Abstract
This study was to explore the association between gene variants and p21 expression and investigate the TP53-independent p21 regulation in hepatitis B virus (HBV) related hepatocellular carcinoma (HCC) patients from Guangxi by genome-wide association study. 426 HBV-related HCC patients were enrolled. Results showed that, after quality control, a total of 21,643 SNPs were identified in 107 p21 positive and 298 p21 negative patients. The variants of epidermal growth factor receptor (EGFR; rs2227983 and rs6950826) and spectrin repeat containing, nuclear envelope 2 (SYNE2; rs8010699, rs4027405 and rs1890908) were associated with p21 expression. Moreover the haplotype block (rs2227983 and rs6950826, r2 = 0.378) in EGFR and the haplotype block in SYNE2 (rs8010699 was in strong LD with rs4027405 and rs1890908 (r2 = 0.91 and 0.70, respectively)) were identified, and the haplotype A-G of EGFR and haplotype G-A-A of SYNE2 were significantly associated with p21 expression (P < 0.01). rs4027405 and rs1890908 were significantly associated with overall survival, and patients with AG/GG genotypes of SYNE2 gene had a worse overall survival (P = 0.001, P = 0.002). Our findings indicate that variants of EGFR and SYNE2 play an important role in p21 regulation and are associated with the clinical outcome of HBV-related HCC in a TP53-indenpdent manner.
Worldwide, the primary liver cancer has been the 5th most common malignancy in men and the 6th in women1. Global Cancer Statistics1 in 2012 shows that approximate 782,500 new liver cancer cases and 745,500 deaths occur worldwide in 2012, about half of which are found in China. Moreover, about 70–90% of primary liver cancer is hepatocellular carcinoma (HCC)2. There is a geographic difference in the incidence of HCC, which is mainly caused by some etiological variations, with the highest incidence in Eastern, South-Eastern Asia and Northern and Western Africa and the lowest incidence in South-Central Asia and Northern, Central, and Eastern Europe. The major causes of HCC include the infection by hepatitis virus B (HBV) and/or hepatitis C (HCV), aflatoxin-B1 (AFB1) exposure, fasciola hepatica infection and alcoholic cirrhosis3,4. Both HBV infection and AFB1 exposure are the prominent etiological factors of HCC in Guangxi, a region in southern China5.
Hepatocarcinogenesis is an extremely complex process, and there is no a specific molecular mechanism that can elucidate this process. Investigations have demonstrated that the dys-regulation of cell cycle plays an important role in the occurrence and progression of HCC6,7. p21 is a major cell cycle regulator. It is a cyclin-dependent kinase (CDK) inhibitor and a downstream factor of TP53. It can causes G1 growth arrest and thus it is also as known as a tumor suppressor8,9. p21 may bind to and inhibit the CDK activity, which mediates its various biological activities and may cause cell cycle arrest at a specific stage10,11. p21 has been found as one of most important and effective effector molecules of TP53. TP53 is able to bind to the promoter of p21 gene, leading to the direct activation of p21 expression8,12. Studies have shown that p21 can also be regulated in TP53-independent manners, such as by MYC, E2A and BRCA113,14. In addition, it is controversial on whether the HBX gene of HBV affects the p21 regulation. Some studies report that HBX gene can suppress p21 expression15,16, but this is not found in other studies17,18.
Although p21 is a tumor suppressor, it may also promote oncogenesis in certain circumstances. p21-deficient mice may spontaneously develop tumors, supporting the tumor suppressor activity of p2119. Previous studies have reported that p21 mRNA or protein expression in HCC tissues is often lower than in adjacent normal tissues20,21,22. These results suggest that p21 might act as a tumor suppressor in HCC. Moreover, the spontaneous development of lymphomas is suppressed in p21-deficient mice with TP53- or ATM-deficiency23,24, indicating that p21 serves as an oncogene. In fact, p21 is over-expressed in a variety of cancers, such as breast cancer, prostate cancer, oesophageal squamous cell carcinoma and cervical cancer, and p21 up-regulation as a poor prognostic biomarker is closely related to the tumor grade, invasiveness and aggressiveness in many cases11,25,26,27.
Kao et al.28 and Zhang et al.29 reported that p21 expression was a favorable survival factor in HCC, but this was not found in our prior study. This may be ascribed to the difference in patients selected. In their study, the HBV-related and non-HBV-related HCC patients were recruited, but only HBV-related HCC patients were enrolled into our study. Furthermore, different from patients in other regions, patients in Guangxi have a high level exposure to AFB1 in their food which is closely associated with hepatocarcinogenesis5. Study has reported that AFB1 exposure leads to a high mutation frequency at TP53 249Ser, which has been considered as a molecular fingerprint of HCC pathogenesis30. However, at present, whether AFB1 exposure is involved in the regulation of p21 is unclear.
In the present study, a genome-wide association study (GWAS) using Illumina HumanExome BeadChip-12-1_A was performed in Chinese HBV-related HCC patients from Guangxi, aiming to detect the gene variants associated with p21 expression and reveal the TP53-independent p21 regulation.
Results
GWAS
Baseline Characteristics
The clinicopathological characteristics are shown in Table 1. There were no significant differences in the age, gender, BMI, race, smoking status, drinking status, Barcelona Clinic Liver Cancer (BCLC) stage, serum alpha fetoprotein (AFP) level, transcatheter arterial chemoembolization (TACE) status, pathological grade and TP53 249Ser mutation between p21 negative group and p21 positive group (P > 0.05). TP53 expression status and hepatic cirrhosis status were significantly different between p21 negative group and p21 positive group (P = 0.005 and 0.001, respectively). A significantly positive relationship was observed between p21 expression status and TP53 expression status (r = 0.144, P < 0.01).
