Abstract
Ribosomal proteins are involved in numerous essential cell activities in plants. However, the regulatory role in specific plant developmental processes has not yet been fully elucidated. Here we identified the new ribosomal protein L18aB, which is specifically involved in sexual reproduction and plays a critical role in male gametophyte development and embryo pattern formation. In rpl18aB mutant plants, the mature pollen grains can germinate normally, but their competitiveness for growing in the style is significantly reduced. More interestingly, RPL18aB is required in early embryogenesis. rpl18aB embryos displayed irregular cell division orientations in the early pro-embryo and arrested at the globular stage with possible, secondary pattern formation defects. Further investigations revealed that the polar transportation of auxin is disturbed in the rpl18aB mutant embryos, which may explain the observed failure in embryo pattern formation. The cell type-specific complementation of RPL18aB in rpl18aB was not able to recover the phenotype, indicating that RPL18aB may play an essential role in early cell fate determination. This work unravels a novel role in embryo development for a ribosomal protein, and provides insight into regulatory mechanism of early embryogenesis.
In plants, ribosomal proteins are encoded by small gene families. The Arabidopsis thaliana genome encodes for approximately 80 cytoplasmic ribosomal proteins, and each protein is encoded by 2–7 family members since different genes within a family share 65–100% amino acid sequence identity1. The presence of multiple genes for each ribosomal protein in plants might be necessary to maintain constant ribosomal protein amounts or some degree of ribosome heterogeneity and functional specialization. Phenotypes resulting from mutations in several different ribosomal protein genes provide strong evidence that ribosomes participate as regulatory components of developmental pathways2.
Although ribosomes are ubiquitously distributed in the plant and are essential for plant development, many ribosomal protein loss-of-function mutants are viable. These mutants typically display a subtle change in leaf shape3,4,5,6,7,8,9. These observations appear to indicate that many ribosomal proteins must be functionally specialized for a precise developmental process, thus their loss-of-function does not affect all cellular processes. Furthermore, it was recently demonstrated that ribosomal proteins control auxin mediated developmental programs by translational regulation of auxin response factors10. Together these findings indicate that the specific role of each ribosomal protein in plant development must be studied individually, despite their general function being well known.
Previous studies also indicated that ribosomes play an important role in sexual plant reproduction. Loss-of-function mutations in single ribosomal protein genes could result in female gametophyte defects. Mutations in the ribosomal protein genes RPL24B, RPL10A, RPS5A and in the ribosomal protein RPL27aC reduce female fertility11,12,13,14. Mutations in seven cytoplasmic ribosomal proteins, S6, S11, L2, L8, L23, L19 L40 and RPL27a, resulted in embryo developmental defects14,15. In these mutants, the earliest divisions of the zygote forming a suspensor and a globular stage embryo appear to be unaltered, indicating that early patterning events are unaffected by the loss of these ribosomal proteins. These ribosomal proteins are required for the development of the embryo beyond the globular stage. Early-embryo development specific ribosomal proteins and ribosomal proteins involved in other processes of sexual plant reproduction remain to be identified.
Here, we isolated the new ribosomal protein gene RPL18aB. The Loss of function of this gene result in weakened function of male gametophyte and abortion of early embryo development, with a striking alteration in early embryo patterning, indicating the essential role of RPL18aB in these developmental processes.
Results
A T-DNA insertion in RPL18aB affects seed development
To screen genes required for embryo development in Arabidopsis thaliana, we established a T-DNA insertion mutant library that contains one insertion site in the genomic DNA of qrt1 plant background16,17. The T-DNA mutants contain the hygromycin resistance gene (Hygr), and the LAT52::GFP gene. LAT52::GFP which is expressed in pollen, allows the identification of homozygous offspring from a given mutant plant17,18. The seed of qrt1 is shown (Fig. 1B) for comparison. A heterozygous mutant with albino seed was isolated (Fig. 1C). The ratio of albino seeds was 16.35% (n = 981) out of the total seeds in the mutant siliques.
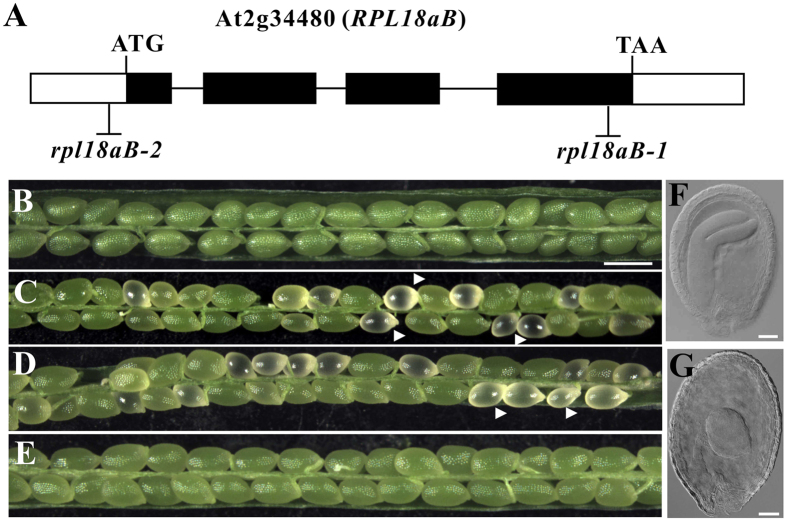
Figure 1. Embryo developmental processes are impaired in rpl18aB plant.
(A) Diagram of the RPL18aB genomic DNA and the insertion sites for the T-DNA. The RPL18aB gene (AT2g34480) has four exons. The T-DNA inserted into the fourth exon in rpl18aB-1 and the T-DNA was inserted into the promoter at 23 bp upstream of the ATG start codon in rpl18aB-2. Micrographs of siliques of wild type (B), rpl18aB-1 plant (C), rpl18aB-2 plant (D) and genomic complemented line (E). Silique of wild type and genomic complemented line show only green seeds (B,E), while rpl18aB plant silique showing normal green seeds and white seeds indicated by arrow head (C,D). (F,G) Micrographs of seeds of rpl18aB-1 plant. In the rpl18aB-1 plant, when wild type embryo develops totorpedo-shaped embryo (F), the mutant embryo shows a globular-shaped embryo (G). Bars = 500 μm for (B) to (E) and 20 μm for (F,G).
