieved by the conventional dish co-culture. Also, dish-based methods lack the ability of using small samples (< 1000 cells), while CTCs and primary samples are more often available in a small sample. Finally, dish-based studies cannot track individual behaviors of heterogeneous cancer populations. They can only characterize the average behavior of entire cell population. This is an issue as some sub-populations in tumor have a different interaction pathway. For example, it is believed that only the aldehyde dehydrogenase positive (ALDH+) cells have strong interaction with MSC.6
There are a number of previous works reporting on microfluidic platforms for cell-to-cell interaction studies.17-28 Most of them still require loading hundreds or thousands of cells in a device; thus, they suffer from the same issues as in the conventional dish-based co-culture.17-23 Droplet based technology can provide a high-throughput combinatorial pairing of cells, but it lacks the capability of long-term cell culture, limiting its applications in practical co-culture assays.24 Recently, three microfluidic devices reported the cell pairing and cell-to-cell interaction at single-cell resolution.25-28 However, they can only achieve the pairing of two different cells at 1:1 ratio due to geometric restrictions in the device structures. Actually, the cell ratios in tissue can matter in differentiation and it is important to screen assays for many different cell ratios to emulate in-vivo niches.29 In this work, we present a reliable microsystem capable of performing cell interaction assays with a specific ratio between the two different cell types. By applying a hydrodynamic cell capture scheme in two different fluidic streams, we can precisely control the number of captured cells in each type. The captured cells interact through a bridge channel by diffusion of secreted cytokines. Although juxtacrine (contact-dependent) signalling also plays a role in cell-cell interaction, we focus on the secretion based interaction, which has been proved to be important in the interaction between cancer cells and MSCs.6
A major innovation of the proposed microfluidic platform is the electrolytic valving, which generates bubbles to isolate paired cells in a chamber. In previous cell isolation works, pneumatically actuated valves formed from thin PDMS membrane deformation were used.22,26 Although pneumatic 30 actuation has been widely used for control in many microfluidic systems, such microfluidic devices need to be continually connected to the pump during the entire isolation process; otherwise, the pressure, and thus the cell isolation, will be released. This weakness limits the applications and usability of pneumatic valving, especially in mammalian cell culture, which may require specialized culture conditions and long term incubation. The electrolytic valving, on the other hand, can maintain the bubbles generated through electrolysis for isolation without continued external connection. In addition, the pneumatic valving is sensitive to channel geometry.30,31 Channel height and width need to be carefully designed and made rounded to guarantee completely sealing. As the electrolytic bubble can fill almost any shape to isolate the chambers, there are fewer design constraints in the use of electrolytic valving. Moreover, the electrolytic actuation circuit can be implemented compactly using ICs, while the programmable pneumatic control cannot be easily miniaturized. Thus, electrolytic actuation has a higher potential for miniaturization and extension to high-throughput as a standalone micro-system, especially for the applications that need long-term and continual isolation control. In this work, we developed an electrolytic bubble generation and removal scheme, which can be used to control cell-to-cell interaction times to within a precision of one minute.
For the proof of feasibility, we demonstrated the interaction between PC3 (prostate cancer) cells and C2C12 (myoblast) cells by secreted growth factors.32-33 We confirmed that the growth of C2C12 could be boosted by the secretion factors from PC3 cells, and the proliferation rate of C2C12 be affected by the number of PC3 cells inside the same co-culture chamber.
Materials and Methods
Microfluidic Device Operation
The presented platform composes of cell culture chambers, interaction bridges for cell-cell interaction, and bubble chambers with gold electrodes on the substrate (Fig. 1(A)). Two key features of the proposed system are the ability to control the number of cells and cell-to-cell interaction time. To address the first task, the cells were loaded from two separate inlets. The number of loaded cells in a chamber can be determined by the number of capture sites in each chamber. To regulate the cell-cell interaction time, we used an electrolystically generated bubble for valve-less isolation of chambers. Each cell culture chamber is sandwiched by two bubble chambers in the upstream and downstream, respectively. When a sufficient potential for electrolysis is applied to the electrodes, a bubble can be generated, sealing the fluidic path. Then, the isolated cells in the chambers can interact through the interaction bridge by diffusion of secreted factors.