Table 1. Clinicopathological characteristics of HBV-related HCC patients after data quality control.
| Variable | GWAS |
Survival Analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| Negative (n = 298) | Positive (n = 107) | OR (95% CI) | P value | Patients (n = 405) | MST (months) | HR* (95% CI) | P* value | |
| Age (years) | 0.052 | 0.708 | ||||||
| ≤46 | 154 | 67 | Ref. | 221 | 52 | Ref. | ||
| >46 | 144 | 40 | 1.56 (1–2.46) | 184 | 44 | 1.06 (0.79–1.42) | ||
| Gender | 0.619 | 0.428 | ||||||
| Male | 268 | 98 | Ref. | 366 | 47 | Ref. | ||
| Female | 30 | 9 | 0.82 (0.38–1.79) | 39 | 82 | 0.80 (0.46–1.39) | ||
| Race | 0.397 | 0.929 | ||||||
| Han | 184 | 71 | Ref. | 255 | 47 | Ref. | ||
| Minority | 114 | 36 | 0.82 (0.52–1.30) | 150 | 50 | 1.01 (0.75–1.38) | ||
| BMI | 0.048 | 0.922 | ||||||
| ≤25 | 229 | 92 | Ref. | 321 | 48 | Ref. | ||
| >25 | 69 | 15 | 0.54 (0.29–0.99) | 84 | 45 | 1.02 (0.72–1.45) | ||
| Smoking status | 0.506 | 0.152 | ||||||
| None | 187 | 71 | Ref. | 258 | 51 | Ref. | ||
| Ever | 111 | 36 | 0.85 (0.54–1.36) | 147 | 39 | 1.22 (0.92–1.67) | ||
| Drinking status | 0.627 | 0.143 | ||||||
| None | 173 | 65 | Ref. | 238 | 51 | Ref. | ||
| Ever | 125 | 42 | 0.89 (0.57–1.40) | 167 | 41 | 1.24 (0.93–1.66) | ||
| Child-pugh class | 0.754 | 0.030 | ||||||
| A | 238 | 84 | 322 | 50 | Ref. | |||
| B | 41 | 16 | 1.11 (0.59–2.07) | 57 | 31 | 1.54 (1.04–2.27) | ||
| Missing | 19 | 7 | 26 | |||||
| BCLC stage | 0.639 | <0.001 | ||||||
| A | 169 | 59 | Ref. | 228 | 95 | Ref. | ||
| B | 52 | 16 | 0.88 (0.47–1.66) | 68 | 39 | 1.89 (1.27–2.81) | ||
| C | 76 | 32 | 1.21 (0.73–2.00) | 108 | 27 | 2.89 (2.09–4.00) | ||
| Missing | 1 | 0 | 1 | |||||
| p21 Expression | NA | 0.443 | ||||||
| Negative | 298 | 0 | 298 | 50 | Ref. | |||
| Positive | 0 | 107 | 107 | 40 | 1.13 (0.83–1.54) | |||
| TP53 Expression | 0.005 | 0.161 | ||||||
| Negative | 122 | 27 | Ref. | 149 | 58 | Ref. | ||
| Positive | 164 | 74 | 2.04 (1.24–3.36) | 238 | 41 | 1.25 (0.92–1.70) | ||
| missing | 12 | 6 | 18 | |||||
| AFB1 exposure | 0.315 | |||||||
| no | 190 | 74 | Ref. | 264 | 50 | Ref. | 0.364 | |
| yes | 108 | 33 | 0.79 (0.49–1.26) | 141 | 40 | 1.15 (0.85–1.55) | ||
| TACE status | ||||||||
| before hepatectomy | 0.383 | 0.769 | ||||||
| No | 235 | 80 | Ref. | 315 | 48 | Ref. | ||
| Yes | 63 | 27 | 1.26 (0.75–2.11) | 90 | 44 | 1.05 (0.75–1.47) | ||
| after hepatectomy | 0.091 | 0.872 | ||||||
| No | 134 | 38 | Ref. | 172 | 76 | Ref. | ||
| Yes | 164 | 69 | 1.48 (0.94–2.34) | 233 | 43 | 1.03 (0.76–1.40) | ||
| Cirrhosis | 0.001 | 0.174 | ||||||
| No | 22 | 21 | Ref. | 43 | 88 | Ref. | ||
| Yes | 276 | 85 | 0.32 (0.17–0.62) | 361 | 45 | 1.40 (0.86–2.28) | ||
| missing | 0 | 1 | 1 | |||||
| Serum AFP | 0.237 | 0.155 | ||||||
| ≤400 (ng/ml) | 158 | 48 | Ref. | 206 | 51 | Ref. | ||
| >400 (ng/ml) | 122 | 49 | 1.32 (0.83–2.1) | 171 | 41 | 1.24 (0.92–1.68) | ||
| missing | 18 | 10 | 28 | |||||
| Radical resection | 0.382 | 0.034 | ||||||
| Yes | 163 | 63 | Ref. | 226 | 73 | Ref. | ||
| No | 127 | 40 | 0.82 (0.52–1.29) | 167 | 40 | 1.37 (1.02–1.84) | ||
| missing | 8 | 4 | 12 | |||||
| Pathological grade | 0.13 | 0.618 | ||||||
| Well | 22 | 2 | Ref. | 24 | 79 | Ref. | ||
| Moderately | 230 | 88 | 4.2 (0.97–18.27) | 318 | 44 | 1.38 (0.71–2.72) | ||
| Poorly | 9 | 2 | 2.44 (0.30–20.12) | 11 | 47 | 1.21 (0.37–3.92) | ||
| missing | 37 | 15 | 52 | |||||
| Antiviral therapy | 0.001 | 0.009 | ||||||
| Yes | 120 | 23 | Ref. | 143 | >117 | Ref. | ||