We identified the flanking sequence of the T-DNA insertion site of the mutated gene by thermal asymmetrical interlaced PCR19,20 and found that the T-DNA was inserted in the fourth exon of AT2g34480, a gene predicted to encode the ribosomal protein L18aB (RPL18aB), which is the member of the ribosomal L18ae/LX family protein1,21. The T-DNA insertion site is 113 bp upstream of the TAA end codon, and we designated the mutant rpl18aB-1 (Fig. 1A). We also obtained an additional AT2g34480 allele (Flag-209B07), which we named rpl18aB-2, with a T-DNA insertion in the 5′ UTR, 23bp upstream of the ATG start codon (Fig. 1A). rpl18aB-2/RPL18aB plants showed a similar phenotype of aborted seeds as rpl18aB-1/RPL18aB during seed development (Fig. 1D). To further confirm that the phenotype of defective seed development was caused by loss of function of AT2g34480, a genomic RPL18aB gene was cloned in a kanamycin resistant vector and introduced into rpl18aB-1/RPL18aB. The seeds of transformed plants were screened on medium containing kanamycin and hygromycin, and seedlings showing both KANr and HYGr were designated as complementation lines, transplanted to soil. Homozygous rpl18aB-1 plants were obtained in the T2 progeny of genomic complementation lines (rpl18aB-1/rpl18aB-1, g-AT2g34480/g-AT2g34480) and normal seed development was restored (Fig. 1E). These results indicated that the AT2g34480 genomic fragments could successfully complement the rpl18aB-1/+ phenotype.
Normal ovules and albino ovules from the same silique in rpl18aB-1/+ were separated and observed by whole-mount clearing technique. In normal ovules, embryos had developed to the cotyledon stage (Fig. 1F), while in albino ovules the embryo remained in an abnormally globular-shaped stage (Fig. 1G). Both rpl18aB-1/rpl18aB-1 and rpl18aB-2/rpl18aB-2 mutant plants could not be recovered, suggesting the rpl18aB-1/rpl18aB-1 and rpl18aB-2/rpl18aB-2 embryos in albino ovules are embryo lethal. Unless specially mentioned, rpl18aB plant from here on means rpl18aB/RPL18aB heterozygous plant.
RPL18aB is expressed in majority development process of Arabidopsis
To analyze the expression pattern of RPL18aB, we fused a 1.5 kb fragment of its native promoter to reporters GUS22 and H2B-GFP22 and produced pRPL18aB::GUS and pRPL18aB::H2B-GFP. The transgenic T2 progeny of pRPL18aB::GUS homozygous lines showed high GUS activity in the stem apex and the root tip of seedlings, mainly in the meristematic region (Fig. 2A). The vascular tissue of leaves also showed GUS activity. As for inflorescence developmental stages, GUS signal was observed in mature flowers but not in younger flowers (Fig. 2B,C). To clearly visualize the expression pattern of RPL18aB in reproductive organs, we studied the distribution of pRPL18aB::H2B-GFP and observed its expression in both male (Fig. 2D) and female gametophytes(Fig. 2E) and during embryo development (Fig. 2F–H).
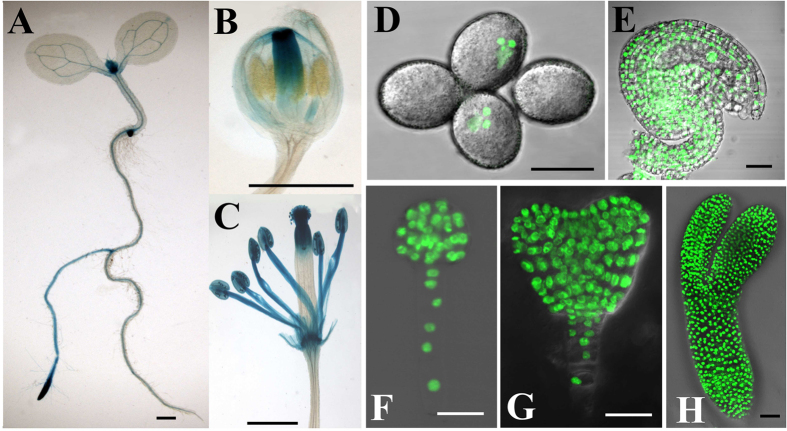
Figure 2. The expression pattern of RPL18aB.
(A) pRPL18aB::GUS lines showed high GUS activity in the stem apex and root tip of seedlings, mainly in themeristem region of both parts. (B) Young flowers showed high GUS activity in pistil. (C) Mature flower showed GUS activity in stamens and upper pistil. (D) pRPL18aB::H2B-GFP lines showed GFP signal in nuclei of sperm cells and vegetative cell. (E) pRPL18aB::H2B-GFP lines showed GFP signal in nuclei of ovule cells, including female gametophytes. (F) pRPL18aB::H2B-GFP lines showed GFP signal in nuclei of globular-shaped embryocells. (G) pRPL18aB::H2B-GFP lines showed GFP signal in nuclei of early heart-shaped embryocells. (H) pRPL18aB::H2B-GFP lines showed GFP signal in nuclei of torpedo-shaped embryocells. Bars = 1 mm for (A) to (C) and 20 μm for (D) to (H).
rpl18aB mutant reduces male/female gametophyte heredity
Our study showed that 16.35% aborted seeds were obtained from the siliques of rpl18aB-1 plants, and 42.79% (n = 1138) of rpl18aB-1 plants progeny seeds showed HYGr resistance. The ratios are lower than the expected 25% and 66.7% ratios, respectively. These results suggest the existence of additional defects beyond the phenotype of abnormal seed development.
To clarify whether the additional defects are caused by the male or female part, we determined the mutant transmission efficiency through male/female gametophyte by reciprocal cross between rpl18aB-1 plants and wild type plants. As shown in Table 1, the Hygr/Hygs ratio of the crossed line’s seeds was 0.73:1 (Hygr accounted for 42.32% of the total) when rpl18aB-1 plant was used as female parent, indicating a weak defect exists in the female gametophyte. However, the ratio dropped to 0.06:1 (Hygr accounted 5.74% of the total) when rpl18aB-1 plant was used as male parent, demonstrating a stronger defect occurred in the male gametophyte.
Table 1. Segregation of the rpl18aB-1 Mutation in Selfed and Reciprocally Backcrossed Offspring Populations.
| Parental Genotype | Progeny Genotype | |||
|---|---|---|---|---|
| Male | Female | Hygr | Hygs | Hygr Ratio |
| wild type | rpl18aB-1/RPL18aB | 482 | 657 | 42.32% |
| rpl18aB-1/RPL18aB | wild type | 38 | 624 | 5.74% |
| rpl18aB-1/RPL18aB | rpl18aB-1/RPL18aB | 1270 | 1856 | 40.63% |
rpl18aB lowers pollen tube competitiveness
We carried out assays to determine what defect occurred in the male gametophyte of RPL18aB mutant. Firstly, pollen grains of rpl18aB-1 and wild-type plants were observed by scanning electron microscopy (SEM) to observe whether mutant pollen grains had structural defects, but no obvious morphological differences were observed (Fig. 3A,B). Secondly, pollen grains of rpl18aB-1 were smeared on solid medium for pollen germination. After being cultured for 12 hours at 22 °C, pollen tubes of both mutant (marked by GFP) and wild type (without GFP) were germinated and grown to a similar length at the same time (Fig. 3C). Thirdly, we pollinated pollen grains of rpl18aB-1 plants on wild type stigma. The pollinated stigma was cut from the style 1 hour later, and cultured on solid pollen germination medium. Pollen tubes could grow out of the cut end for several millimeters 4–8 hours after pollination, but few of them were rpl18aB-1 pollen tubes (marked by GFP) (Fig. 3D). These results indicated that wild type pollen tube growth is much faster than mutant pollen tubes.
