Abstract
Considerable evidence suggests that cancer stem-like cells (CSCs) are critical in tumor pathogenesis, but their rarity and transience has led to much controversy about their exact nature. Although CSCs can be functionally identified using dish-based tumorsphere assays, it is difficult to handle and monitor single cells in dish-based approaches; single cell-based microfluidic approaches offer better control and reliable single cell derived sphere formation. However, like normal stem cells, CSCs are heavily regulated by their microenvironment, requiring tumor-stromal interactions for tumorigenic and proliferative behaviors. To enable single cell derived tumorsphere formation within a stromal microenvironment, we present a dual adherent/suspension co-culture device, which combines a suspension environment for single-cell tumorsphere assays and an adherent environment for co-culturing stromal cells in close proximity by selectively patterning polyHEMA in indented microwells. By minimizing dead volume and improving cell capture efficiency, the presented platform allows for the use of small numbers of cells (<100 cells). As a proof of concept, we co-cultured single T47D (breast cancer) cells and primary cancer associated fibroblasts (CAF) on-chip for 14 days to monitor sphere formation and growth. Compared to mono-culture, co-cultured T47D have higher tumorigenic potential (sphere formation rate) and proliferation rates (larger sphere size). Furthermore, 96-multiplexed single-cell transcriptome analyses were performed to compare the gene expression of co-cultured and mono-cultured T47D cells. Phenotypic changes observed in co-culture correlated with expression changes in genes associated with proliferation, apoptotic suppression, tumorigenicity and even epithelial-to-mesechymal transition. Combining the presented platform with single cell transcriptome analysis, we successfully identified functional CSCs and investigated the phenotypic and transcriptome effects induced by tumor-stromal interactions.
Keywords: Cell-cell interaction, Co-culture, Suspension culture, Tumor sphere, Cancer associated fibroblast, PolyHEMA, Single cell analysis
Introduction
There is now a wealth of data supporting the presence of a sub-population of highly potent, stem-like cells in a variety of cancers, including breast cancers [1–8]. Often called the cancer stem-likes cells (CSCs), these cells are part of a subset of cells that retain the ability to differentiate and initiate new tumors [4, 7]. Thus, CSCs have been implicated in metastasis, radiation and chemotherapy resistance, and relapse after therapy, making them important clinical targets [3, 6, 9–10]. Despite this evidence, the nature and even the existence of these cells remains controversial. Conventionally, CSCs have been characterized across different cancer types and in different patients utilizing only a handful of markers (e.g. CD133, CD44, CD24), but it is unclear whether these markers identify the same or separate CSC populations or if their function is preserved across types. Even within similar cancers, markers that denote ‘stemness’ in one, may be absent in another [11–13].
Approaches that functionally select CSC populations can be used instead to overcome the limitations and ambiguity of marker-based identification. First utilized for the identification of neural stem cells, clonal sphere formation has since been adapted and validated in breast cancer to enrich for CSC populations [14]. For normal differentiated cells, adhesion to an extracellular matrix (ECM) scaffold is essential for maintenance of cellular homeostasis. Disruption of cell attachment leads to anoikis, a form of programmed cell death [15]. When breast cancer cells are cultured in suspension, bulk non-stem cells undergo anoikis, while only cells with more stem-like characteristics survive and proliferate to form spheres, as they are anoikis resistant and capable of differentiation and proliferation afterwards [15]. As such, the formation of tumor spheres from a populations of breast cancer cells can be used to functionally identify cells with these stem-like characteristics.
However, deployment and control of single cells using traditional methods is challenging. Fluorescence-activated cell sorting (FACS) approaches help in the deployment of single cells but are also widely known to hurt cell viability and have a small, but significant, contamination rate. As such, we developed a user friendly, high throughput microfluidic-based tumorsphere assay [16, 17] based on our previous single cell capture devices [18]. These microfluidic approaches are ideal for precise fluid handling and single cell deployment. With our microfluidic tumorsphere assay, we can functionally assess CSC in cancer cell populations by simply pipetting our sample (primary or cell lines) into the device and monitoring the resulting sphere formation, making it an ideal approach for large scale screening applications.
Though mechanisms and response of CSC can be studied in isolation, CSC in vivo are regulated by a complex microenvironment, much like their normal counterparts [7, 19–22]. Stromal cells, such as cancer associated fibroblasts, promote CSC phenotypes and tumorigenecity through many pathways and mechanisms. As such, adaptable high throughput assays capable of dissecting CSC behavior within a physiologically relevant tumor microenvironment are needed. Although tumor-stromal interaction experiments have been performed using conventional dish based approaches, these co-cultures lack single cell isolation for selecting functional CSC [22–24]. There are also a number of previous works reporting microfluidic platforms for cell-to-cell interaction studies as well [18, 25–34], but these devices also don’t provide single cell isolation in suspension. [25–31]. While droplet based technology can provide high-throughput combinatorial pairings of cells, it lacks capabilities for long-term cell culture, which is required to perform weeks long tumorsphere assays [32]. Recently, several microfluidic devices reported cell pairing and cell-to-cell interaction at single-cell resolutions [18, 33–35], but those works are still restricted to adherent cell co-culture alone.
To elucidate the effect of tumor-stromal interactions on functionally selected CSC, there is a need to combine both a suspension environment for single cancer cell for CSC identification and an adherent substrate for stromal cells to survive. The two different culture environments should be connected in close proximity for cell-cell interactions. As such, we developed a novel co-culture platform combining both single cell suspension and adherent culture in close proximity. The design minimizes dead volume and retains all loaded cells to achieve better high capture efficiency as compared to previous single-cell platforms [36–39]. The platform provides the suspension environment for tumorsphere assays to functionally select CSCs and the adherent environment for stromal cells (e.g. fibroblast cells, endothelial cells) [40]. As a proof concept, we successfully demonstrated elevated stemness and EMT-like expression in cancer stem cells co-culture with primary cancer associated fibroblasts.
Materials and Methods
Device Fabrication
The device is fabricated from two separately patterned PDMS layers. These two PDMS layers (the channel layer and the substrate layer) were fabricated using standard soft lithography processes separately and then aligned and bonded as shown in Supplementary Fig. 1–2. For the channel layer, two masks were used to fabricate a SU8 (SU-8 2010, Microchem) master mold: the first for narrow interaction channels (3 μm height) between the two culture environments and the second mask for the main microfluidic channels and the adherent cell culture chambers (40 μm height). Only a single mask was used to fabricate the SU8 master mold for the substrate layer and consisted of indentations (40um depth) for the suspension cell culture wells. To make the non-adherent microwells, polyHEMA (P3932, Sigma-Aldrich) was filled in the suspension culture chambers on the PDMS mold via a stamping process developed in our lab. Prior to stamping, polyHEMA was dissolved in an ethanol solution (60 mg/mL in 95% ethanol) and then coated on the PDMS substrate [16]. A piece of blank PDMS was pressed on top to squeeze out the excess solution leaving the polyHEMA only in the indented micro-wells [17]. To improve the coating quality, the indented PDMS substrate was plasma treated to increase the hydrophilicity. This increases the affinity of the polyHEMA solution to the patterned PDMS substrate. Then, the substrate and the blank PDMS stamp were put on a hot plate at 110 degrees for 2 hours under pressure, in order to facilitate evaporation of the ethanol through the PDMS. As the ethanol evaporates it deposits the polyHEMA into the microwells. To remove any undesired residual polyHEMA on the surface that might inhibit adherent culture on the rest of the PDMS surface, 30 seconds of 800 Watt plasma etching was performed using a YES polymer striper (the expected etching depth is 0.3 μm). This results in a clean PDMS surface with polyHEMA only left in the suspension culture chambers. The fabricated substrate is the then aligned and bonded to the other PDMS fluidic layer containing the outer chambers and interaction channels. Before alignment, the two layers of PDMS were exposed to 100 Watt plasma for 60 seconds. These layers were then aligned on a KARL SUSS MJB3 Mask Aligner. The alignment error could be controlled to under 25 μm, while the alignment tolerance in the device itself is designed to be 60 μm. Thus, we can achieve a yield of approximately 90%. A fabricated device is shown in Fig. 1(c).