Fig. 1.
Proposed microfluidic chip for paired cell-to-cell interaction assays. (A) The cell culture chamber is surrounded by two bubble chambers in the upstream and downstream. (B) Two different cells are loaded in each cell culture chamber, respectively. (C) After cells are attached on the substrate, bubbles are generated by electrolysis from electrical signals applied to the electrodes. The bubbles seal the microfluidic channels to completely isolate cell culture chambers. The cytokines secreted by cells can diffuse through an interaction bridge, inducing cell-to-cell interaction between the cells in two adjacent cell culture chambers. (D) Since the PDMS is gas permeable, bubbles can be gradually removed by applying negative pressure to the bubble removal channels. Thus, the time of cell isolation can be precisely controlled.
Fig. 1 illustrates the operation of the presented microfluidic platform for paired cell-to-cell interaction. In the cell loading phase (Fig. 1 (B)), cell A is loaded from the left inlet and captured in the left chamber, and cell B is loaded and captured in the right chamber. Since the pressure is balanced between the two fluidic streams, cell loading can be carried out without interference between the two laminar fluids. The number of captured cells (for both type A and type B cells) can be determined by the number of cell capture sites in each chamber (Fig. 4). After 4-6 hours, the captured cells will adhere to the substrate and remain viable and then proliferate within the microfluidic chambers.
Fig. 4.
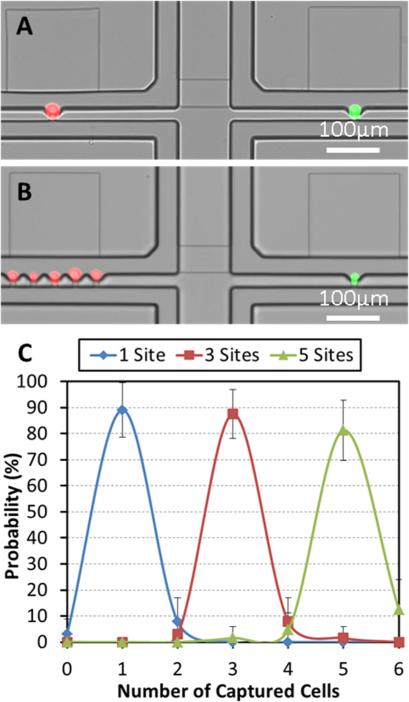
Ratio controlled cell pairing: (A) One of each PC3 (red fluorescent labelled) and C2C12 (green fluorescent labelled) cells, (B) Five PC3 cells and one C2C12 cell, (C) Capture rate for various different numbers in capture sites: one, three, and five, respectively. (N = 4 devices)
In the electrolytic isolation phase (Fig. 1 (C)), voltage is applied to the gold electrodes, and bubbles are generated by electrolysis to seal the cell culture chambers. The cytokines secreted by the cells can be accumulated and diffused through the interaction bridge, so the cells captured in two separate sides can communicate with each other by protein signals. The interaction bridge is narrow (10 μm in width and height) and long (100 μm), so that it will prevent cells from migrating into the other side. The bubbles generated in the bubble chamber are stable over more than three hours. If a longer cell-to-cell interaction time is required, the electrolysis electrodes can be activated again to generate additional bubbles to maintain the isolation. The interaction can be stopped at any time by removing the bubble (Fig. 1 (D)). When negative pressure is applied to the bubble removal channel, the bubble can diffuse out through the PDMS (50 μm thick) from the bubble chamber since PDMS is gas permeable. After the bubble is completely removed, media perfusion is resumed and the secreted proteins are washed away. Thus, the interaction stops.