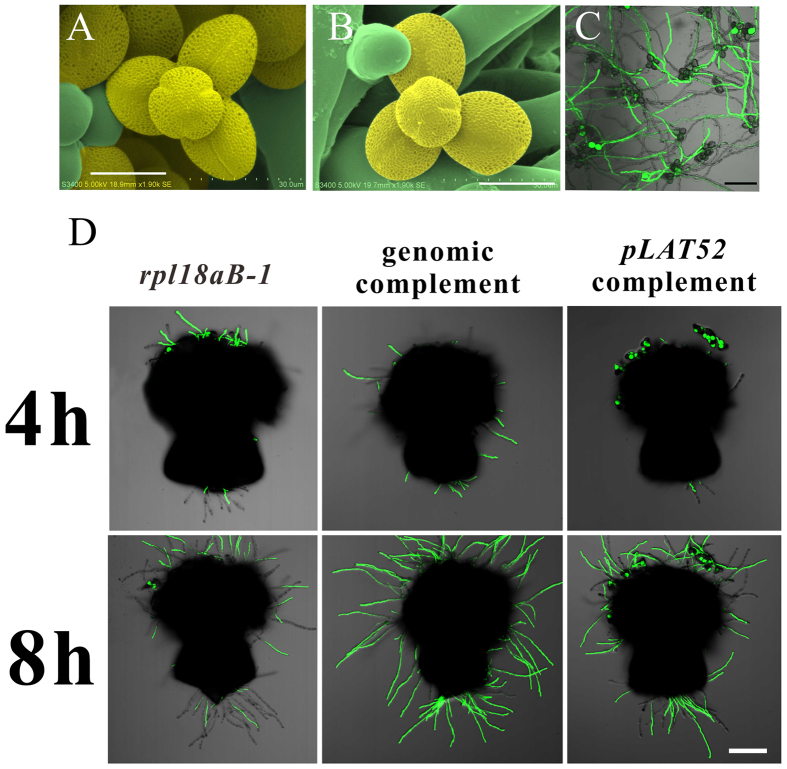
Figure 3. rpl18aB-1 lowers pollen tube competitiveness.
(A) Scanning electron micrographs showing pollen of wild type; the pollen was false-colored with yellow. (B) Scanning electron micrographs showing pollen of rpl18aB-1; the pollen was false-colored with yellow. (C) A CLSM image showing pollen tubes of both rpl18aB-1 (marked by GFP) and wild type (without GFP) were germinated and grown to a similar length in rpl18aB-1. (D) CLSM images of pollen tubes growth for 4 h and 8 h on semi-in vitro conditions in rpl18aB-1, genomic complemented line and pLAT52 complemented line respectively. rpl18aB-1 pollen tubes were marked by GFP. Bars = 20 μm for (A) to (B) and 100 μm for (C) to (D).
To test whether the decreased competitiveness of mutant pollen tubes could be recovered if specifically compensated for the defect of the pollen, we transformed rpl18aB-1 plants with the pLAT52::RPL18aB construct. T1 pLAT52 complementation lines (rpl18aB-1/RPL18aB, pLAT52::RPL18aB/+) were screened for further pollination experiments. The genomic complementation line served as a positive control. As expected, the genomic complementation line could recover the growth defect of mutant pollen tubes of rpl18aB-1 (Fig. 3D). The ratio of GFP pollen tubes (rpl18aB-1 background) that penetrated through style was higher in pLAT52 complementation line compared with rpl18aB-1 mutant (Fig. 3D).
It is possible that in rpl18aB-1 plants few rpl18aB-1 pollen tubes can enter the ovules because most ovules have been already occupied by wild type pollen tubes, leading to the very low observed transmission efficiency of Hygr through the male gametophyte. If this were true, decreasing the number of pollen grains used for pollination to ensure there are enough ovules to attract rpl18aB-1 pollen tubes, the transmission efficiency of Hygr through the male gametophyte would be expected to increase. We tested this by the limited pollination technique and found that this ratio increased to 0.26:1 (Hygr accounted for 20.50% of the total) (Table 2, data of the first arrow). We further crossed wild type with the pollens of the four pLAT52 complementation lines, respectively, to determine the transmission efficiency of Hygr through the male gametophyte. The results showed that the male transmission efficiency of Hygr increased from 5.74% (Table 1, data of the second arrow) to about 30% in all four pLAT52 complementation lines (Table 2, data of the second to the fifth arrows). Together, these results confirmed that rpl18aB lowers pollen tube competitiveness in the style.
Table 2. Segregation of the rpl18aB-1 Mutation in Backcrossed Offspring Populations under Limited Pollination Conditions and pLAT52 complementation lines.
| Parental Genotype | Progeny Genotype | |||
|---|---|---|---|---|
| Male | Female | Hygr | Hygs | Hygr Ratio |
| rpl18aB-1/RPL18aB, L | WT | 230 | 892 | 20.50% |
| LAT52::RPL18aB-1 | WT | 174 | 411 | 29.74% |
| LAT52::RPL18aB-2 | WT | 159 | 412 | 27.85% |
| LAT52::RPL18aB-3 | WT | 178 | 425 | 29.72% |
| LAT52::RPL18aB-4 | WT | 262 | 533 | 32.96% |
L refers to limited pollination. LAT52::RPL18aB series are pLAT52 complementation lines.
Both radial and apical-basal patternings are disturbed in rpl18aB-1/rpl18aB-1 embryo
To study embryogenesis in mutant ovules, a time series of sections of whole siliques were carried out with rpl18aB-1 plants. In wild type ovules, embryogenesis developed from the globular stage, heart-shaped stage to torpedo-shaped stage, and finally to the matured cotyledon stage (Fig. 4A–D). During this process the radial pattern was first recognized with a clear protoderm layer surrounding the embryo (Fig. 4A). In the same silique, rpl18aB-1/rpl18aB-1 embryos underwent a delayed pattern formation or failed to form this pattern (Fig. 4E–H). The cell division orientation was greatly altered, and thus the cell organization in rpl18aB-1/rpl18aB-1 embryos was irregular, resulting in abnormal shapes of embryos with an undulated protoderm-like layer or without a recognizable protoderm cell layer (Fig. 4F,H). The rpl18aB-1/rpl18aB-1 embryo development was arrested at the globular stage and never went through the transition stage for apical-basal pattern formation (Fig. 4F,H). Thus, in the mutant embryos both the radial and the apical-basal pattern formation are altered, and this leads to the abortion of embryogenesis and seed formation.