| No | 178 | 84 | 2.46 (1.47–4.13) | 262 | 41 | 1.60 (1.12–2.29) | ||
| Oncological behavior | ||||||||
| Tumor size | 0.052 | <0.001 | ||||||
| ≤5 cm | 106 | 27 | Ref. | 133 | 123 | Ref. | ||
| >5 cm | 192 | 80 | 1.64 (1–2.69) | 272 | 40 | 2.08 (1.45–2.96) | ||
| No. of tumors | 0.576 | 0.003 | ||||||
| Single (n = 1) | 220 | 76 | Ref. | 296 | 58 | Ref. | ||
| Multiple (n > 1) | 78 | 31 | 1.15 (0.70–1.88) | 109 | 34 | 1.61 (1.17–2.20) | ||
| Regional invasion | 0.963 | 0.129 | ||||||
| Absence | 254 | 91 | Ref. | 345 | 52 | Ref. | ||
| Presence | 44 | 16 | 1.02 (0.55–1.89) | 60 | 40 | 1.37 (0.91–2.06) | ||
| Intrahepatic metastasis | 0.47 | 0.001 | ||||||
| Absence | 155 | 60 | Ref. | 216 | 76 | Ref. | ||
| Presence | 143 | 47 | 1.18 (0.76–1.84) | 190 | 36 | 1.66 (1.23–2.23) | ||
| Vascular invasion | 0.748 | <0.001 | ||||||
| Absence | 241 | 85 | Ref. | 326 | 76 | Ref. | ||
| Presence | 57 | 22 | 1.09 (0.63–1.90) | 79 | 20 | 2.98 (2.16–4.13) | ||
| PVTT | 0.979 | |||||||
| No | 250 | 88 | Ref. | 338 | 71 | Ref. | <0.001 | |
| vp1 | 8 | 3 | 1.07 (0.28–4.10) | 11 | 28 | 1.85 (0.82–4.23) | ||
| vp2 | 12 | 4 | 0.95 (0.30–3.01) | 16 | 18 | 2.76 (1.52–5.00) | ||
| vp3 | 22 | 10 | 1.29 (0.59–2.83) | 32 | 22 | 2.25 (1.44–3.50) | ||
| vp4 | 6 | 2 | 0.95 (0.19–4.78) | 8 | 8 | 5.11 (2.37–11.02) | ||
OR = odds ratio, 95% CI = 95% confidence intervals, MST = median survival time, HR = Hazard Ratio, Ref. = Reference.
*HR and P value for univariate survival analysis.
Quality Control
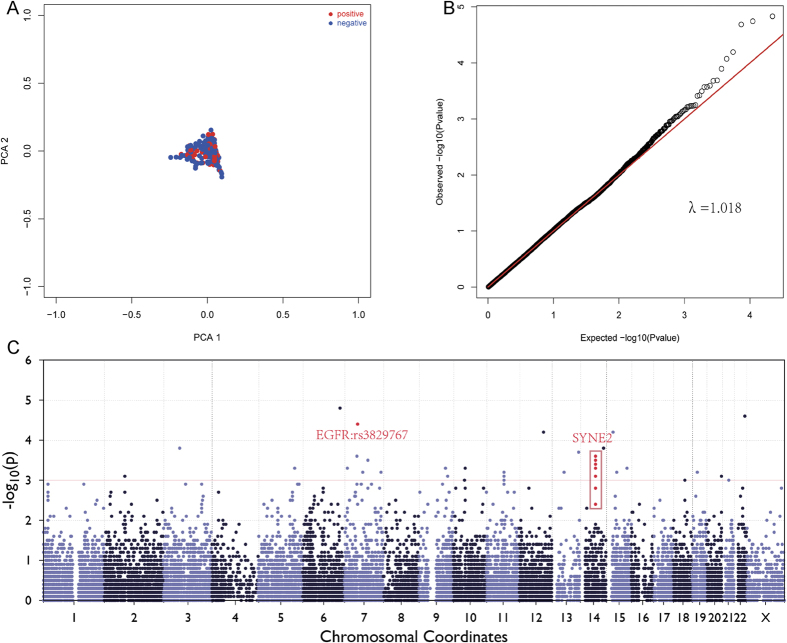
After quality control (QC), a total of 21,643 SNPs were identified in 107 p21 positive patients and 298 p21 negative patients. The total genotyping rate in remaining individuals was 98.34%. The principal component analysis (PCA) plot (Fig. 1A) showed that no or mild population stratification in this study population, as indicated by the Quantile-Quantile (Q-Q) plot (Fig. 1B). The genomic control inflation factor (λ) was 1.018.
Figure 1.
(A) Principal components analysis (PCA) for ancestry and population stratification implemented in EIGENSOFT package. The blue dot represents controls and red dot represents patients. No or mild population stratification was found in this study population. (B) Quantile-Quantile (Q-Q) plots from Single Variant Test. Reported genomic inflation factor was calculated by MATLAB 7.0, based on P value, and the genomic control inflation factor (λ) was 1.018. (C) Manhattan plots for association analysis. Results from Single Variant Test (−log10 P values) were plotted against genomic position (GRCh37/hg19).
Association Analysis
Due to the limitation of sample size, we focused on the loci with minor allele frequencies (MAF) > 0.05. Manhattan plot is shown in Fig 1C. 30 SNPs in the association analysis are reported in Supplementary Table 2. The SNPs in epidermal growth factor receptor (EGFR) and spectrin repeat containing, nuclear envelope 2 (SYNE2) were associated with p21 expression in HBV-related HCC (Table 2). rs2227983 (MAF = 0.49, P = 3.61 × 10−5) was located in the exon 13 of EGFR gene and rs6950826 (MAF = 0.32, P = 4.06 × 10−3) in the intron 7 of EGFR gene. rs8010699 (MAF = 0.19, P = 2.41 × 10−4), rs3829767 (MAF = 0.20, P = 2.94 × 10−4), rs4027402 (MAF = 0.22, P = 3.60 × 10−4), rs9944035 (MAF = 0.10, P = 5.46 × 10−4), rs4902264 (MAF = 0.19, P = 8.03 × 10−4), rs4027405 (MAF = 0.16, P = 1.71 × 10−3) and rs1890908 (MAF = 0.32, P = 3.63 × 10−3) were found in the SYNE2 gene.