Device Fabrication
The presented platform composes of PDMS microfluidic layers and the substrate with patterned gold electrodes (Fig. S1). For the microfluidic layer, two masks were used to fabricate the SU8 master. One was used to make the shallow (10 μm) interaction bridge and capture sites, and the other to fabricate the deep (40 μm) channel. The patterned PDMS (PDMS, Sylgard 184, Dow Corning) was formed by the standard soft lithography process (Fig. S1). The Au/Cr layer (400 nm/10 nm) was deposited on the glass wafer by evaporation (EnerJet Evaporator), and patterned by gold etchant (Transene GE-8148 Gold Etch). To seal the device, both the PDMS and the glass substrate were treated by oxygen plasma and then aligned and bonded together.26 The microphotograph of the fabricated device is shown in Fig. 2.
Fig. 2.
Microphotograph of the fabricated device. The light grey color denotes the microfluidic layers including culture chambers, bubble chambers, and bubble removal channels. The cell culture chamber is surrounded by two bubble chambers for isolation. The gold electrode has an inter-digitized comb shape for uniform electrolysis.
Cell Culture and Experiment
C2C12 cells were cultured with DMEM (Gibco 11965), 10% FBS (Gibco 10082), and 1% Pen/Strep (Gibco 15140), and PC3 cells were cultured with RPMI (Gibco 21870), 10% FBS, and 1% Pen/Strep. In the device preparation, the substrate was coated with collagen (BD 354236) overnight before cell loading to enhance cell adhesion. Trypsin/EDTA (Gibco 25200) was used to detach cells from polystyrene culture dishes, and the detached cells were re-suspended in culture media. Then, the cell solution was diluted to 2×105 cells/mL, and 100 μL of cell solution was pipetted into the inlet of the microfluidic platform. The cell solution of one cell type was pipetted in the left inlet first, and the same amount of media was added to the right inlet to balance the pressure. The cells were captured hydrodynamically by gravity flow in the left side. After five minutes, we loaded the other cell type into the right inlet to achieve cell pairing. After loading cell, the cell solution was removed, and both inlets were washed with culture media three times. The culture media (1:1 mixture of the culture media of PC3 and C2C12) was pipetted into the inlets, and the device was placed into the incubator.
After the cells adhered to the glass substrate, the electrodes were connected to a function generator (Agilent 33250A), and a pulse signal (3 V, 0.1 Hz, 1 μs pulse) was applied for electrolysis of the media for 60 seconds. Bubbles filled the whole bubble chambers in the chip and thus completely sealed the culture chambers above and below. The sealing could be maintained for more than three hours until the bubbles gradually disappeared by diffusion through PDMS. The electrolytic isolation can be elongated to adjust the cell-to-cell interaction time, if desired, by applying another pulse signal to the electrolysis electrodes. Cell interaction can be terminated early by applying the negative pressure to the bubble removal channels. It took about one minute to completely remove the bubble. After three days interaction, the number of living cells were determined by LIVE/DEAD staining (Life Technologies), so the cell viability and proliferation rate can measured.
Result and Discussion
Cell Capture Mechanism
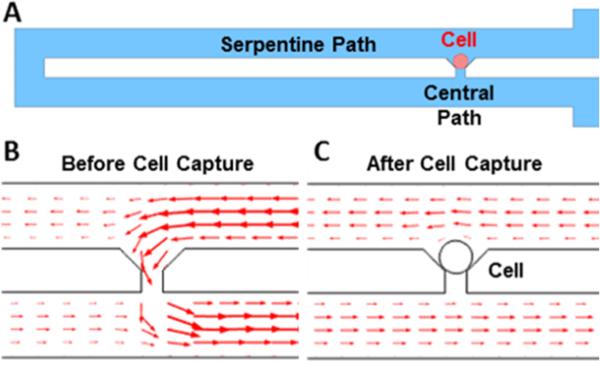
In order to capture the specific number of cells in each culture chamber reliably and reproducibly, cellular valving mechanism has been adopted to deploy the cells hydrodynamically at each capture site (Fig. 3 (A)).34-37 To capture cells at each designated capture site, two paths are created in the design: a central path and a serpentine path. The hydraulic resistance of each path is inversely proportional to its flow rate. As a result, the long serpentine structure has a higher hydrodynamic resistance than the central path. Thus, the cells, following the major flow stream, are likely to be guided to the central path (Fig. 3 (B)). Since the opening of the central path is slightly smaller (Height: 10 μm, Width: 10 μm) than the size of PC3 (human prostate cancer) cells and C2C12 (mouse fibroblast) cells, the cells are sterically 25 captured and plugs the gap, blocking the flow through the central path. Thus, the next cells will be guided through the serpentine path and captured in the downstream capture sites (Fig. 3 (C)). After optimizing the serpentine length, a capture rate of ~90% has been achieved for C2C12, PC3, and Skov3 (ovarian cancer) cells (Table 1).