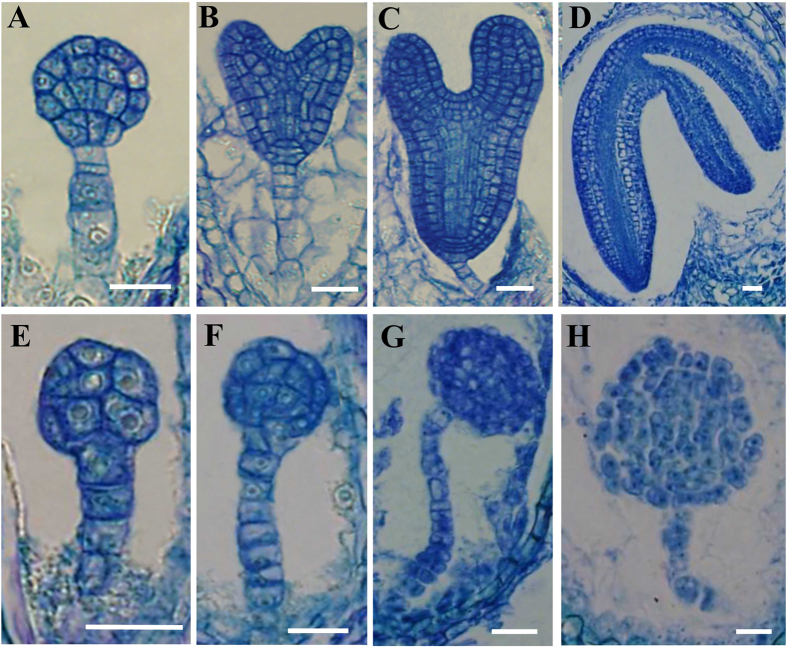
Figure 4. Pattern formation is perturbed in rpl18aB-1/rpl18aB-1 embryos.
(A–D) embryo development of wild type. In wild type ovules, embryogenesis developed from the globular stage (A), heart-shaped stage (B) torpedo-shaped stage (C), and finally to the matured cotyledon stage (D). (E–H) embryo development of rpl18aB-1/rpl18aB-1 is delayed compared with wild-type embryos in the same slique of rpl18aB-1. Mutant embryos with undulated protoderm-like layer (F) or without recognizable protoderm cell layer (G). The rpl18aB-1/rpl18aB-1 embryos were arrested at the globular stage and never underwent the transition stage for apical-basal pattern formation (G,H). Bars = 20 μm for (A) to (H).
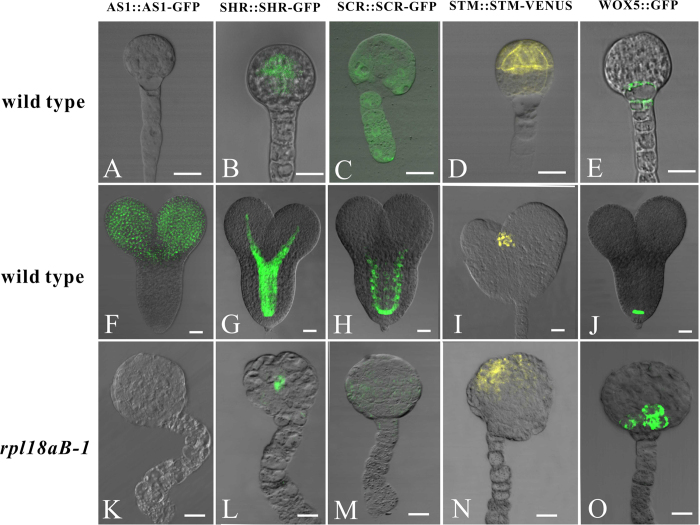
Although morphologically the rpl18aB-1/rpl18aB-1 embryos were blocked at the globular stage, the embryos actually continued to undergo different rounds of cell division, resulting in big globular-shaped embryos of approximately one hundred cells (Fig. 4H). To further detect whether the pattern formation is indeed disturbed in rpl18aB-1/rpl18aB-1, we constructed embryo markers and transformed them into rpl18aB-1 plants. AS1::AS1-GFP, which is expressed mainly in the cotyledon primordia and growing point23 (Fig. 5B), SHR::SHR-GFP24 and SCR::SCR-GFP25, which are expressed in the central area of embryo proper (Fig. 5D,E,G,H), In addition, a shoot apical meristem marker line, STM::STM-VENUS26 (Fig. 5J,K) and the root tip marker line WOX5::GFP line27 (Fig. 5M,N) were crossed with rpl18aB-1 respectively. All positive transgenic lines and hybrids were screened. When wild type embryos developed to the heart-shaped embryo stage, both normal and mutant embryos in the same silique were isolated to observe the expression pattern of the tissue specific markers. In addition, wild type globular embryos in these marker lines were isolated. In wild type, AS1::AS1-GFP was not expressed in the globular embryo but expressed in heart-shaped embryos, while there was no AS1::AS1-GFP expression in rpl18aB-1/rpl18aB-1 embryos (Fig. 5A–C). These results further indicated that the rpl18aB-1/rpl18aB-1 embryos were arrested at the globular stage and never underwent the transition stage for apical-basal pattern formation. As shown in Fig. 5, the other cell-type specific expression pattern of marker genes were disrupted in rpl18aB-1/rpl18aB-1 embryos (Fig. 5F,I,L,O). These observations further suggested that pattern formation in the mutant embryo was disturbed.
Figure 5. Cell-type specific expression pattern of marker genes were disrupted in rpl18aB-1/rpl18aB-1 embryos.
The upper arrow showed the globular embryos isolated from different marker lines. Both normal ovules and abnormal ovules in the same silique of different marker lines in rpl18aB-1 could be found. The middle arrow showed isolated heart-stage embryos in normal ovules and the under arrow showed isolated rpl18aB-1/rpl18aB-1 embryos in abnormal ovules.(A–C) AS1::AS1-GFP expression in wild-type globular embryo (A), heart-stage embryos (B) and rpl18aB-1/rpl18aB-1 embryos (C). (D–F) SHR::SHR-GFP expression in wild-type globular embryo (D), heart-stage embryos (E) and rpl18aB-1/rpl18aB-1 embryos (F). (G–I) SCR::SCR-GFP expression in wild-type globular embryo (G), heart-stage embryos (H) and rpl18aB-1/rpl18aB-1 embryos (I). (J–L) STM::STM-VENUS expression in wild-type globular embryo (J), heart-stage embryos (K) and rpl18aB-1/rpl18aB-1 embryos (L). (M–O) WOX5::GFP expression in wild-type globular embryo (M), heart-stage embryos (N) and rpl18aB-1/rpl18aB-1 embryos (O). Bars = 20 μm for (A) to (O).