Table 2. SNPs of EGFR and SYNE2 genes are associated with p21 expression in HBV-related HCC.
| SNP | Chr | Position | Gene | Allele | Negative group | Positive group | MAF | P value* |
|---|---|---|---|---|---|---|---|---|
| rs2227983 | 7 | 55229255 | Nonsynonymous: EGFR | G/A | 71/135/89 | 42/52/12 | 0.49 | 3.61 × 10−5 |
| rs6950826 | 7 | 55223176 | Intron: EGFR | G/A | 158/103/34 | 34/55/15 | 0.32 | 4.06 × 10−3 |
| rs8010699 | 14 | 64522843 | Nonsynonymous: SYNE2 | A/G | 9/77/204 | 10/38/55 | 0.19 | 2.41 × 10−4 |
| rs3829767 | 14 | 64519455 | Nonsynonymous: SYNE2 | A/G | 9/80/201 | 10/38/53 | 0.20 | 2.94 × 10−4 |
| rs4027402 | 14 | 64496749 | Nonsynonymous: SYNE2 | C/T | 12/85/189 | 15/34/51 | 0.22 | 3.60 × 10−4 |
| rs9944035 | 14 | 64447776 | Nonsynonymous: SYNE2 | T/C | 198/86/8 | 56/41/9 | 0.10 | 5.46 × 10−4 |
| rs4902264 | 14 | 64491695 | Nonsynonymous: SYNE2 | T/C | 7/82/202 | 9/37/55 | 0.19 | 8.03 × 10−4 |
| rs4027405 | 14 | 64498037 | Nonsynonymous: SYNE2 | G/A | 4/72/215 | 8/30/61 | 0.16 | 1.72 × 10−3 |
| rs1890908 | 14 | 64519035 | Nonsynonymous: SYNE2 | A/G | 3/64/227 | 6/29/68 | 0.14 | 3.63 × 10−3 |
*Adjustment for age, gender, smoking status, drinking status, BMI, BCLC stage, TP53 expression status, TACE status before hepatectomy, pathological grade and hepatic cirrhosis.
SNP = Single nucleotide polymorphism, OR = odds ratio, 95% CI = 95% confidence intervals. Chr = chromosome; MAF = minor allele frequency.
Genetic model analysis
Table 3 shows the genetic model considering rs2227983, rs6950826, rs8010699, rs4027405 and rs1890908. The genetic model of rs2227983 was an additive model (AG vs. AA, P = 1.26 × 10–3, OR = 3.23, 95%CI = 1.58–6.58; GG vs. AA, P = 6.44 × 10–6, OR = 5.65, 95%CI = 2.66–12.00), the G allele was strongly associated with positive p21 expression. The genetic model of rs6950826 was over dominant model (AA/GG vs. AG, P = 4.64 × 10−3, OR = 2.08, 95%CI = 1.25–3.45). The genetic model of rs8010699 was an additive model (AG vs. GG, P = 0.016, OR = 1.83, 95%CI = 1.12–2.99; AA vs. GG, P = 3.43 × 10−3, OR = 4.12, 95% CI = 1.60–10.64). The genetic model of rs4027405 was a dominant model (AG+GG vs. AA, P = 0.018, OR = 1.84, 95% CI = 1.60–10.64). The genetic model of rs1890908 was also a dominant model (AG/AA vs. GG, P = 7.8 × 10−3, OR = 2.00, 95% CI = 1.20–3.33).
Table 3. Genetic model of rs2227983, rs6950826, rs8010699, rs4027405 and rs1890908.
| SNPs | Group |
OR* (95% CI) | P value* | |
|---|---|---|---|---|
| Negative | Positive | |||
| rs2227983 Additive | ||||
| AA | 89 | 12 | Ref. | 3.73 × 10−5 |
| AG | 135 | 52 | 3.23 (1.58–6.58) | 1.26 × 10−3 |
| GG | 71 | 42 | 5.65 (2.66–12.00) | 6.44 × 10−6 |
| rs6950826 Overdominant | ||||
| AA/GG | 192 | 49 | Ref. | |
| AG | 103 | 55 | 2.08 (1.25–3.45) | 4.64 × 10−3 |
| rs8010699 Additive | 129 | 51 | ||
| GG | 204 | 55 | Ref. | 2.29 × 10−3 |
| GA | 77 | 38 | 1.83 (1.12–2.99) | 0.0155 |
| AA | 9 | 10 | 4.12 (1.60–10.64) | 3.43 × 10−3 |
| rs4027405 Dominant | ||||
| AA | 215 | 61 | Ref. | |
| AG/GG | 76 | 38 | 1.84 (1.11–3.04) | 0.0177 |
| rs1890908 Dominant | ||||
| GG | 227 | 68 | Ref. | |
| GA/AA | 67 | 35 | 2.00 (1.20–3.33) | 7.79 × 10−3 |
SNP = single nucleotide polymorphism, OR = odds ratio, 95% CI = 95% confidence intervals. Chr = chromosome; MAF = minor allele frequency, Ref. =reference.
*Adjustment for age, gender, smoking status, drinking status, BMI, BCLC stage, TP53 expression status, TACE status before hepatectomy, pathological grade and cirrhosis.