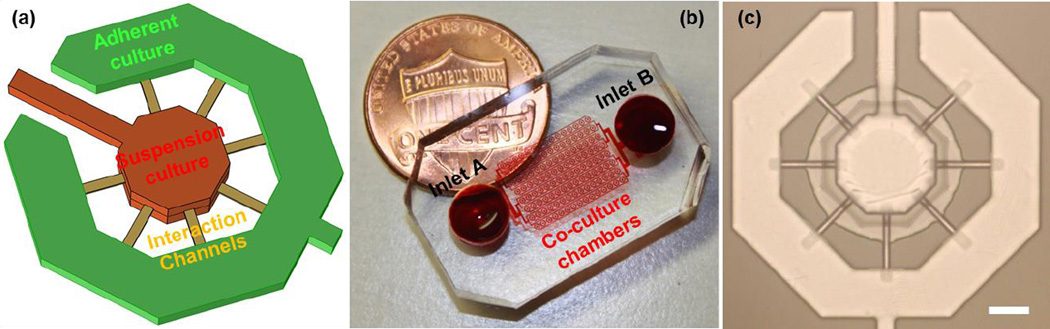
Fig. 1.
The adhesion/suspension co-culture chip: (a) The schematics of a co-culture chamber (b) a fabricated device with 120 chambers, and (c) a fabricated co-culture chamber (scale bar: 100 μm).
Harvesting of Cancer Associated Fibroblast Cells
The primary breast cancer associated fibroblasts (CAFs) were obtained from the lab of Dr. Max Wicha. Breast tumor specimens were obtained according to the approved IRB (HUM00042204) from the patients diagnosed with breast cancer and operated at the University Hospital of University of Michigan. Samples were anonymously coded and a written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. First, primary tumor tissue was obtained from ER+/PR+ patients and then minced into fine pieces using a scalpel. The minced tissue was transferred into a 50 mL conical tube, and a 1X Collagenase/hyaluronidase solution (Stem Cell Technologies, Cat # 07912) was added in a 1:1 ratio. The tissue was then gently dissociated on a rotary shaker (200 RPM) for 30–60 minutes at 37 °C or until all larger tissue fragments were digested. After dissociation, the solution was filtered through a 100 micron filter into a new 50 mL conical tube and centrifuged at 40 × g for 2 minutes. Then, the supernatant containing the fibroblasts was pipetted carefully into a new 50 mL conical tube. The fibroblasts were spun down at 250 × g for 5 minutes. After spinning down, the pellet was washed with HBSS and spun down again at 250 × g for 5 minutes. After centrifugation, the pellet was re-suspended in fibroblast growth media (Science Cell Research Laboratories, Cat # 2301), plated in a cell culture flask, and incubated at 37 °C and 5% CO2. The media was changed after 1.5–2 hours to separate the adhered fibroblasts from other floating blood or epithelial cells. The adhered cancer associated fibroblasts were then expanded in fibroblast media for up to 3–4 passages for use in experiments.
Cell Culture
Several cell lines, including MDA-MB-231 (human breast cancer cell line), C2C12 (mouse myoblast cell line), and T47D (human breast cancer cell line), were cultured for the adhesion-suspension co-culture cell experiments. MDA-MB-231 cells were obtained from Dr. Gary Luker’s Lab (University of Michigan, MI, USA). C2C12 cells were obtained from the Dr. Ken Pienta’s Lab (the University of Michigan, MI, USA, currently at Johns Hopkins University). T47D cells were obtained from Dr. Max Wicha’s Lab (University of Michigan, MI, USA). MDA-MB-231 and C2C12 cells were cultured in DMEM (Gibco 11965) with 10% FBS (Gibco 10082) and 1% penicillin/streptomycin (Gibco 15070). T47D cells were cultured in RPMI (Gibco 11875) with 10% FBS (Gibco 10082) and 1% penicillin/streptomycin (Gibco 15140). Cancer associated fibroblasts were obtained from Dr. Max Wicha’s Lab and cultured in Fibroblast Medium (ScienCell Cat. No. 2301), which is composed of of 500 ml of basal medium, 10 mL of fetal bovine serum (FBS, Cat. No. 0010), 5 mL of fibroblast growth supplement (FGS, Cat. No. 2352), and 5 mL of penicillin/streptomycin solution (P/S, Cat. No. 0503). All the cells were cultured in polystyrene culture dishes and passaged at or before cells reached 80% confluency.
Cell Loading
The fabricated microfluidic devices were put in a low pressure desiccator (0.4 atm) for 30 minutes, and then culture media was pipetted to the inlet and outlet to prime the device. Before the experiment, the devices were examined under microscope to make sure that there were no bubbles trapped in the device after priming. CAF cells were first harvested from a petri-dish with 0.05% Trypsin/EDTA and centrifuged at 100 × g for 5 minutes. Then, cells were re-suspended at 1 × 106 cells/mL in culture media, and 100 μL of the cell solution was pipetted into the adherent cell port. Flow for cell loading (10 μL/min) was generated by applying negative pressure to the suspension port using a Pasteur pipette bulb. The merit of our design is that it is a self-adjusting system. When CAFs are loaded into a chamber, the cells will block the interaction channels connecting the adherent and suspension chambers, and the flow resistance in this chamber will increase. As such, the next coming CAF is less likely to enter this chamber and instead will flow into a lower resistance chamber (one with fewer CAFs already inside). Therefore, we achieve uniform cell loading (10 ± 2 CAFs per chamber) of CAFs in a robust manner. Within 1–2 minutes, the CAFs cells were loaded in the outer (adherent) chambers, and the cell solution was replaced with CAF culture media (ScienCell Cat. No. 2301). After two days of culture, CAF cells had completely adhered and formed a mono-layer in the outer chamber. At this time, the single T47D (breast cancer) cells were loaded. T47D cells were harvested from a petri-dish with 0.05% Trypsin/EDTA and centrifuged at 100 × g for 5 minutes. Then, cells were re-suspended at 1 × 104 cells/mL in culture media, and 100 μL of the cell solution was pipetted into the suspension cell port. Then, negative pressure generated by a Pasteur pipette bulb was applied to the adherent cell port for 1–2 minutes to generate flow. Cells were loaded into the inner suspension chambers, mostly as single cells following a Poisson distribution based on the initial cell concentration. For samples with small numbers of cell, cells were re-suspended in 20 μL. With our approach, the whole sample will flow through the device to minimize the loss of cells in the dead volume of the ports. Losses are further minimized by the channel and capture layout. The ports are only connected via the narrow interaction channels, so that cells are not lost in an outlet port and are maintained on the side loaded.