Auxin gradient is altered in rpl18aB-1/rpl18aB-1 embryo
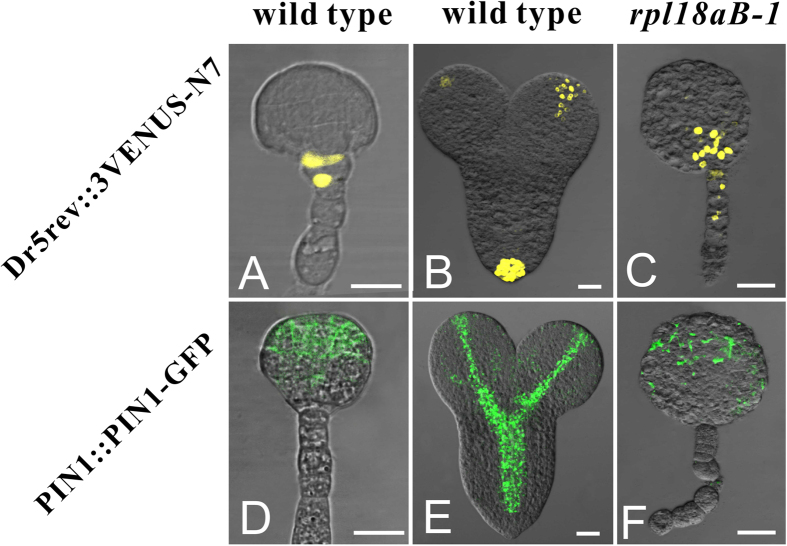
It is well established that auxin is important for pattern formation of embryo28,29,30,31,32,33. As rpl18aB-1 embryo could not develop into heart stage, we reasoned that the auxin distribution might be impaired in these mutants. Firstly a cellular auxin levels marker line DR5rev::3VENUS-N734, was crossed with rpl18aB-1. Fluorescence was observed at the base of embryo proper during the globular stage and mainly at the tips of cotyledons and in the quiescent center during the heart stage in wild-type (Fig. 6A,B). However, in globular-shaped rpl18aB-1/rpl18aB-1 embryos, the distribution of DR5rev::3VENUS-N7 was extended from the embryo proper bottom to uppermost suspensor. These results indicate that the auxin gradient is altered in rpl18aB-1/rpl18aB-1 embryos (Fig. 6C).
Figure 6. Auxin gradient was altered in rpl18aB-1/rpl18aB-1 embryos.
(A–C) DR5rev::3VENUS-N7 expression in wild-type globular embryo (A), heart-stage embryos (B) and rpl18aB-1/rpl18aB-1 embryos (C). (D–F) PIN1::PIN1-GFP expression in wild-type globular embryo (D), heart-stage embryos (E) and rpl18aB-1/rpl18aB-1 embryos (F). Bars = 20 μm for (A) to (F).
The Arabidopsis auxin polar transport carrier PIN-FORMED 1 (PIN1) is responsible for the establishment of an auxin gradient and expressed in the apical part of embryo27,29,31,35. To visualize the localization of PIN1 in the embryos, we used PIN1::PIN1-GFP plants27 and crossed them with rpl18aB-1 plants. In wild type embryos, PIN1 mainly localized in the embryo proper at the globular stage, and in vasculature and hypocotyls of developing cotyledons at the heart stage (Fig. 6D,E). In rpl18aB-1/rpl18aB-1 late globular embryos, PIN1 presented a disorganized distribution (Fig. 6F). These findings indicated the auxin polar transportation pathway is affected in rpl18aB-1/rpl18aB-1 embryos, leading to a disordered auxin distribution and abnormal pattern formation.
Specific expression of RPL18aB in different tissues of mutant embryo proper could not rescue the phenotype
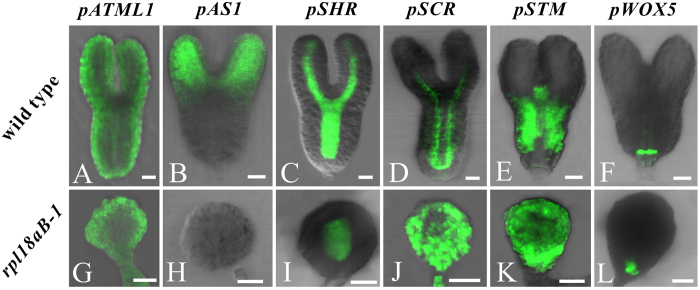
As described above, specific tissue layers of mutant embryos were abnormal. We reasoned that the abortion of rpl18aB homozygous embryos may be the result of the suppression of a certain tissue layer responsible for the formation of heart-shape embryo. If it were true, specifical rescue of the relevant cell layers would recover the defect in pattern formation. To test this hypothesis, we constructed seven expression vectors and transformed them to rpl18aB-1 plants independently. These constructs include pATML1::RPL18aB-EGFP, which use the promoter of epidermal cell layer specific gene ARABIDOPSIS THALIANA MERISTEM LAYER1 (ATML1)36,37 to drives the expression of RPL18aB in the epidermal cell layer of the embryo (Fig. 7A); pAS1::RPL18aB-EGFP, which drives the expression of RPL18aB in the growing ends of the cotyledon (Fig. 7B); pSHR::RPL18aB-EGFP and pSCR::RPL18aB-EGFP, which drive the expression of RPL18aB in the central area of embryo proper (Fig. 7C,D); pSTM::RPL18aB-EGFP, which drives the expression of RPL18aB in the shoot apical meristem (Fig. 7E); pWOX5::RPL18aB-EGFP, which drives the expression RPL18aB in the hypophysis (Fig. 7F). All the six constructs were unable to recover the aberrant embryogenesis mutant phenotype (Fig. 7G–L). Among them, pATML1::RPL18aB-EGFP, pSHR::RPL18aB-EGFP, pSCR::RPL18aB-EGFP, pSTM::RPL18aB-EGFP and pWOX5::RPL18aB-EGFP were detected in the rpl18aB-1/rpl18aB-1 embryo proper but with ectopic expression (Fig. 7G,I–L). For the construct pAS1::RPL18aB-EGFP, it was not expressed in the predicted embryo proper domain in rpl18aB-1/rpl18aB-1 embryo (Fig. 7H). These results indicate that recovery of the specific tissue or cell layer are not sufficient to support normal pattern formation.
Figure 7. Specific expression of RPL18aB in different tissues of mutant embryo proper could not rescue the phenotype.