Linkage disequilibrium (LD) and Haplotype Analysis
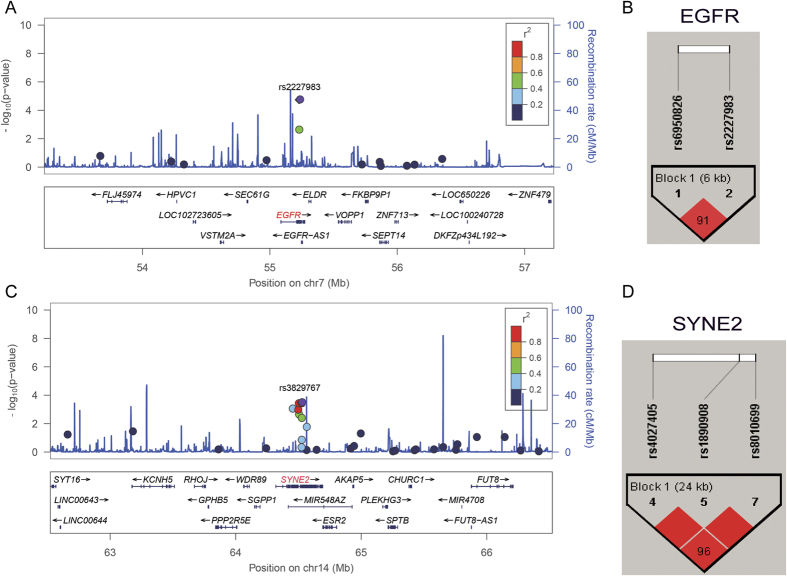
Analysis was performed for LD and haplotype about 2 Mb nearby EGFR and SYNE2 genes (Fig. 2). The haplotype block (rs2227983 and rs6950826, r2 = 0.378) in EGFR and the haplotype block in SYNE2 (rs8010699 was in strong LD with rs4027405 and rs1890908 (r2 = 0.91 and 0.7, respectively)) were identified, and the haplotype A-G of EGFR and haplotype G-A-A of SYNE2 were significantly associated with p21 expression (P = 2.43 × 10−5 and 8.63 × 10−4, respectively; Table 4).
Figure 2.
LocusZoom plot for analysis of local linkage disequilibrium (LD) and recombination patterns nearby EGFR (A) and SYNE2 (B) about 2 Mb. The left vertical axis shows association P-values on the −log10 scale, the right vertical axis shows the recombination rate and the horizontal axis shows the chromosomal position. The bottom of plot shows the near gene. The purple diamond is the most significant SNP at each plot. LD (r2) and recombination rate are estimated from the 1000 Genomes Project ASN data (March2012, build GRCh37/hg19). (C) Haploview LD graph of SNPs in EGFR was visualized using the Haploview software 4.2. (D) Haploview LD graph of SNPs in SYNE2 was visualized using the Haploview software 4.2. Haploview linkage disequilibrium plots display using the standard color scheme, the number on the cell is the LOD score of D’.
Table 4. Associations of haplotypes of EGFR (rs6950826, rs2227983) and SYNE2 (rs4027405, rs1890908 and rs8010699) SNPs with p21 expression.
| Haplotypes | Positive (2n) | Negative (2n) | OR (95% CI) | P value |
|---|---|---|---|---|
| EGFR | ||||
| G-A | 76 | 308 | Ref. | 1.16 × 10−4 |
| A-G | 88 | 164 | 0.46 (0.32–0.86) | 2.43 × 10−5 |
| Othersa | 50 | 125 | 0.62 (0.41–0.93) | 0.0219 |
| SYNE2 | ||||
| A-G-G | 153 | 486 | Ref. | 3.53 × 10−3 |
| G-A-A | 45 | 71 | 0.49 (0.33–0.75) | 8.63 × 10−4 |
| Othersb | 16 | 39 | 0.76 (0.41–1.40) | 0.383 |
OR = odds ratio, 95% CI = 95% confidence interval, Ref. =reference.
aOthers include haplotypes G-G and A-A.
bOthers include haplotypes A-G-A and G-G-G.
Pathway Analysis and Correlation analysis in mRNA
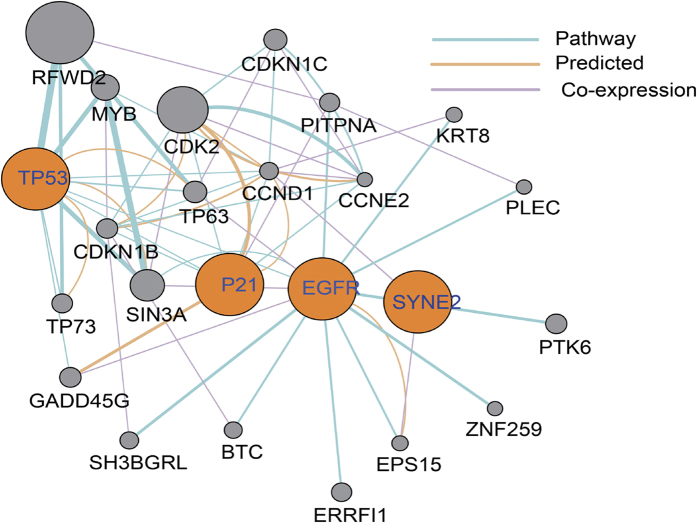
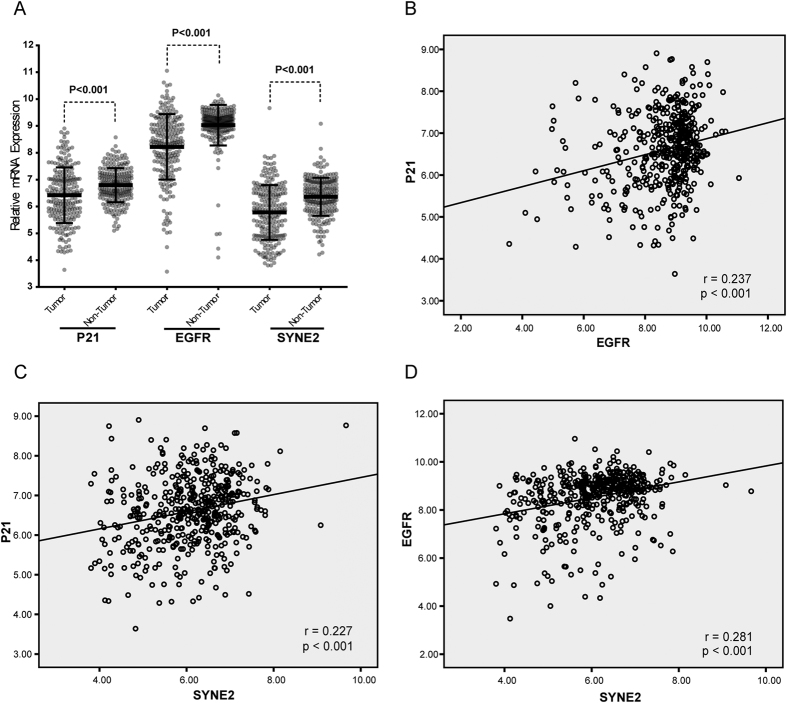
After above analysis, GeneMania Software31 was employed (see URLs) to predict the interaction between genes shown in Table 2. Signaling pathway analysis (Fig. 3) showed that p21, SYNE2 and EGFR were in a common pathway or co-expressed. Furthermore, the mRNA expression of p21, SYNE2 and EGFR was compared between HCC tissues and adjacent normal tissues. Data from Gene Expression Omnibus (GEO accession: GSE14520) showed, when compared with adjacent normal tissues, down-regulated expression of p21, SYNE2 and EGFR was observed in HCC tissues (Fig. 4A). Correlation analysis was used to analyze the correlation among p21, SYNE2 and EGFR (Fig. 4B–D). Results showed that there were positive correlations among three genes (SYNE2 vs. p21, P < 0.001, r = 0.227; SYNE2 vs. EGFR, P < 0.001, r = 0.281; EGFR vs. p21, P < 0.001, r = 0.237).