Fig. 3.
Mechanism of single cell capture scheme: (A) schematic of cellular valving in a serpentine microfluidic channel, (B) Simulated flow direction and velocity before cell capture. The simulated flow distribution suggests that the cells are likely to be guided to a capture site and get captured, (C) Simulated flow direction and velocity after cell capture. The captured cell blocks the flow and the rest of incoming cells will be guided to take a serpentine path.
Table 1.
Optimized geometric parameters of capture sites for C2C12, PC3 and Skov3 cells and the corresponding capture rates.
| Cell Types | C2C12 | PC3 | Skov3 |
|---|---|---|---|
| Cell Diameter Average (μm) | 12.9 | 16.3 | 14.1 |
| Cell Diameter S.D. (μm) | 2.8 | 3.7 | 3.2 |
| Serpentine Length (μm) | 600 | 600 | 600 |
| Central Path Height (μm) | 10 | 10 | 10 |
| Central Path Width (μm) | 10 | 12 | 12 |
| Capture Rate (%) | 92 | 89 | 94 |
By utilizing a high capture rate of single cells over 90% in each capture site, we can extend our design to capture multiple cells simply by adjusting the number of capture sites in each chamber. We can also deploy an arbitrary number (up to five) of two cell types in separate flow streams and study the effect of cell ratio in cell-to-cell interaction during co-culture. Fig. 4 (A, B) shows the ratio-controlled cell capture: pairing one PC3 cell and one C2C12 cell (Fig. 4. (A)), and pairing five PC3 cells and one C2C12 (Fig. 4 (B)). We can vary the combination of pairing from 1:1 to up to 5:1 (or even higher). When the number of capture sites is equal to or smaller than five, more than 80% of the chambers are filled with the captured cells as shown in Fig. 4 (C).
Electrolytic Isolation
In continual media perfusion culture, secreted proteins will be washed away. Therefore, it is important for cell-to-cell interaction studies to control and sustain the isolation of cell culture chamber to accumulate the secreted proteins and cytokines inside the chamber. In this work, we achieve this by generating electrolytic air bubbles in the bubble chambers located between the adjacent cell culture chambers. Fig. 5 (A) shows a bubble chamber with gold electrodes and bubble removal channels. As the PDMS is gas permeable, small bubbles can easily diffuse out through the PDMS membrane within several minutes. To maintain stable channel isolation, we incorporate a large bubble chamber of 100 μm by 100 μm and 40 μm in height (shown with a blue outline in Fig. 5) to facilitate the creation of a larger isolation bubble, which can sustain for more than 3 hours. If longer isolation is required, electrolysis can be performed again after 3 hours to sustain the bubble. Comb finger-shaped electrodes (20 μm in width for each comb finger and 20 μm in spacing between fingers) can generate bubbles evenly inside the whole bubble chamber. Thus, the generated bubble can quickly and completely seal the whole chamber in a minute.
Fig. 5.