(A,G) pATML1::RPL18aB-EGFP expression in wild-type torpedo-shaped embryo (A) and rpl18aB-1/rpl18aB-1 embryos (G). (B,H) pAS1::RPL18aB-EGFP expression in wild-type torpedo-shaped embryo (B) and rpl18aB-1/rpl18aB-1 embryos (H). (C,I) pSHR::RPL18aB-EGFP expression in wild-type torpedo-shaped embryo (C) and rpl18aB-1/rpl18aB-1 embryos (I). (D,J) pSCR::RPL18aB-EGFP expression in wild-type torpedo-shaped embryo (D)and rpl18aB-1/rpl18aB-1 embryos (J). (E,K) pSTM::RPL18aB-EGFP expression in wild-type torpedo-shaped embryo (E) and rpl18aB-1/rpl18aB-1 embryos (K). (F,L) pWOX5::RPL18aB-EGFP expression in wild-type torpedo-shaped embryo (F) and rpl18aB-1/rpl18aB-1 embryos (L). Bars = 20μm for (A) to (L).
Discussion
Embryogenesis is the process of pattern formation. In Arabidopsis, the zygote divides asymmetrically following fertilization, produces an apical cell and a basal cell, and then further develops into the embryo proper and suspensor domains38,39. In the embryo proper domain, a radial pattern could be observed as early as at the 16-celled embryo stage, which is indicated by a clear protoderm and the enclosed provescular and ground tissue39,40. When the embryo develops into the globular stage, the primordia of cotyledons are generated, which morphologically marks the establishment of apical-basal pattern41.
The differentiation of protoderm cells involves some tissue specific expressed genes. ATML1is specifically expressed in the apical cell lineage, and after the 16-cell embryo stage it only expresses in protoderm cells37. PROTODERMAL FACTOR2 (PDF2) has a similar expression pattern. Both genes encode START domain containing transcription factors and positively regulate epidermal cell differentiation42,43. Although other genes are also involved in radial pattern formation of the embryo, such as FASS, SCR and WOL24,25,44, the mechanisms determining the cell fate of the protoderm and other radial layers of the embryo remain to be elucidated.
Apical-basal pattern is gradually established after the globular embryo stage. WUS is detected first in the 16-cell embryo, and is critical for apical meristem formation45. PINOID and ENHANCER OF PINOID are required for the auxin accumulation in apical cells and cotyledon formation46,47,48,49. CUP SHAPED COTYLEDON is critical to suppress the apical meristem between cotyledon primordia and thus to separate them50,51,52,53. These studies revealed the existence of a very complex regulatory mechanism for embryo pattern formation, and highlight the remaining need for future investigations to unravel this regulatory network.
In our study, we found that rpl18aB embryos undergo developmental arrest at the globular stage, with possible, secondary pattern formation defects. In fact, as early as at the 8-celled embryo stage irregular cell division orientations could be observed, resulting in disorganization of embryo cells and the differentiation of different domains in both radial and apical-basal structures of embryos, suggesting that an early step is interrupted and this early developmental process is critical for both the radial and apical-basal pattern formation. Interestingly, based on previous works, we carried out cell type-specific complementation of RPL18aB in mutant embryos. We used pATML1::RPL18aB-EGFP for epidermal cell layer, pAS1::RPL18aB-EGFP for cotyledon primordia and growing end, pSTM::RPL18aB-EGFP for shoot apical meristem, pWOX5::RPL18aB-EGFP for hypophysis, pSHR::RPL18aB-EGFP and pSCR::RPL18aB-EGFP for the central area of the embryo proper specific complementation respectively. These complementations were expected to recover different cell types and thus offer the opportunity to test the role of each of these cell types in embryo pattern formation. However, the results showed that none of these complementations could recover the failed embryo pattern formation phenotype. These results indicated that RPL18aB might play an essential role in the cell fate determination during early embryogenesis. Since most of these marker genes are not expressed at the very early stage, our results suggest that the first several divisions of apical cell lineage are critical for the subsequent embryo pattern formation. Even though some of the cell types were recovered by the cell type specific supplementation and the later embryo development could be improved in some way, the pattern formation phenotype could not be fully recovered since early cell division was irregular. How these early cell divisions could change cell fate is an interesting question and should be elucidated in future studies.
Recently, ribosomal proteins were found to play a multifunctional role in plant development3,4,5,6,7,8,9, and their role in sexual plant reproduction has drawn a great attention11,12,13,14. During embryogenesis, mutations in several cytoplasmic ribosomal proteins, such as S6, S11, L2, L8, L23, L19 and L40, results in embryo lethality, indicating their critical role in embryogenesis. These ribosomal proteins mainly function in the later stages of embryogenesis15. In the present study, we report a new ribosomal protein RPL18aB that also functions in the early stage of development and specifically involved in early cell division orientations. The mutant rpl18aB offers an opportunity to test the influence of early cell division on the cell fate determination and subsequent embryo pattern formation. Future studies should focus on elucidating the relationship between this ribosomal protein, the control of cell division orientation and to the cell fate decision.
Materials and Methods
Plant materials and growth conditions
The rpl18aB-1 allele was isolated from our mutant library with Hygr (Wu et al.17). The rpl18aB-2 (Flag-209B07) was obtained from INRA (Versailles, France), STM::STM-VENUS (CS-67930) and DR5rev::3VENUS-N7(CS-67931) were obtained from ABRC, Ohio, USA. Marker lines WOX5::GFP and PIN1::PIN1-GFP were provided by Dr. Jian Xu (Temasek Life Sciences Laboratory, Singapore). Seeds were surface sterilized with 20% bleach for 10 min, and washed three times with sterile distilled water. Seeds were stratified for 3 days at 4 °C, and then sown on 1/2 MS plates with 1.0% (w/v) sucrose. Agar plates were placed in a growth room with a photoperiod of 16 h light/8h dark. For kanamycin selection, 50 mg/L of kanamycin (Sigma) was supplemented to the media. Similarly, 50 mg/L of hygromycin (Roche) was added for hygromycin selection. Plants were grown in soil in a greenhouse under long-day conditions (16 h light/8 h dark) at 22 °C.
Cloning of the T-DNA flanking sequence of the rpl18aB-1
The T-DNA flanking sequence in the rpl18aB-1 mutant was cloned by TAIL-PCR (Liu et al.19). The authenticity of the cloned sequence was confirmed by PCR using two pairs primers located around the T-DNA border (rpl18aB-1-T1: GCCCCTCTCTTGACTAATGTAATC; LB-S: CCAAAATCCAGTACTAAAATCCAG) and (rpl18aB-1-T2: CATGAACAAGTTGGGCTTGTTGG; LB-S: CCAAAATCCAGTACTAAAATCCAG) respectively.