Figure 3. Co-expression/pathway/predicated analysis of p21, SYNE2, EGFR and TP53 according to human expression data in GeneMANIA.
Figure 4.
(A) Scatter diagram for mRNA expression of p21, SYNE2 and EGFR between HCC tissues and adjacent normal tissues (all P < 0.001). (B) Correlation analysis for the mRNA expression between p21 and EGFR (P < 0.001, r = 0.237). (C) Correlation analysis for the mRNA expression between p21 and SYNE2 (P < 0.001, r = 0.227). (D) Correlation analysis for the mRNA expression between EGFR and SYNE2 (P < 0.001, r = 0.281). All data were from Gene Expression Omnibus (GEO accession: GSE14520).
Survival Analysis
Patient’s characteristics and clinical predictors
The clinical and pathologic characteristics of patients are shown in Table 1. Overall, the median follow-up duration was 41 months (range: 3–123 months) and median survival time (MST) was 47 months. In this study, 118 (46.4%) patients died. The 2-year, 5-year and 10-year overall survival (OS) rate was 69.3%, 46.2% and 21%, respectively. As shown Table 1, univariate analysis indicated that patients with BCLC stage B and C (HR = 1.89 and 2.89, respectively), child-pugh class B (HR = 1.54), non-radical resection (HR = 1.37), non-antiviral therapies (HR = 1.6) and portal vein tumor thrombus (PVTT) (PV1–4, HR = 1.85, 2.76, 2.25 and 5.11, respectively) had higher risk for death when compared with patients with BCLC stage A, child-pugh class A, radical resection, antiviral therapies and non-PVTT, respectively. The biobehaviors of HCC, including tumor size (>5 cm), number of tumors (multiple), intrahepatic metastasis and vascular invasion, had adverse effects on the OS.
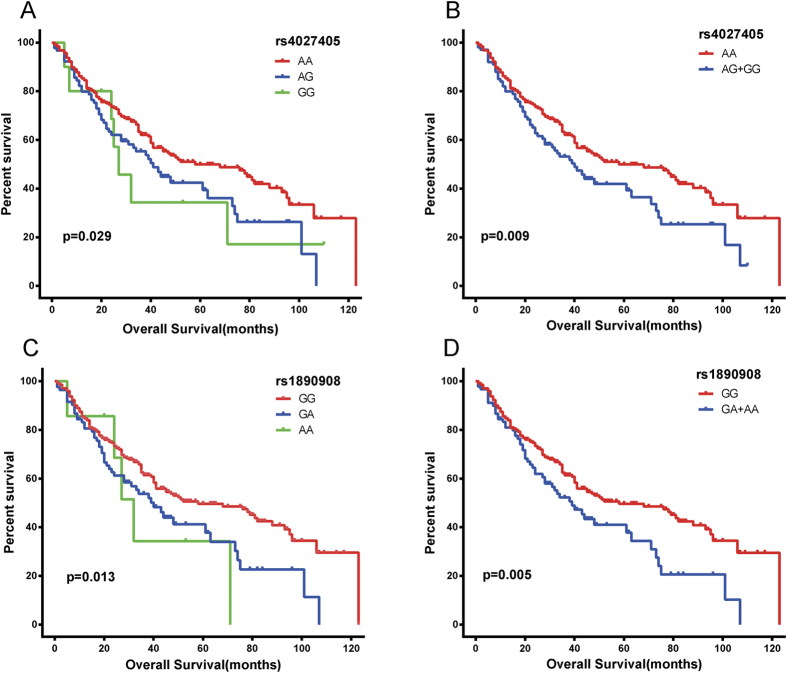
Association of SYNE2 SNPs (rs4027405 and rs1890908) with survival
Cox proportional hazard regression analysis showed rs4027405 and rs1890908 were significantly associated with OS after adjustment for age, body mass index (BMI), smoking status, drinking status, TACE status, serum AFP, radical resection, pathological grade, cirrhosis, intrahepatic metastasis, vascular invasion, antiviral therapies, child-pugh class, BCLC stage and PVTT (Table 5 and Fig. 5). rs4027405 was significantly associated with OS, and patients with AG/GG genotypes had a worse OS (P = 0.001, HR = 1.86, 95%CI = 1.28–2.69). Similar findings were also found in rs1890908 (P = 0.002, HR = 1.84, 95%CI = 1.26–2.68). In addition, rs2227983, rs6950826 and rs8010699 were not related to the OS.