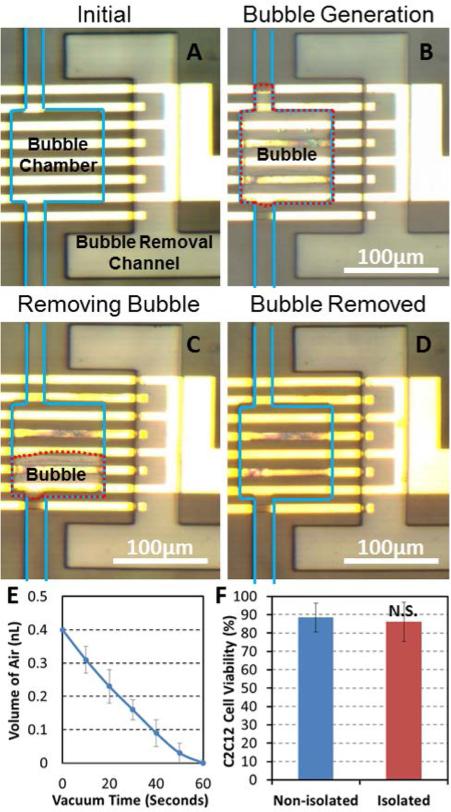
Process of electrolytic bubble generation and removal. (A) Before electrolysis the bubble chamber was filled with cell culture media and the bubble removal channel was filled with air at atmospheric pressure. The blue line delineates microfluidic channels. (B) After 60 seconds of electrolysis, a bubble completely filled the bubble chamber and blocks the flow. The red dotted line is the outline of a bubble. (C) To remove the bubble, we applied negative pressure to the bubble removal channel. After 30 seconds, a bubble shrank in half. (D) After 60 seconds, the bubble was completely removed. (E) The volume of residual air in a bubble chamber as a function of time after applying negative pressure to the bubble removal channel. (F) The day 3 cell viability with and without the operation of electrolytic valve. No significant difference was observed, suggesting electrolysis has negligible effect on cell viability. (N = 4 devices)
The electric field as well as heat generated during electrolysis can be harmful to cells. We have implemented several techniques to minimize these effects. First, we tried to minimize electrolytic voltage. Since the minimum potential required for water electrolysis is around 1.23 Volts, the applied voltage should be higher than this to overcome extra potential drops in the metal wires and media. However, the higher the voltage we use, the more heat we generate. We chose 3 V as an optimal voltage for on-chip electrolysis that can balance electrolytic efficiency and cell viability, based upon design parameters and preliminary experiments. With 3V as a peak operating voltage, we optimize the waveform. Compared to applying a DC voltage, a pulsed waveform can help dissipate the accumulated heat, and a large single bubble can be easily formed from multiple bubble generations from the comb finger-shaped electrodes. For these reasons, we chose to use a pulse wave (3 Volts, 0.1 Hz, 1 μs pulse) for electrolytic bubble generation. The average power is less than 1 nW, and the resulting temperature increase in the cell culture chamber should be less than 0.01°(from simulation). After electrolysis for 60 seconds, a bubble can occupy the whole bubble chamber, and the culture chamber can be completely sealed and isolated (Fig. 5 (B)). In addition, we located the bubble chamber 500 μm away from the cell culture chambers to minimize the effect of any excessive heat generation and E-fields.
Bubble Removal
To precisely control the cell isolation time, the electrolytic bubble can be immediately removed at the end of interaction cycles. We achieve this by applying a negative pressure to the bubble removal channel. As the PDMS is gas permeable, the bubble can diffuse through the PDMS membrane.38-39 We minimized the distance (50 μm) between the bubble chamber and the air removal channel to achieve rapid removal of bubbles in less than a minute. Fig. 5 (C, D) demonstrates the process of bubble removal. The red dotted line is the outline of the bubble, which occupied the whole bubble chamber after electrolysis. After applying negative pressure to the bubble removal channel for 30 seconds, the bubble roughly shrank by half (Fig. 5 (C)); and after another 30 seconds, the bubble completely disappeared (Fig. 5 (D)). Using a negative pressure of around 7 psi, the bubble removal rate was measured as 0.4 nL per minute (Fig. 5 (E)). The bubble removal rate matches well with the predication from other reports.38-39 We confirmed full functionality of the total 14 bubble 55 chambers after ten cycles of bubble generation and removal, demonstrating the reliability and robustness of electrolytic sealing.