Vector construction and plant transformation
Plasmid P092 and P094 were produced as previous described17. To generate the genomic complementation construct, A 3250-bp wild-type genomic sequence containing the AT2g34480 gene, 1690-bp upstream of the ATG codon and 314-bp downstream of the TAG codon sequences, was PCR-amplified with primers RPL18aB-F1: NNNNTCTAGATTGGTGATCAAAGTCAATACTCATG and RPL18aB-R1: NNNNGCGGCCGCGTTTGAAGGCAAAATTACAGTAGAG from genomic DNA and then was cloned into the P092 plasmid. To investigate the expression pattern of RPL18aB, we first cloned the GUS fragment with the NOS terminator from pCAMBIA1305 and then introduced them into the plasmid PART27 to generate P093. We then cloned the H2B coding fragment (AT5G22880) and cloned it into P094to generate P095. The RPL18aB promoter was amplified with primers RPL18aB-F1 and RPL18aB-F1-R2: NNNNAAGCTTTTCGTCTGGAGAGAGACAATG and put upstream of GUS in P093 and H2B-GFP in P095 to generate pRPL18aB::GUS and pRPL18aB::H2B-GFP. To produce the embryo markers of AS1::AS1:GFP, SHR::SHR:GFP and SCR::SCR:GFP constructs, the promoter and CDs sequences of AS1, SHR, SCR were amplified with primers shown in Table 3 and cloned into p094 respectively. To specifically express RPL18aB in the pollens and embryos respectively, the RPL18aB ORF was amplified with primers RPL18aB-CDS1: NNNNGGTACCGCCACCATGGGTGCTTTCAGGTTTCACC and RPL18aB-CDS2: NNNNAAGCTTCATGAACAAGTTGGGCTTGTTG from the cDNA of flower and cloned into p094 to generate a RPL18aB-EGFP construct. Then the promoter of AS1, ATML1, SHR, SCR, WOX5, STEM and LAT52 were amplified with primers shown in Table 3 and fused upstream of RPL18aB-EGFP respectively.
Table 3. Primers.
| Name | Primers (5′ to 3′) |
|---|---|
| SCR CDS-1 | NNNNCCTAGGATGGCGGAATCCGGCG |
| SCR CDS-2 | NNNNGAATTCAGAACGAGGCGTCCAAGCT |
| SHR CDS-1 | NNNNGCGGCCGCATGGATACTCTCTTTAGACTAGTCAGTCTCC |
| SHR CDS-2 | NNNNGAATTCACTAGCCCAAACCACCGGCT |
| AS1 CDS-1 | NNNNGCGGCCGCATGAAAGAGAGACAACGTTGGAGTG |
| AS1 CDS-2 | NNNNGAATTCGGGGCGGTCTAATCTGCAAC |
| LAT52 promoter-1 | NNNNGAGCTCTGTCGACATACTCGACTCAGAAG |
| LAT52 promoter-2 | NNNNGGTACCTTTAAATTGGAATTTTTTTTTTTGG |
| SCR promoter-1 | NNNNGCGGCCGCGATTTTGCTGGTGTTGAATGGAT |
| SCR promoter-2 | NNNNCCTAGGGGAGATTGAAGGGTTGTTGGTC |
| ATML1 promoter-1 | NNNNCCAACGCGTTGGGTGTTTACATTGATTAGTAGGCTGC |
| ATML1 promoter-2 | NNNNGCGGCCGCGATGATGATGGATGCCTATCAAT |
| SHR promoter-1 | NNNNCCAACGCGTTGGATGGGGAAGATGTTTTCTCCAAGTA |
| SHR promoter-2 | NNNNGCGGCCGCTAATGAATAAGAAAATGAATAGAAG |
| AS1 promoter-1 | NNNNCCAACGCGTTGGGAGAAATATATTAGGCAATCAAACGG |
| AS1 promoter-2 | NNNNGCGGCCGCCTCCTACTCCTCCTGACATCACTTC |
| WOX5 promoter-1 | NNNNCCAACGCGTTGGCTTGCAAAATTTGATCGTCGTAC |
| WOX5 promoter-2 | NNNNGCGGCCGCGTTCAGATGTAAAGTCCTCAACTGTT |
| STM promoter-1 | NNNNGCGGCCGCACGTTATCCTAATTTTGTTTATCCTA |
| STM promoter-2 | NNNNCCTAGGCTTCTCTTTCTCTCACTAGTATTATTATTCA |
All constructs were transformed into Agrobacterium tumefaciens strain GV3101, and then transformed into Arabidopsis plants by the floral dip method54.
Phenotype Characterization of embryo development
To prepare cleared whole-mount seeds, seeds were dissected from siliques and cleared using chloral hydrate solution55. The cleared seeds were placed under a microscope (Olympus) fitted with differential interference contrast optics for imaging.
Fertilized siliques at different developmental stages were fixed overnight in 4% (v/v) formaldehyde, dehydrated in a graded ethanol series (10%, 30%, 50%, 70%, 80%, 90%, 95% and 100% twice), transferred to xylene and embedded in Sigma’s paraplast. Semi-thin sections (10 μm) were cut on a rotary microtome (American optical, USA), and heat fixed to glass slides. Sections were stained with toluidine blue before removal of paraffin with xylene (20 min twice). Bright-field images of the ovule cross-sections were taken using an Olympus microscope.
Pollen germination assays
In vitro pollen germination was performed according to Ye et al.56. Pollen was harvested from newly opened flowers and placed onto the pollen germination medium consisting 1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 0.01% (w/v) H3BO3, and 18% (w/v) sucrose solidified with 0.8% (w/v) agar, pH 7.0. The plates were cultured at 28 °C at high humidity. Semi-in vitro pollen growth assay was performed as previously described57. After hand-pollination, pistils were cut at the shoulder region of the ovaries. Cut pistils were incubated on pollen tube growth medium at 28 °C.
Scanning Electron Microscopy
For scanning electron microscopy examination, the opened flowers of the qrt1 and rpl18aB-1 were dissected and the dehiscent anthers were coated with 8-nm gold. Observations and images were performed with a S-3400N scanning electron microscope (HITACHI).
Histochemical analysis of GUS activity
The histochemical analysis of GUS activity was performed according to Vielle-Calzada et al.58. Plant tissues were incubated at 37 °C in GUS staining solution [2 mM 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) in 50 mM sodium phosphate buffer, pH 7.0] containing 0.1% Triton X-100, 2 mM K4Fe(CN)6 and 2 mM K3Fe(CN)6. The stained tissues were then transferred to 70% (v/v) ethanol solution. Samples were mounted with traditional clearing solution and placed under a microscope (Olympus) fitted with differential interference contrast optics for imaging.
Embryo separation and CLSM microscopy
For fluorescent marker line analysis, ovules of Arabidopsis at specific development periods were collected and put on a 30 mm diameter culture plate with a drop of 10% glycerin added with 80 mM sorbitol. A sharp capillary glass tube was used as the dissection tool to extract the embryos from maternal tissues. The isolated embryos were visualized using a FV1000 confocal laser-scanning microscope (CLSM; Olympus). GFP fluorescence was detected with excitation at 488 nm.
Additional Information
How to cite this article: Yan, H. et al. Ribosomal protein L18aB is required for both male gametophyte function and embryo development in Arabidopsis. Sci. Rep. 6, 31195; doi: 10.1038/srep31195 (2016).