Cell Culture On-chip
After loading of the cancer cells, the media in the device was replaced with standard tumor sphere assay media. This serum-free media contains MEBM (CC-3151, Lonza) supplemented with B27 (Gibco 17504-044), 20 ng/ml bFGF (BD 354060), 20 ng/ml EGF (BD 354052), 5 μg/mL insulin (Sigma I6634), 1 mM lipid concentrate (Gibco 11905-031), 1 μg/mL hydrocortisone (Sigma H4001), 7.8 μg/mL mercaptoethanol (Sigma M3148), 3.9 μg/mL cholesterol (Sigma C4951), and 1% penicillin/streptomycin (Gibco 15070) [40, 41]. To exchange the media each day, we first take out all the residual media in the adherent port and suspension port, and then we add 100 μL of fresh media into the adherent port. The total volume in a 120-well device is around 1.5 μL, so the media in all of the chambers can be completely replaced. The media flows from adherent port to suspension port driven by gravity flow. Initially, the flow rate is around 8 μL/hr. In the first 12 hours of each day, the media, and thus the secreted proteins, flows from the CAF chamber to the cancer cell chambers downstream. Gravity flow ceases after 12 hours; therefore, diffusion enables mutual interaction between CAFs and cancer cells during this time. Simulations show that the concentration distribution reaches steady-state within 8 hours when flow is present in the device and within 4 hours when no flow is present (Supplementary Fig. 3–6). According to simulation, the fibroblast secreted protein concentration is around 20 nM concentration, which is more than enough for effective cell-cell interaction because this number is much higher than the concentration used to induce EMT in literature [42]. Though PDMS can absorb some secreted proteins, the high cell density (>1.7 cell/nL volume) and the resulting high concentration of secreted proteins in our microfluidic chamber allow cells to adequately interact with each other via secretion. The cells were cultured on-chip for 14 days, and spheres larger than 50 μm in diameter were counted and harvested for further analysis.
Image Acquisition
The microfluidic chips were imaged using an inverted microscope (Nikon). The bright-field and fluorescent images were taken with a 10x objectives and a charge-coupled device (CCD) camera (Coolsnap HQ2, Photometrics). A FITC/TRITC filter set was used for the fluorescent imaging of Cell tracker Green and Orange (Invitrogen, C2925, C2927). Bright field imaging was performed using an exposure time shorter than 10 ms, and the fluorescent imaging was performed using an exposure time shorter than 100 ms, minimizing the phototoxic effect on cells. The microfluidic cell chamber array was scanned with a motorized stage (ProScan II, Prior Scientific). Before each scanning, the stage was leveled to ensure the image remained in the focus throughout the whole imaging area.
Sphere Retrieval, Dissociation and Single Cell Gene Expression Analysis
After 2 weeks of culture, spheres formed from single cells at a rate characteristic of the abundance of CSCs in the loaded cell population. The devices were dissembled, and the spheres formed inside the chambers were retrieved as previously described [16]. The retrieved spheres were spun down and then re-suspended in 1mL of 0.05% Trypsin/EDTA (Gibco 25200). The suspension was pipetted up and down 20 times to enhance the dissociation of the spheres. The tube was incubated for 5 minutes and then pipetted up and down 20 times again. The dissociated cells were loaded onto the C1 (Fluidigm) chip and processed by the C1 instrument to isolate the single cells. All the chambers of C1 chip were examined under the IX83 fluorescent microscope to record the status of captured cells in each chamber. Single cells underwent lysis, RNA release, reverse transcription, and finally cDNA pre-amplification for 96 target gene transcripts in the C1 chip. The pre-amplified cDNAs from each single cell were analyzed using the BioMark HD instrument that generates nearly 10,000 qPCR data-points in a single run using a 96 × 96 chip and TaqMan assays. Technical and biological replicate experiments were performed to show convincing reproducible data (Supplementary Fig. 7).
Data Analysis and Processing
Statistical analyses were performed using R (version 3.0). One-way ANOVA tests were used for all comparisons and significance level of p < 0.05 was used to consider statistical significance. * refers to P < 0.05, ** refers to P < 0.01, and *** refers to P < 0.001. Results are presented as mean ± SD. Measurements with high variability (such as gene expression levels) were compared on the log-scale. The gene expression data were normalized to GAPDH, a common housekeeper gene in the cell, and the un-normalized data were provided as supplementary information. For single-cell qRT-PCR data generated from the Fluidigm Biomark HD system, we used SINGuLAR v3.0 for data analysis, such as outlier detection, hierarchical clustering and principal component analysis. R package SingleCellAssay was used for its improved statistical power in detecting differentially expressed genes.
Results and Discussion
Design of the Adherent/Suspension Co-Culture Platform
The presented co-culture platform is composed of an inner suspension culture chamber, an outer adherent culture chamber, and narrow interaction channels connecting them (Fig. 1 (a)). The whole device consists of 120 (a 15 columns by 8 rows array) co-culture units (Fig. 1 (b)), and each unit is composed of the two culture chambers connected by 7 narrow (cross-section 3 μm by 20 μm and 100 μm long) channels. To facilitate suspension and adherent culture on the same device, two layers of PDMS are used in fabrication. The bottom layer was patterned with indented microwells that were selectively coated with Poly-hydroxyethylmethacrylate (polyHEMA), which has been extensively used as a cell adhesion blocking coating material [16]. The top channels layer is patterned with microfluidic channels for flow control and chambers for co-culture.