Cytokine Diffusion through Bridge Channels
In the fabricated platform, cell-cell interaction is induced from diffusion of secreting proteins through a narrow channel (10 μm by 40 μm, 100 μm long). In order to verify whether this interaction bridge channel can provide adequate diffusion of secreted signalling proteins and cytokines for cell-to-cell communication, we simulated the diffusion of molecules similar to the secreted proteins (Fig. S2). The simulation results show that the two chambers reach almost steady state after 3 hours of isolation. The difference in protein concentrations between the two chambers connected by the interaction bridge channel is only 11% (Fig. S2 (E)), supporting that our platform can provide efficient diffusion flow for cell interaction in the given geometry of our design. In addition, the non-isolated chamber was simulated (Fig. S3). As the secreted proteins are washed away by continual flow, the protein concentration of the receiving side is only 3.3% of that in the isolated chamber (Fig. S3 (E)).
Cell-to-Cell Interaction Assays from Co-Culture of PC3 and C2C12 cells
Before carrying out biological assays in the fabricated devices, we evaluated the effect of electrolysis on cell viability. Since the air generated from electrolysis has more than 1,000 times the volume of the liquid (media), only a small fraction of the media (< 0.005%) in the chamber is consumed. Thus, the change in the media concentration is negligible. Also, the culture media is a buffered media; therefore, the pH of the solution is resistant to possible fluctuations that may be introduced by electrolysis. Three-day cell viability experiments of C2C12 cells were carried out to confirm that there was indeed no effect on cell conditions. After electrolytic sealing for three hours once a day for three days, 86 % of the captured single cells were still viable in the culture chamber. The cell viability was comparable to 89% viability observed in the control without electrolysis (Fig. 5 (F)). Fig. 6 (A) shows that the proliferation of C2C12 cells with electrolytic bubble sealing is similar to that of the control cells without electrolysis.
Fig. 6.
Cell interaction between C2C12 and PC3 cells: (A) Proliferation rates of C2C12 cells with and without bubble isolation. The result confirms that the electrolytic isolation has negligible effect on proliferation. (B) C2C12 and PC3 cell-interaction assays with and without bubble isolation after 3 days. Co-culturing of one C2C12 cell and five PC3 cells significantly enhanced the proliferation of C2C12 cells when electrolytically isolating the culture chambers. Data points represent means ± standard deviations (N = 4 devices), * refers to P < 0.05. (C) With 3 hours of chamber isolation per day for three days, comparing the proliferation of C2C12 cells co-cultured with 1 PC3 and 5 PC3 cells, respectively, the latter showed a significantly higher proliferation rate, confirming that cell ratios are critical for cell interaction studies. Data points represent means ± standard deviations (N = 4 devices), ** refers to P < 0.05. (D) Without chamber isolation, no significant difference was observed in proliferation rates. Data points represent means ± standard deviations (N = 4 devices).
As a proof of concept, we demonstrated the interaction between PC3 cancer cells and C2C12 myoblast cells. PC3 are known to secrete a number of growth factors to enhance the growth of tumors.32 In the previous work, it has been demonstrated that co-culture of PC3 cells and C2C12 cells can enhance the proliferation of C2C12 cells, but whether the ratio of two cell types can affect the interaction is not clear.26,33 Using the fabricated prototype platforms, we loaded both PC3 cells and C2C12 cells in the same device with different ratios on Day 0. For simplicity, we compared only the two different ratios: “1:1” and “5:1” in this experiment. After cell loading, the bubbles were generated to seal the culture chamber for three hours once a day for three days.
We verified the effect of chamber isolation on cell-to-cell interaction assays. We loaded one C2C12 cell and five PC3 cells in the devices. We cultured cells for three days. In one device, we did not apply any electrolytic isolation (control). In the other device, we generated bubbles to isolate the culture chambers for cell-to-cell interaction for three hours per day. The culture chamber sealed by electrolysis (Fig. S4 (A)) has significantly more C2C12 cells than the unsealed one (Fig. S4 (B)). This result implies that the growth factor secreted by the PC3 cells can enhance the proliferation of C2C12 cells. Fig. 6 (B) compares the number of C2C12 cells after three day culture between the control (non-isolated, non-interaction) and the cell-interaction assay.