Table 5. Survival on the basis of genotypes of rs4027405 and rs1890908.
| SNP | Patients | MST (months) | Crude HR (95% CI) | P value | Adjusted HR* (95% CI) | P value* |
|---|---|---|---|---|---|---|
| rs4027405 | ||||||
| AA | 276 | 58 | Ref. | 0.029 | Ref. | 0.003 |
| AG | 102 | 39 | 1.48 (1.08–2.04) | 0.016 | 1.81 (1.22–2.67) | 0.003 |
| GG | 12 | 27 | 1.72 (0.84–3.53) | 0.141 | 2.40 (1.05–5.50) | 0.039 |
| AG+GG | 114 | 38 | 1.51 (1.11–2.05) | 0.009 | 1.86 (1.28–2.69) | 0.001 |
| Missing | 15 | |||||
| rs1890908 | ||||||
| GG | 295 | 52 | Ref. | 0.013 | Ref. | 0.006 |
| GA | 93 | 39 | 2.10 (0.93–4.78) | 0.076 | 2.04 (0.84–4.92) | 0.115 |
| AA | 9 | 27 | 1.52 (1.10–2.11) | 0.011 | 1.81 (1.22–2.70) | 0.003 |
| GA+AA | 102 | 38 | 1.57 (1.15–2.14) | 0.005 | 1.84 (1.26–2.68) | 0.002 |
| Missing | 8 | |||||
HR = hazard ratio, 95% CI = 95% confidence interval, MST = median survival time, Ref. =reference.
*Adjustment for age, gender, BMI, race, smoking status, drinking status, child-pugh class, cirrhosis, BCLC stage, pathological grade, TACE status post hepatectomy, antiviral therapy after hepatectomy, radical resection, intrahepatic metastasis, vascular invasion and PVTT.
Figure 5. Kaplan-Meir survival curves for SNPs of EGFR and SYNE2.
(A) rs4027405 (AA vs. AG and GG). (B) rs4027405 (AA vs. AG/GG), AG and GG were included into one group. (C) rs1890908 (GG vs. AG and AA). (D) rs1890908 (GG vs. AG/AA), AG and AA were included into one group.
Stratified analysis on the association of SYNE2 SNPs (rs4027405 and rs1890908) with survival
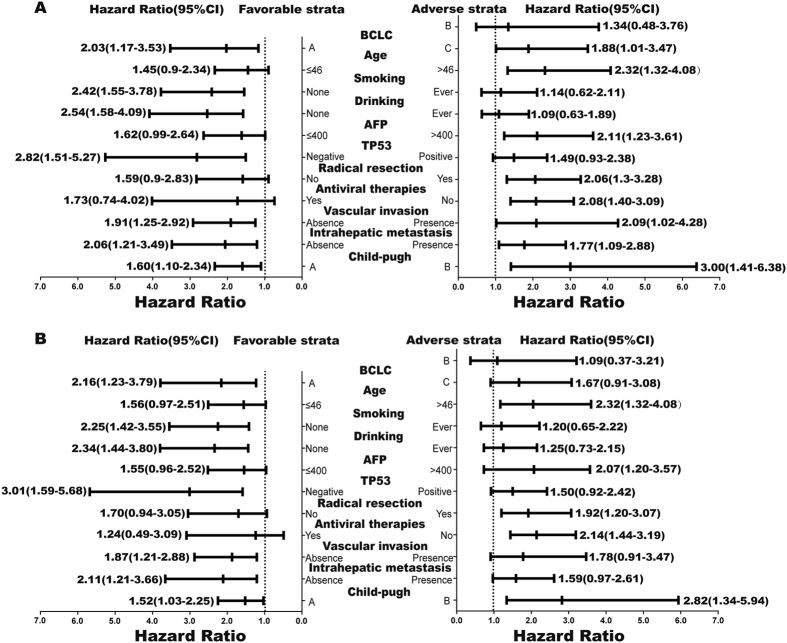
A stratified analysis was performed to evaluate the associations of rs4027405 and rs1890908 genotypes with HBV-related HCC survival by age, smoking status, drinking status, serum AFP level, radical resection, TP53 expression status, intrahepatic metastasis, vascular invasion, antiviral therapies, child-pugh class and BCLC stage (Fig. 6). Patients with rs4027405 AG/GG had a poorer OS when they were older than 45 years, or had AFP > 400 ng/ml, non-smoking status, non-drinking status, negative TP53 expression, child-pugh class B or BCLC stage C. Specifically, a significantly increased risk for death conferred by SNP rs4027405 genotypes (AG/GG) was observed in patients negative for TP53 expression (P = 0.001, HR = 2.82, 95%CI = 1.51–5.27) or child-pugh class B (P = 0.004, HR = 3.00, 95%CI = 1.41–6.38). Similar results were also observed in rs1890908.
Figure 6.
Stratification analysis on the association of rs4027405 (A) and rs1890908 (B) with clinical outcome of HBV-related HCC patients. HR was indicated for overall survival, stratified by the favorable and adverse outcomes.