Characterization of the Engineered Surface
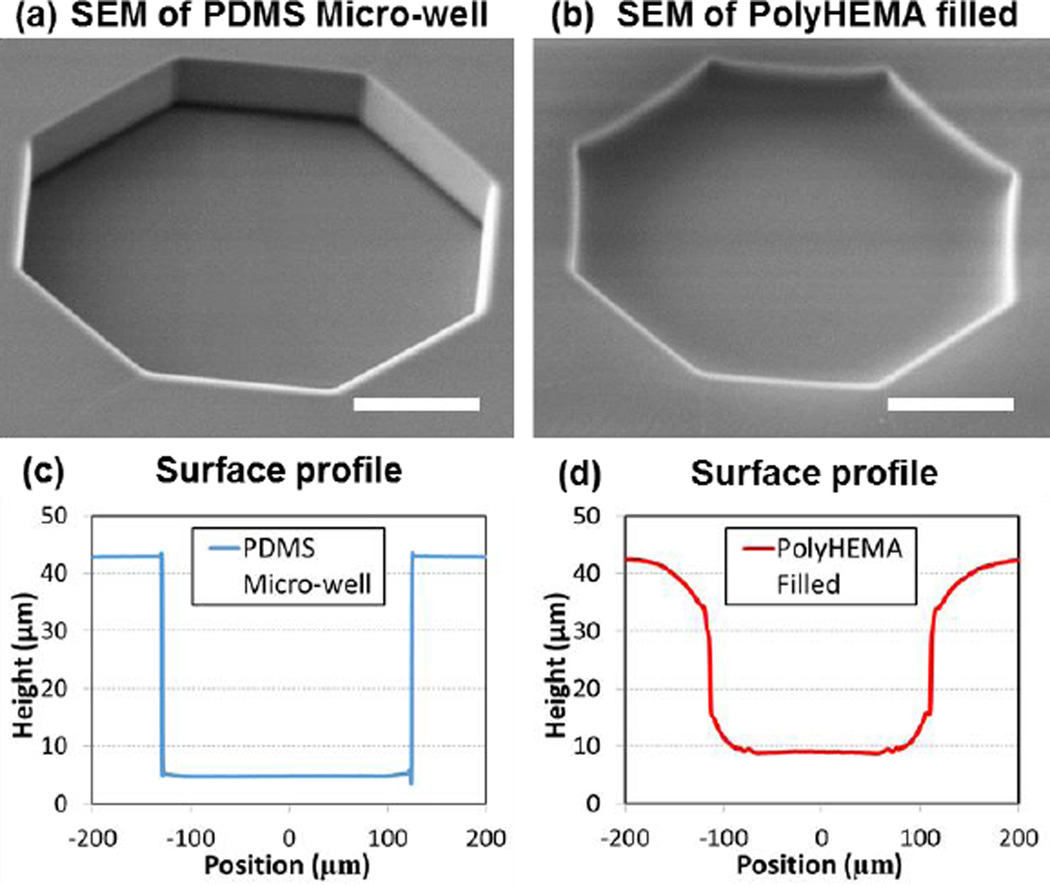
First, we examined the surface profile of the fabricated substrates using scanning electron microscopy (SEM). As shown in Fig. 2(a), we can clearly see the vertical side wall of the indented micro-well before it is coated with polyHEMA. After polyHEMA filling, the side wall of the indented microwell becomes smooth, showing (Fig. 2(b)). If the polyHEMA is over-etched, exposed PDMS on the bottom of the micro-well can be clearly visualized by SEM (Supplementary Fig. 8). This was used as an indicator to determine the proper etching time (30 seconds) during fabrication optimization. In addition, we measured the surface profiles using a laser interference microscope (LEXT, Olympus) as shown in Fig. 2 (c, d)). Due to better affinity to the PDMS surface, more polyHEMA deposited at the corners of the microwell, and thus smoothened the cross-section of the microwell. Based on a comparison of the profiles, the polyHEMA coating depth at the center of the chamber is approximately 4 μm. Based on our previous experiments, this is a sufficient coating thickness to generate a non-adherent culture surface.
Fig. 2.
Fabrication of non-adherent microwell: (a, b) SEM of microwell before and after filling polyHEMA and (c, d) surface profile of microwell before and after filling polyHEMA measured by LEXT interferometer. PolyHEMA was measured to be 4 μm thick in the center of the well. Due to the difference in coefficient of thermal expansion between PDMS and polyHEMA, polyHEMA shrinks more than PDMS after coating on the hotplate and cooling down. The PDMS sidewall is actually “dragged down” by polyHEMA coated in the microwell. The curved surface is therefore formed due to this stress. (scale bar: 100 μm)
Cell Culture on the Engineered Surface
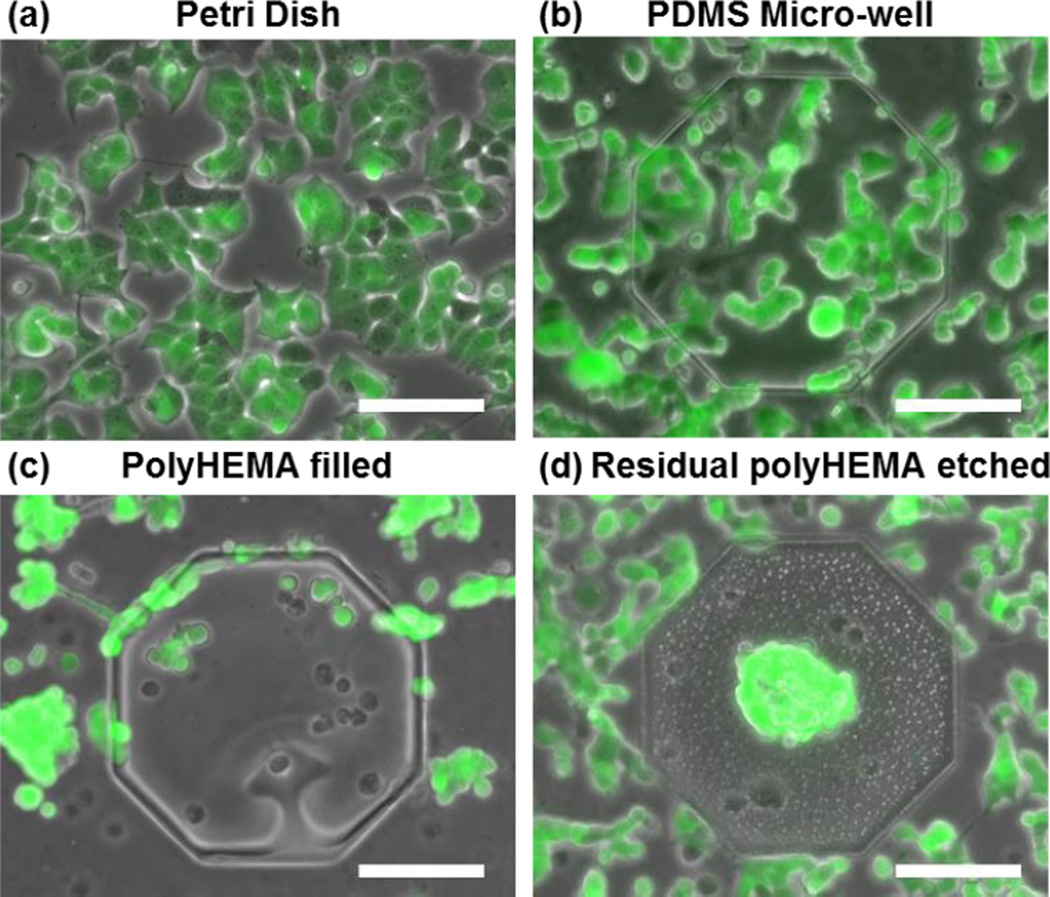
To verify the effect of polyHEMA coatings for suspension culture, T47D breast cancer cells were cultured on the selectively coated substrate, an uncoated PDMS substrate, and a standard tissue culture plastic dish. (Fig. 3). Cells were clearly attached on both the PS dish and the uncoated PDMS substrate. The uncoated PDMS was fabricated following the same protocol and using the same master as the polyHEMA coated substrate (40 μm deep wells). The cells adhered both inside and around the wells (Fig. 3 (b)).
Fig. 3.
T47D cells grow on: (a) Petri dish (all adherent), (b) micro-well without coating (all adherent), (c) surface coated with polyHEMA (all suspension) and (d) selectively polyHEMA coated substrate (suspension in the microwell, but adherent elsewhere). (scale bar: 100 μm)
In contrast, polyHEMA coatings successfully blocked cell adhesion (Fig. 3 (c, d)). Without the plasma etching process, a thin polyHEMA residue remains on the top surface of the polyHEMA coated substrate. As such in Fig. 3 (c), we observed that cells remain rounded and aggregated on both the top surface and within the microwells, demonstrating that the T47D could not adhere. To remove this residue from the surface that will be used for adherent culture, a short plasma etch was performed. As the residual poylHEMA is much thinner (< 0.5 μm) than the polyHEMA deposited in the wells (~ 4 μm), the polyHEMA coating inside micro-wells can be preserved while removing all polyHEMA on the non-indented surface. Fig. 3 (d) demonstrates successfully non-adherent culture within the microwell with adherent culture in close proximity around the well. In addition to the luminal breast cancer cell line (T47D) shown here, a basal breast cancer cell line (MDA-MB-231) and a mouse myoblast cell line (C2C12) were tested on the substrate (Supplementary Fig. 9). The selectivity was observed in all these cell lines, indicating that the fabrication process is robust and reliable for suspension/adherent cell culture.