The effect of different cell pairing ratios was investigated and compared. As five PC3 cells secrete more growth factors than single PC3 cell, it is expected that proliferation enhancement would be more significant where C2C12 cells are paired with five PC3 cells. Fig. 6 (C) supports this hypothesis. In order to confirm that these effects indeed come from cell interaction as a result of building up of secreted factors in the isolated chambers, we conducted the same assay without electrolytic isolation as a control. Fig. 6 (D) shows that the pairing ratio does not give any effect on proliferation behavior without isolation of culture chambers. These experiments confirmed that both chamber isolation and cell ratio control are critical parameters that make significant effects on cell-to-cell interaction assays. These preliminary results successfully demonstrated the capability of our microfluidic prototype devices as a potential platform for high-throughput cell-to-cell interaction assays.
Conclusions
A novel microfluidic cell-to-cell interaction chip has been developed for precise control of cell-pairing ratios and cell-to-cell interaction time. Using hydrodynamic capture schemes in a dual streams in laminar flow, we achieved a high cell capture rate over 80% in pairing cell ratios from 1:1 to 1:5. We implemented a cell isolation scheme based on electrolytic bubble generation and removal without using any on-chip microvalves or external pneumatic pumps. As the bubbles can be generated and removed within a minute, the presented platform can precisely control the cell interaction time for the paired cells inside a chamber. We confirmed that cell viability and proliferation rates are not affected by electrolysis and bubble removal operations. As a proof of the concept, we have performed the cell interaction assays by co-culturing C2C12 and PC3 cells in different cell-pairing ratios using the fabricated chip. Experimental results showed that proliferation rate was enhanced where C2C12 cells were co-cultured with higher pairing ratios of PC3 cells. This demonstrated the capability of our microfluidic prototype devices as a potential platform for high-throughput cell-to-cell interaction assays, and the compatibility of electrolysis for spatial temporal microenvironment control during cell culture.
Supplementary Material
Acknowledgements
This work was supported in part by the Department of Defense (W81XWH-12-1-0325) and in part by the National Institute of Health (1R21CA17585701). We acknowledge the Microfluidics in Biomedical Science Training Program (MBSTP) at the University of Michigan supported by the NIH. C2C12 cells were obtained from the Dr. Ken Pienta's Lab (the University of Michigan, MI, USA, currently at Johns Hopkins University).
References
- 1.Polyak K, Haviv I, Campbell IG. Trends Genet. 2009;25(1):30–8. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Peerani R, Zandstra PW. J. Clin. Invest. 2010;120:60–70. doi: 10.1172/JCI41158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Langley RR, Fidler IJ. J Cancer. 2011;128(11):2527–35. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fidler IJ. Nat Rev Cancer. 2003;3(6):453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, Jung Y, Dontu G, Taichman R, Wicha MS. Cancer Res. 2011;71(2):614–24. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solinas G, Germano G, Mantovani A, Allavena P. J Leukoc Biol. 2009;86(5):1065–73. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 8.Quatromoni JG, Eruslanov E. Am J Transl Res. 2012;4(4):376–89. [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, Maru DM, Hawke DH, Rak J, Mani SA, Zweidler-McKay P, Ellis LM. Cancer Cell. 2013;23(2):171–85. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franses JW, Baker AB, Chitalia VC, Edelman ER. Sci Transl Med. 2011;3(66):66. doi: 10.1126/scitranslmed.3001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Y, Zhou H, Krueger J, Kaplan C, Lee S,H, Dolman C, Markowitz D, Wu W, Liu C, Reisfeld R,A, Xiang R. J Clin Invest. 2006;116(8):2132–2141. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miki Y, Ono K, Hata S, Suzuki T, Kumamoto H, Sasano H. J Steroid Biochem Mol Biol. 2012;131(3-5):68–75. doi: 10.1016/j.jsbmb.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N Engl J Med. 2004;351(8):781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 14.Patricia SS. Nature Rev. Cancer. 2006;12:895–904. [Google Scholar]