Acknowledgments
We thank Dr. Jian Xu for providing the transgenic line WOX5::GFP and PIN1::PIN1-GFP27. This work was supported by the National Natural Science Foundation (grant No. 31570317, 31270362).
Footnotes
Author Contributions H.Y., D.C., Y.W., Y.S., J.Z. and X.P. performed the experiments; H.Y., M.S. and X.P. wrote the manuscript; D.C., Y.W., Y.S., J.Z. and M.S. reviewed the manuscript; X.P. designed the experiments.
References
- Barakat A. et al. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127, 398–415 (2001). [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E. A role for the ribosome in development. Trends Plant Sci 14, 512–9 (2009). [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M. et al. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13, 3378–88 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U., Horiguchi G., Ponce M. R., Micol J. L. & Tsukaya H. Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J 59, 499–508 (2009). [DOI] [PubMed] [Google Scholar]
- Stirnberg P., Liu J. P., Ward S., Kendall S. L. & Leyser O. Mutation of the cytosolic ribosomal protein-encoding RPS10B gene affects shoot meristematic function in Arabidopsis. BMC Plant Biol 12, 160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi D. & Byrne M. E. Involvement of ribosomal protein RPL27a in meristem activity and organ development. Plant Signal Behav 6, 712–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Kim G. T. & Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 22, 257–64 (2000). [DOI] [PubMed] [Google Scholar]
- Pinon V. et al. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–24 (2008). [DOI] [PubMed] [Google Scholar]
- Horiguchi G. et al. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 65, 724–36 (2011). [DOI] [PubMed] [Google Scholar]
- Rosado A., Li R., van de Ven W., Hsu E. & Raikhel N. V. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA 109, 19537–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D. et al. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–99 (2001). [DOI] [PubMed] [Google Scholar]
- Nishimura T., Wada T., Yamamoto K. T. & Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17, 2940–53 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A., Komura M., Kawano E., Kuwashiro Y. & Takahashi T. A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56, 881–90 (2008). [DOI] [PubMed] [Google Scholar]
- Szakonyi D. & Byrne M. E. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65, 269–81 (2011). [DOI] [PubMed] [Google Scholar]
- Tzafrir I. et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135, 1206–20 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S. Y. & Davis R. W. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264, 1458–1460 (1994). [DOI] [PubMed] [Google Scholar]
- Wu J. J. et al. Mitochondrial GCD1 dysfunction reveals reciprocal cell-to-cell signaling during the maturation of Arabidopsis female gametes. Dev Cell 23, 1043–58 (2012). [DOI] [PubMed] [Google Scholar]
- Twell D., Wing R., Yamaguchi J. & McCormick S. Isolation and expression of an anther-specific gene from tomato. Molecular and General Genetics MGG 217, 240–245 (1989). [DOI] [PubMed] [Google Scholar]
- Liu Y. G., Mitsukawa N., Oosumi T. & Whittier R. F. Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463 (1995). [DOI] [PubMed] [Google Scholar]
- Liu Y.-G. & Whittier R. F. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681 (1995). [DOI] [PubMed] [Google Scholar]
- Carroll A. J., Heazlewood J. L., Ito J. & Millar A. H. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Molecular & cellular proteomics 7, 347–369 (2008). [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C. et al. Dynamic analyses of the expression of the HISTONE:: YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. The Plant Cell 13, 495–509 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E. et al. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–71 (2000). [DOI] [PubMed] [Google Scholar]
- Helariutta Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–67 (2000). [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L. et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–33 (1996). [DOI] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I. & Barton M. K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 (1996). [DOI] [PubMed] [Google Scholar]
- Blilou I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005). [DOI] [PubMed] [Google Scholar]
- Friml J. et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- Liu C., Xu Z. & Chua N.-H. Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. The Plant Cell 5, 621–630 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B. & Weijers D. Auxin control of embryo patterning. Cold Spring Harbor Perspectives in Biology 1, a001545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Vernoux T., Furutani M., Traas J. & Tasaka M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965–3974 (2002). [DOI] [PubMed] [Google Scholar]
- Hadfi K., Speth V. & Neuhaus G. Auxin-induced developmental patterns in Brassica juncea embryos. Development 125, 879–887 (1998). [DOI] [PubMed] [Google Scholar]
- Smit M. E. & Weijers D. The role of auxin signaling in early embryo pattern formation. Current opinion in plant biology 28, 99–105 (2015). [DOI] [PubMed] [Google Scholar]
- Heisler M. G. et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current biology 15, 1899–1911 (2005). [DOI] [PubMed] [Google Scholar]
- Gälweiler L. et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 (1998). [DOI] [PubMed] [Google Scholar]
- Takada S. & Jürgens G. Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134, 1141–1150 (2007). [DOI] [PubMed] [Google Scholar]
- Lu P., Porat R., Nadeau J. A. & O’Neill S. D. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–68 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T., Würschum T. & Breuninger H. Genetic regulation of embryonic pattern formation. The Plant Cell 16, S190–S202 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik P. D., Gillmor C. S. & Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 23, 207–236 (2007). [DOI] [PubMed] [Google Scholar]
- Colette A., Lu K.-J. & Weijers D. Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142, 420–430 (2015). [DOI] [PubMed] [Google Scholar]
- Jürgens G. Apical–basal pattern formation in Arabidopsis embryogenesis. The EMBO journal 20, 3609–3616 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M., Takahashi T. & Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J 26, 487–94 (2001). [DOI] [PubMed] [Google Scholar]
- Abe M., Katsumata H., Komeda Y. & Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635–43 (2003). [DOI] [PubMed] [Google Scholar]
- Mahonen A. P. et al. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14, 2938–43 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F. et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–15 (1998). [DOI] [PubMed] [Google Scholar]
- Christensen S. K., Dagenais N., Chory J. & Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478 (2000). [DOI] [PubMed] [Google Scholar]
- Furutani M. et al. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030 (2004). [DOI] [PubMed] [Google Scholar]
- Treml B. S. et al. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132, 4063–74 (2005). [DOI] [PubMed] [Google Scholar]
- Friml J. et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306, 862–865 (2004). [DOI] [PubMed] [Google Scholar]
- Aida M., Ishida T. & Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–70 (1999). [DOI] [PubMed] [Google Scholar]
- Takada S., Hibara K., Ishida T. & Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–35 (2001). [DOI] [PubMed] [Google Scholar]
- Vroemen C. W., Mordhorst A. P., Albrecht C., Kwaaitaal M. A. & de Vries S. C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–77 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K. et al. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18, 2946–57 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–43 (1998). [DOI] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M. & Laux T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Developmental cell 14, 867–876 (2008). [DOI] [PubMed] [Google Scholar]
- Ye J. et al. Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21, 3868–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R. & Preuss D. Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol 6, 7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada J. P., Baskar R. & Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature 404, 91–4 (2000). [DOI] [PubMed] [Google Scholar]