Single Cell Loading
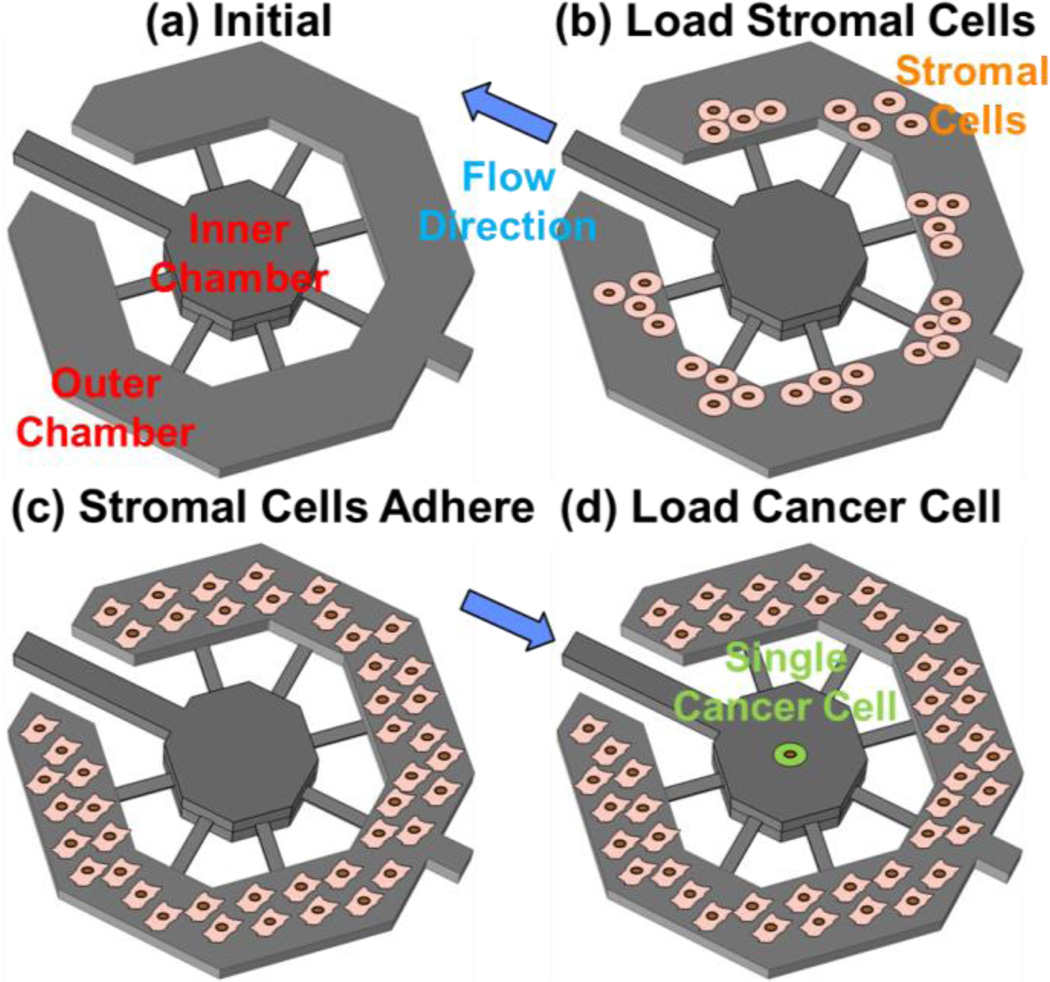
During cell loading, the stromal cells are loaded first in the outer ring and then allowed to adhere to the substrate. The single cancer cells are then loaded in the suspension culture microwells for sphere formation. When a single cell is loaded into the indented well, it settles to the bottom of the well. As demonstrated by the fluidic simulations shown in Supplementary Fig. 10, the flow rate at the bottom of the indented well is slow, preventing the non-adherent cell from being washed away. The interaction channels are 3 μm in height, preventing migration between the inner and outer chambers while allowing paracrine based interactions between the two populations. As a proof of concept, we demonstrated cell loading using T47D (breast cancer) and C2C12 (mouse myoblast) cells as the suspension and adherent populations, respectively. The myoblasts were first loaded in outer chamber (Fig. 4 (b)). These cells were then cultured in fibroblast media, which contains serum that encourage cell adhesion. After 2 days, cells attached and grew to monolayer (Fig. 4 (c)). Then, the cancer cells were loaded from the suspension cell port. As the interaction channels are much narrower than the size of cell, all loaded single cancer cells were captured in the inner chambers (Fig. 4 (d)) and not able to flow out to the port. One example of single tumor cell co-cultured with C2C12 fibroblast is shown in Fig. 5 (a).
Fig. 4.
Cell loading process: (a) schematics of the a co-culture chamber, (b) stromal cells loaded in the outer culture chamber on day 0, (c) stromal cells adhere and grow into a monolayer by day 2, and (d) single cancer cell loaded in the inner chamber by revsersing the flow.
Fig. 5.
Single cell loading efficiency: (a) co-culturing of adherent stromal cells and suspended single cancer cell (scale bar: 100 μm), (b) distribution of the number of captured cancer cells per chamber when loading 50 cells into a 120-well device (N=3), and (c) the number of chambers with exactly one cell when loading different number of cells into a 120-well device (N=3).
Although conventional hydrodynamic capture schemes can have higher capture rates (60–90%), they are not ideal for small samples such as primary cells or CTC, due to lower cell capture efficiency (typically less than 10%). This lower efficiency is caused by large dead volumes in channels and loss of cells in the inlet and outlet ports. We attained a higher capture efficiency in the presented platform by (1) collecting all incoming cells (narrow interaction channel prevent cells from flowing out to the ports) and (2) minimizing the dead volume in the inlet (suspension cell port). Though the presented probabilistic capture scheme cannot guarantee single cell capture in each chamber, the distribution of cells in the chambers should follow Poisson's distribution. This can be used to tune single cell capture characteristics. When the number of incoming cells is much smaller than the number of the chamber, it is likely single cells will be distributed per chamber. As a proof of concept, we loaded 50 cells in 10 μL into a device having 120 chambers; 37 single cells (> 70% capture efficiency) were captured in the wells. The experimental results matched well with a Poisson distribution model (Fig. 5 (b)), showing that the capture scheme is tunable and suitable for studies using small numbers (< 100) of cells. Different numbers of cells were loaded into devices containing 120 chambers, and the number of wells capturing exactly single cells is shown in Fig. 5 (c). Due to the nature of Poisson distribution, minor change in cell number will not significantly affect the capture efficiency, leading to robust operation and capture.