- 15.Mina JB, William CH. Nature Medicine. 2011;17:320–329. [Google Scholar]
- 16.Mignatti P, Morimoto T, Rifkin DB. Proc Natl Acad Sci U S A. 1991;1588(24):11007–11. doi: 10.1073/pnas.88.24.11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heneweer M, Muusse M, Dingemans M, de Jong PC, van den Berg M, Sanderson JT. Toxicol Sci. 2005;83(2):257–63. doi: 10.1093/toxsci/kfi025. [DOI] [PubMed] [Google Scholar]
- 18.Hsiao AY, Torisawa YS, Tung YC, Sud S, Taichman RS, Pienta KJ, Takayama S. Biomaterials. 2009;30(16):3020–7. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J, Koito H, Li J, Han A. Biomed Microdevices. 2009;11(6):1145–53. doi: 10.1007/s10544-009-9331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer M, Su G, Beebe DJ, Friedl A. Integr Biol. 2010;2(7-8):371–8. doi: 10.1039/c0ib00001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H, Liu T, Qin J, Lin B. Electrophoresis. 2010;31(10):1599–605. doi: 10.1002/elps.200900776. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Majumdar D, Jovanovic B, Shaifer C, Lin PC, Zijlstra A, Webb DJ, Li D. Biomed Microdevices. 2011;13(3):539–48. doi: 10.1007/s10544-011-9523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majumdar D, Gao Y, Li D, Webb DJ. J Neurosci Methods. 2011;196(1):38–44. doi: 10.1016/j.jneumeth.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tumarkin E, Tzadu L, Kumacheva E. Integr. Biol. 2011;3:653–662. doi: 10.1039/c1ib00002k. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y-C, Cheng Y-H, Kim HS, Ingram PN, Nor JE, Yoon E. Lab Chip. 2014;14(16):2941–2947. doi: 10.1039/c4lc00391h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingram P, Kim YJ, Bersano-Begey T, Lou X, Asakura A, Yoon E. Proceedings of MicroTAS, Groningen. 2010:277–279. [Google Scholar]
- 27.Frimat J, Becker M, Chiang Y, West J. Lab Chip. 2011;11:231–237. doi: 10.1039/c0lc00172d. [DOI] [PubMed] [Google Scholar]
- 28.Hong S, Pan Q, Lee LP. Integr. Biol. 2012;4:374–80. doi: 10.1039/c2ib00166g. [DOI] [PubMed] [Google Scholar]
- 29.Stine RR, Matunis EL. Trends in Cell Biology. 2013;23(8):357–364. doi: 10.1016/j.tcb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fordyce PM, Diaz-Botia CA, DeRisi JL, Gomez-Sjoberg R. Lab Chip. 2012;12(21):4287–95. doi: 10.1039/c2lc40414a. [DOI] [PubMed] [Google Scholar]
- 31.Chiu SH, Liu CH. Lab Chip. 2009;9:1524–1533. doi: 10.1039/b900139e. [DOI] [PubMed] [Google Scholar]
- 32.Hofer DR, Sherwood ER, Bromberg WD, Mendelsohn J, Lee C, Kozlowski JM. Cancer Res. 1991;51(11):2780–5. [PubMed] [Google Scholar]
- 33.Christov C, Chrétien F, Abou-Khalil R, Bassez G, Vallet G, Authier FJ. Mol. Biol. Cell. 2007;18:1397–1409. doi: 10.1091/mbc.E06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan W-H, Takeuchi S. Proc Natl Acad Sci U S A. 2007;104(4):1146–1151. doi: 10.1073/pnas.0606625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung J, Kim YJ, Yoon E. Appl. Phys. Lett. 2011;12:3701–3703. doi: 10.1063/1.3565236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YC, Lou X, Ingram P, Yoon E. Proceedings of MicroTAS, Seattle. 2011:1409–1411. [Google Scholar]
- 37.Chen Y-C, Ingram Pa., Lou X, Yoon E. Proceedings of MicroTAS, Okinawa. 2012:1241–1244. [Google Scholar]
- 38.Eddings MA, Gale BK. J. Micromech. Microeng. 2006;16:2396. [Google Scholar]
- 39.Lochovsky C, Yasotharan S, Günther A. Lab Chip. 2012;12:595–601. doi: 10.1039/c1lc20817a. [DOI] [PubMed] [Google Scholar]
- 40.VanBerkel GJ, Zhou F, Aronson JT. International journal of mass spectrometry and ion processes. 1997;162(1):55–67. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.