Single Cell Derived Sphere Formation under the Influence of CAFs
The single cell derived sphere formation rate is an indicator of tumorigenic potential within the cell population, and these clonal spheres are derived from CSC [39]. Thus, in order to investigate the effects of stromal interactions on CSC populations, we quantify single T47D sphere formation rates in mono-culture and co-culture with cancer associated fibroblasts. After co-culturing single tumor cells with CAFs for 14 days, we quantified the cell-cell interaction effect by counting the number of single-cell-derived spheres present throughout a 500-well array [16]. Compared to mono-cultured single cancer cells (Fig. 6 (a)), the cancer cells co-cultured with CAFs (Fig. 6 (b)) have doubled the sphere formation rate (Fig. 6 (c)), indicating an increased tumorigenic potential. As compared to T47D/CAF co-culture, T47D/T47D co-culture did not significantly increase sphere formation rate. These results suggest that the interaction with CAFs rather than the same cancer cells is critical in enhancing tumorigenic potential. In addition, CAF co-cultured spheres were observed to be larger as well (Fig. 6 (d)), indicating a higher cell proliferation rate induced by cancer-stromal interaction. This experiment demonstrates the feasibility of performing co-culture tumor-sphere assays using the presented dual suspension and adherent co-culture environment. The results demonstrate quantifiable differences in cancer sphere formation and sphere proliferation when cultured in close proximity to a stromal microenvironment.
Fig. 6.
Cancer associated fibroblast boost the sphere formation of T47D cancer cells: Representative cancer spheres on day 14 from (a) mono-cultured or (b) co-cultured. (c) Sphere formation rate of mono-cultured, T47D co-cultured, and CAF co-cultured T47D cells after 14-day. (N = 5), ** P < 0.01. (d) Average size of mono-cultured, T47D co-cultured, and CAF co-cultured T47D cells (N = 5), * P < 0.05. (scale bar: 100 μm)
Single Cell mRNA Expression
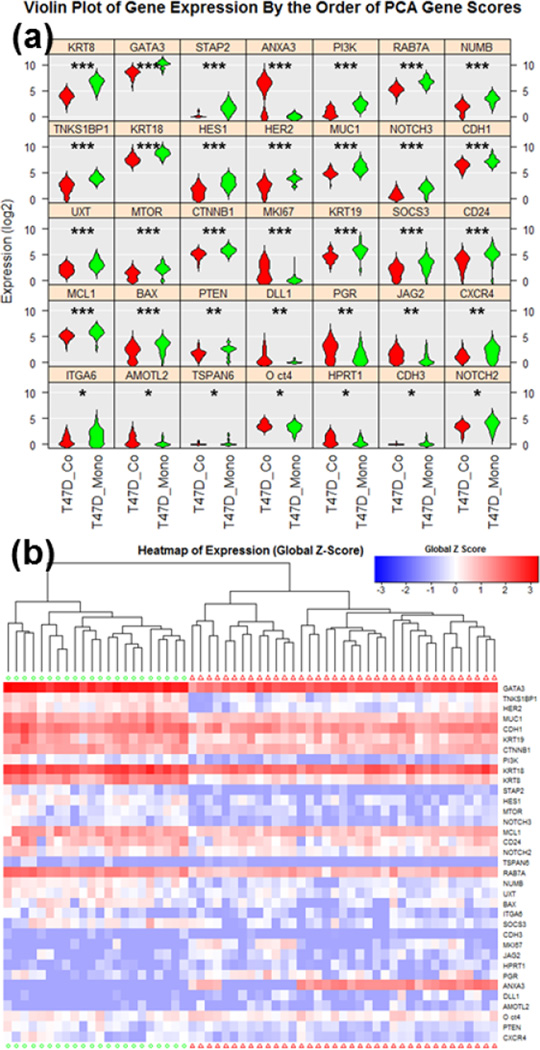
In addition to the phenotypic observation, the platform allows for the retrieval of mono-cultured and co-cultured cells for downstream genotypic analysis. To decipher the genetics mechanism causing the observed behavioral changes, we chose to perform transcriptome analysis by multiplexed single cell qRT-PCR. After 14 days of culture, spheres from different culture conditions were dissociated into single cells via trypsinization and mechanical force (pipetting) and loaded into the Fluidigm C1/Biomark HD system for multiplexed transcriptome analysis. The 96-gene panel (Supplementary Table 1) was chosen to identify the oncogenic signature of breast cancer cells and CSC [21]. At the time of sphere retrieval, there is no risk of non-sphere forming cells (non-CSCs) contaminating the retrieved sample; the non-adherent environment induces anoikis and cell death in the cells not able to form spheres. If dead cells are retained, they are unlikely to be captured in the Fluidigm C1 chip due to their shrunken size, and even if captured, the dead cells can be identified under microscope and then excluded in the data analysis. Due to capture rate and capture site limitations (96 capture sites), the Fluidigm C1 chip will not capture each cell loaded from a single sphere. However, testing has shown that C1 can capture cells with different sizes and morphology without significant bias or preference toward certain population of single cells (Supplementary Fig. 11). Expression levels were normalized to GAPDH, a common housekeeping gene, but the un-normalized data is provided in Supplementary Fig. 12–14. The genes that are statistically distinct between the two culture conditions are shown as violin plots in Fig. 7 (a) and as a heatmap in Fig. 7 (b). P-values, based on one-way ANOVA, are shown in Supplementary Table 2. The principal component analysis (PCA) clustering showing clear separation between the two culture conditions (with and without stroma). The violin plots and heatmap of genes that are not significantly different are shown in Supplementary Fig. 15–17.
Fig. 7.
Single cell gene-expression data of co-cultured and mono-cultured T47D cells utilizing Fluidigm C1/Biomark HD for multiplexed gene expression analysis. (a) Violin plots of genes that were scored as statistically different between co-cultured (red, N = 37) and mono-cultured (green, N = 22) cells. * refers to P < 0.05, ** refers to P < 0.01, and *** refers to P < 0.001. (b) Heatmap hierarchical clustering of single cell expression analysis for between co-cultured and mono-cultured cells.
The single cell expression data obtained has great explanatory power for interpreting our functional sphere co-culture assay. When examining the differences between mono-cultured and co-cultured conditions, we first noticed the significant differences in MKI67, STAP2 (Signal-Transducing Adaptor Protein 2) and BAX (BCL2-Associated X Protein). MKI67 and STAP2 are proliferation indices; the cells under co-cultured condition have significantly higher expression. This matches with the larger sphere size observed (indicating more cell proliferation) after 14 days of culture [41, 42–44]. BAX, on the other hand, is known to be a pro-apoptotic marker. The mono-cultured cells have higher expression, correlating with the lower sphere formation and anoikis (pro-apoptotic) in the suspension environment [45]. More interestingly, we observed direct evidence that cancer associated fibroblasts facilitated induction of EMT during suspension culture for the formation of spheres from CSC. EMT is a critical process in both cancer progression and metastasis. Epithelial markers CDH1 (E-CAD), KRT8 (Keratin 8), and KRT18 (Keratin 18) were down-regulated in the co-cultured cells [46–47]. At the same time, AMOTL2 (Angiomotin-like protein 2) and Oct-4 (octamer-binding transcription factor 4), which are upstream controllers of EMT, are up-regulated in the co-cultured group [48–50], while GATA3 (GATA Binding Protein 3), which can reverse EMT, is down-regulated [51]. In addition, we saw a down regulation of tumor suppressor SOCS3 (Suppressor of Cytokine Signaling 3) in the co-culture group. Upregulation of SOC3 in tumors has been associated with poor cancer prognosis, so correlates well with the phenotypic and genotypic changed observed during co-culture with CAFs [52]. Using the 96-multiplexed single cell qRT-PCR, we can clearly link increased sphere formation and sphere size to CAF-induced enhanced proliferation, malignancy, and EMT transition.
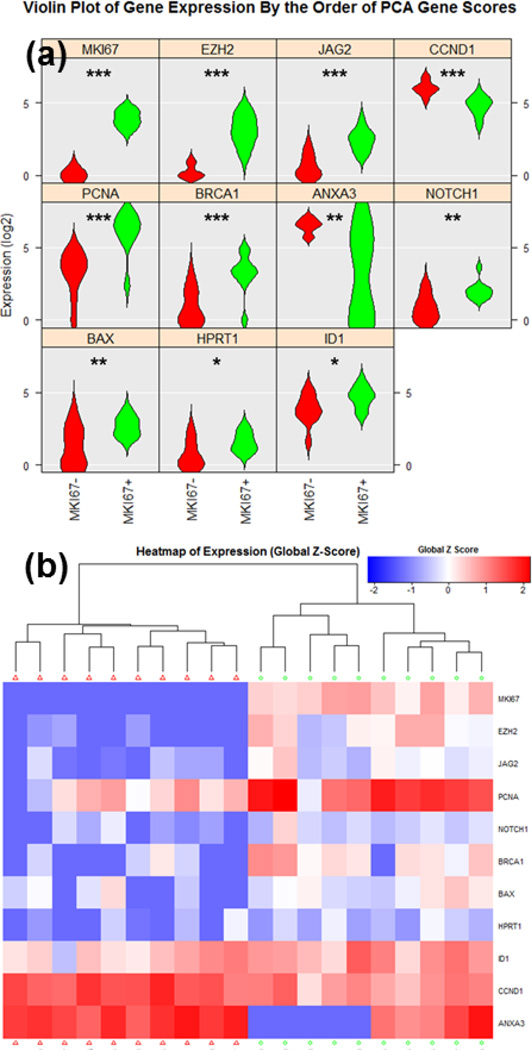
In addition to comparing the expression differences between culture conditions, with single cell resolution, we can investigate the cellular heterogeneity in gene expression within the clonal sphere populations. As a proof of concept, we examined heterogeneity in MKI67 expression. Though the co-cultured cells generally have higher expression of MKI67 than mono-cultured, significant heterogeneity exists between single cells in the developed spheres. Comparing the 10 cells with the highest MKI67 expression to the 10 with the lowest within a single co-cultured sphere, we observed markedly different expression of other genes (Fig. 8). P-values for the significant genes can been seen in Supplementary Table 3. PCA clustering and the results from the entire gene panel are shown in Supplementary Fig. 18–20. Using the same MKI67 expression threshold that was used for the highest cells in the co-culture condition, only 1 cell out of 22 cells were scored as MKI67 high in the mono-culture group. This obviously tracks well with the lower proliferative index observed in mono-culture of the spheres. The comparison between the MKI67 high cell and 10 MKI67 low cells from mono-culture group is also shown in Supplementary Fig. 21–23. For the MKI67 high cell, cancer proliferation genes EZH2 (Enhancer Of Zeste Homolog 2), PCNA (Proliferating Cell Nuclear Antigen), and ID1 (Inhibitor Of Differentiation 1) genes [53–55] are all significantly upregulated as compared to MKI67 low cells, demonstrating good correlation between MKI67 and these genes even at a single cell level. In addition, we found the elevated expressions in NOTCH1 and JAG2, which is a ligand of NOTCH signaling, in the MKI67 high group indicating that up-regulation of NOTCH pathways correlate with cell proliferation (sphere size) and increased stemness and tumorigenicity (sphere formation) in a cancer cell population [56]. Combining our novel microfluidic approach with single cell PCR, we (1) can functionally select CSC in the form of clonal spheres, (2) co-culture spheres within a stromal (CAF) microenvironement, (3) observe phenotypic effects directly prior to multiplex gene expression analysis, and (4) investigate the heterogeneity within a clonal CSC derived population, which has not been previously possible.
Fig. 8.
Single cell gene-expression data of MKI67 high and MKI 67 low T47D cells in the co-cultured group utilizing Fluidigm C1/Biomark HD for multiplexed gene expression analysis. (a) Violin plots of genes that were scored as statistically distinctive between MKI67 high (green, N = 10) and MKI67 low (red, N = 10) cells. * refers to P < 0.05, ** refers to P < 0.01, and *** refers to P < 0.001. (b) Heatmap hierarchical clustering of single cell expression analysis for MKI67 high and MKI67 low cells.
Conclusion
We have successfully demonstrated a dual adherent-suspension co-culture platform, implemented by selectively patterning polyHEMA in indented PDMS microwells. Utilizing the innovative substrate patterning, single cell suspension culture, which is ideal for investigating the tumorigenic potential of cancer cells via CSC selection, and adherent culture, which is favorable for the survival of stromal cells, can be implemented in close proximity. The adhesion-suspension dual culture micro-environment has been proven reliable for both luminal (T47D) and basal (MDA-MB-231) breast cancer cell lines in addition to a variety of fibroblast cells indicating its broad potential. Combining the patterned substrate and the fluidic scheme, the platform achieves high cell capture efficiency (>70% capture efficiency), making the device suitable for studying rare cell populations or for use with small clinical samples. As a proof of concept, we have performed a cell-cell interaction study combining tumorsphere formation with T47D in close proximity co-culture with primary cancer associated fibroblast cells in the fabricated chip. Experimental results showed that both tumor sphere formation rate and the size of the single cell derived spheres were both enhanced under co-culture conditions. In addition by using the Fluidigm C1/Biomark HD system, single cell transcriptome analysis was performed to compare mono-cultured and co-cultured CSC formed spheres. The gene expression was found to support the observed elevated sphere formation rate and increased proliferation. Also, based on the gene expression of related genes, the CAFs were found to induce EMT, a critical process in tumor progression and metastasis. These observation supports the importance of the tumor microenvironment and co-culture during the investigation of cancer pathology.
Supplementary Material
Acknowledgments
This work was supported in part by the Department of Defense (W81XWH-12-1-0325) and in part by the National Institute of Health (1R21CA17585701, 1R21CA195016-01A1). The Fluidigm C1/Biomark HD system was supported by SIG-NIH (S10OD16187). The Lurie Nanofabrication Facility of the University of Michigan (Ann Arbor, MI) are greatly appreciated for device fabrication. The authors also thank Dr. Gary Luker’s lab, Dr. Ken Pienta’s Lab, and Dr. Max Wicha's lab for providing cells for the experiments.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Bonnet D, Dick JE. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 2.Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Rosen JM. Cancer res. 2008;68(12):4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maitland NJ, Collins AT. J Clin. Oncol. 2008;26(17):2862–2870. doi: 10.1200/JCO.2007.15.1472. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Visvader JE. Genes Dev. 2009;23(22):2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J, Zhang Y. Cell Cycle. 2008;7(10):1360–1370. doi: 10.4161/cc.7.10.5953. [DOI] [PubMed] [Google Scholar]
- 7.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Phillips HS. Cancer cell. 2010;17(4):362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 8.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. J Cell Mol Med. 2009;13(8b):2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dexter DL, Leith JT. J Clin Oncol. 1986;4(2):244–257. doi: 10.1200/JCO.1986.4.2.244. [DOI] [PubMed] [Google Scholar]
- 10.Geisler JP, Rose SL, Geisler HE, Miller GA, Wiemann MC. Gynecol. Oncol. 2002;7(1):25–28. [Google Scholar]
- 11.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. J Cell Mol Med. 2009;13(8b):2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Mol. Cancer Res. 2012;10(11):1403–1418. doi: 10.1158/1541-7786.MCR-12-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WJ, Ho CC, Chang YL, Chen HY, Lin CA, Ling TY, Yang PC. Nat. Commun. 2014;5 doi: 10.1038/ncomms4472. [DOI] [PubMed] [Google Scholar]
- 14.Dontu G, Wicha MS. J Mammal. 2005;10(1):75–86. doi: 10.1007/s10911-005-2542-5. [DOI] [PubMed] [Google Scholar]
- 15.Mehlen P, Puisieux A. Nat. Rev. Cancer. 2006;6(6):449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y-C, Ingram P, Fouladdel S, McDermott SP, Azizi E, Wicha MS, Yoon E. Scientific Reports. 2016;6:27301. doi: 10.1038/srep27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingram PN, Chen Y-C, Yoon E. Proceeding of MicroTAS. 2013:1451–1453. [Google Scholar]
- 18.Chen YC, Cheng YH, Kim HS, Ingram PN, Nor JE, Yoon E. Lab Chip. 2014;14(16):2941–2947. doi: 10.1039/c4lc00391h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prestegarden L, Svendsen A, Wang J, Sleire L, Skaftnesmo KO, Bjerkvig R, Enger PØ. Cancer Res. 2010;70(11):4274–4279. doi: 10.1158/0008-5472.CAN-09-3904. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, Hong S, Adams A, D’Angelo R, Ginestier C, Charafe-Jauffret E, Clouthier SG, Birnbaum D, Wong ST, Zhan M, Chang JC, Wicha MS. Stem cell reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalluri R, Zeisberg M. Nat. Rev. Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 22.Peerani R, Zandstra PW. J Clin. Invest. 2010;120(1):60–70. doi: 10.1172/JCI41158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Hornsby PJ. Cancer Res. 2007;67(7):3117–3126. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- 24.Heneweer M, Muusse M, Dingemans M, de Jong PC, van den Berg M, Sanderson JT. Toxicol. Sci. 2005;83(2):257–263. doi: 10.1093/toxsci/kfi025. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao AY, Torisawa YS, Tung YC, Sud S, Taichman RS, Pienta KJ, Takayama S. Biomaterials. 2009;30(16):3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, Koito H, Li J, Han A. Biomed Microdevices. 2009;11(6):1145–1153. doi: 10.1007/s10544-009-9331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer M, Su G, Beebe DJ, Friedl A. Integr. Biol. 2010;2(7–8):371–378. doi: 10.1039/c0ib00001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma H, Liu T, Qin J, Lin B. Electrophoresis. 2010;31(10):1599–1605. doi: 10.1002/elps.200900776. [DOI] [PubMed] [Google Scholar]
- 29.Gao Y, Majumdar D, Jovanovic B, Shaifer C, Lin PC, Zijlstra A, Li D. Biomed. Microdevices. 2011;13(3):539–548. doi: 10.1007/s10544-011-9523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majumdar D, Gao Y, Li D, Webb DJ. J Neurosci. Meth. 2011;196(1):38–44. doi: 10.1016/j.jneumeth.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tumarkin E, Tzadu L, Csaszar E, Seo M, Zhang H, Lee A, Kumacheva E. Integr. Biol. 2011;3(6):653–662. doi: 10.1039/c1ib00002k. [DOI] [PubMed] [Google Scholar]
- 32.Ingram P, Kim YJ, Bersano-Begey T, Lou X, Asakura A, Yoon E. Proceedings of MicroTAS. 2010:277–279. [Google Scholar]
- 33.Frimat JP, Becker M, Chiang YY, Marggraf U, Janasek D, Hengstler JG, West J. Lab Chip. 2011;11(2):231–237. doi: 10.1039/c0lc00172d. [DOI] [PubMed] [Google Scholar]
- 34.Hong S, Pan Q, Lee LP. Integr. Biol. 2012;4(4):374–380. doi: 10.1039/c2ib00166g. [DOI] [PubMed] [Google Scholar]
- 35.Gracz AD, Williamson IA, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza RJ, Sims CE, Galanko JA, Li L, Allbritton NL, Magness ST. Nature Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan WH, Takeuchi S. P. Natl. Acad. Sci. USA. 2007;104(4):1146–1151. doi: 10.1073/pnas.0606625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Nature methods. 2009;6(2):147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung J, Kim Y-J, Yoon E. Appl. Phys. Lett. 2011;98(12):3701. doi: 10.1063/1.3565236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y-C, Ingram P, Yoon E. Analyst. 2014;139:6371–6378. doi: 10.1039/c4an01282h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inwald EC, Klinkhammer-Schalke M, Hofstädter F, Zeman F, Koller M, Gerstenhauer M, Ortmann O. Breast Cancer Res. Tr. 2013;139(2):539–552. doi: 10.1007/s10549-013-2560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. Respir Res. 2005;6(1):56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAllister SD, Murase R, Christian RT, Lau D, Zielinski AJ, Allison J, Desprez PY. Breast Cancer Res. Tr. 2011;129(1):37–47. doi: 10.1007/s10549-010-1177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda O, Sekine Y, Mizushima A, Nakasuji M, Miyasaka Y, Yamamoto C, Matsuda T. J Biol. Chem. 2010;285(49):38093–38103. doi: 10.1074/jbc.M110.162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturm I, Papadopoulos S, Hillebrand T, Benter T, Lück HJ, Wolff G, Daniel PT. Int. J. Cancer. 2000;87(4):517–521. doi: 10.1002/1097-0215(20000815)87:4<517::aid-ijc9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 46.Coradini D, Ambrogi F, Oriana S, Biganzoli E, Boracchi P. Adv. Breast Cancer Res. 2012;1:12–19. [Google Scholar]
- 47.Tomaskovic-Crook E, Thompson EW, Thiery JP. Breast Cancer Res. 2009;11(6):213. doi: 10.1186/bcr2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Genes Dev. 2011;25(1):51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mojallal M, Zheng Y, Hultin S, Audebert S, van Harn T, Johnsson P, Holmgren L, et al. Nat. Commun. 2014;5:4557. doi: 10.1038/ncomms5557. [DOI] [PubMed] [Google Scholar]
- 50.Wang D, Lu P, Zhang H, Luo M, Zhang X, Wei X, Liu C. Oncotarget. 2014;5(21):10803–10815. doi: 10.18632/oncotarget.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. J Biol. Chem. 2010;285(18):14042–14051. doi: 10.1074/jbc.M110.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barclay JL, Anderson ST, Waters MJ, Curlewis JD. Int. J. Cancer. 2009;124(8):1756–1766. doi: 10.1002/ijc.24172. [DOI] [PubMed] [Google Scholar]
- 53.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Akslen LA. J Clin. Oncol. 2006;24(2):268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 54.Kubben FJ, Peeters-Haesevoets A, Engels LG, Baeten CG, Schutte B, Arends JW, Blijham GH. Gut. 1994;35(4):530–535. doi: 10.1136/gut.35.4.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McAllister SD, Murase R, Christian RT, Lau D, Zielinski AJ, Allison J, Desprez PY. Breast Cancer Res. Tr. 2011;129(1):37–47. doi: 10.1007/s10549-010-1177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grudzien P, Lo S, Albain KS, Robinson P, Rajan P, Strack PR, Foreman KE. Anticancer Res. 2010;30(10):3853–3867. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.