Combined effect of SYNE2 SNPs (rs4027405 and rs1890908) and serum AFP on OS
The combined effect of SNPs and serum AFP on the OS of HBV-related HCC patients was further analyzed (Table 6). Patients were classified into 4 groups according to the rs4027405 status and serum AFP level: AFP ≤ 400 ng/mL with AA genotype, AFP > 400 ng/mL with AG/GG genotypes, AFP ≤ 400 ng/mL with AA genotype and AFP > 400 ng/mL with AG/GG genotypes. Multivariate Cox regression analysis indicated that, as compared to patients with AA genotype and low serum AFP (AFP ≤ 400 ng/ml), patients with AG/GG genotype or a high serum AFP (AFP > 400 ng/mL) had a significantly higher risk for death (P = 0.003, HR = 2.25, 95%CI = 1.31–3.87). Similar results were observed when the rs1890908 and serum AFP level were considered simultaneously (Table 6).
Table 6. Combined effects of SNPs (rs4027405, rs1890908) and serum AFP level.
| SNP | MST (months) | Crude HR (95% CI) | P | Adjusted HR* (95% CI) | P* |
|---|---|---|---|---|---|
| rs4027405 | |||||
| AFP ≤ 400+AA | 68 | Ref. | 0.005 | Ref. | 0.012 |
| AFP ≤ 400+AG/GG | 52 | 1.27 (0.87–1.87) | 0.220 | 1.09 (0.69–1.72) | 0.703 |
| AFP > 400+AA | 39 | 1.63 (1.05–2.52) | 0.029 | 1.63 (1.00–2.65) | 0.048 |
| AFP > 400+AG/GG | 28 | 2.25 (1.40–3.61) | 0.001 | 2.25 (1.31–3.87) | 0.003 |
| rs1890908 | |||||
| AFP ≤ 400+GG | 68 | Ref. | 0.006 | Ref. | 0.018 |
| AFP ≤ 400+AG/AA | 52 | 1.24 (0.86–1.79) | 0.254 | 1.09 (0.71–1.69) | 0.683 |
| AFP > 400+GG | 39 | 1.62 (1.04–2.52) | 0.031 | 1.63 (1.00–2.66) | 0.050 |
| AFP > 400+AG/AA | 28 | 2.23 (1.40–3.78) | 0.001 | 2.21 (1.26–3.88) | 0.006 |
HR = hazard ratio, 95% CI = 95% confidence interval, MST = median survival time, Ref. = reference.
*Adjustment for age, gender, BMI, race, smoking status, drinking status, child-pugh class, cirrhosis, BCLC stage, pathological grade, TACE status post hepatectomy, antiviral therapy after hepatectomy, radical resection, intrahepatic metastasis, vascular invasion and PVTT.
Discussion
In this study, GWAS was performed to investigate the p21 regulation in Chinese HBV-related HCC patients in Guangxi. Results showed several SNPs in EGFR gene (rs6950826, rs2227983) and SYNE2 gene (rs4027405, rs1890908 and rs8010699) were associated with p21 expression, and the mRNA expression of p21, EGFR and SYNE2 had positive correlations between each other. Furthermore, the p21 expression was significantly associated with the cirrhosis status and TP53 expression, which was consistent with previous results11,32,33, but not related to TP53 Ser249 mutation (AFB1 exposure).
According to the NCBI database (NG_007726), EGFR gene is mapped to chromosome 7p12 spanning 188.3 kb and contains 30 exons. EGFR is also known as ErbB1 or HER-1 and is a transmembrane receptor belonging to the family of receptor tyrosine kinases (RTK) including ErbB2/HER2, ErbB3/HER3 and ErbB4/HER434. Structurally, these ligands can bind to EGFR system which contains EGF-like domains, including epidermal growth factor (EGF), transforming growth factor α (TGF-α), amphiregulin (AR), epiregulin (EREG), betacellulin (BTC), heparin-binding EGF (HB-EGF) and epigen (EPGN)35. The EGFR system is not only essential for the cell proliferation, survival and migration, but closely related to the occurrence, growth and outcome of HCC36,37,38. In addition, in transgenic animals, results showed the over-expression of different EGFR ligands lead to different incidence of HCC, such as TGF-α or EGF over-expression leads to a higher incidence of HCC39,40,41. There is evidence showing that EGFR expression is related to p21 expression42, which was confirmed in the present study that there was correlation between EGFR and p21 mRNA expression (Fig. 4). In addition, our results showed rs2227983 (G > A, Arg 521 Lys) and rs6950826 (G > A) of EGFR gene were associated with p21 expression. rs2227983 (G > A, R521K) is located in the exon 13 of EGFR gene. Previous study showed, in metastatic colorectal cancer patients treated with EGFR inhibitor, patients harboring at least one minor allele of rs2227983 G > A were more likely to show a higher tumor response as compared to those with homozygous wild-type allele43. In our study, the genetic model of rs2227983 was additive. Patients with allele G had a higher p21 expression. This may be explained as that rs2227983 of EGFR gene may result in an Arg (R) to Lys(K) substitution, leading to the decrease in EGFR activity44. Besides, several studies have shown that rs2227983 is also associated with the interstitial lung disease45 and gastric cancer46. rs6950826 is located in the intron 7 of EGFR gene. To date, no study has been conducted to investigate the association of rs6950826 with the susceptibility to disease or its influence on the clinical outcome of a specific disease. In our study, rs2227983 and rs6950826 were found to be not associated with OS of HBV-related HCC patients. However, how these two loci regulate the expression of EGFR and p21 is still unclear. According to the available findings, we speculate that both loci are likely to affect the function or structure of EGFR gene to regulate the p21 expression, but it is required to be further studied.
In our study, results showed that SNPs (rs4027405, rs1890908 and rs8010699) of SYNE2 gene were associated with p21 expression, and these three loci had strong linkage disequilibrium. Haplotype analysis for the association of SNPs (rs4027405, rs1890908, rs8010699) with p21 expression indicated that patients with haplotype A-G-G had a lower p21 expression in HBV-related HCC. SYNE2, also known as EDMD5, NUANCE and Nesprin-2, is mapped to chromosome 14p23.2 and consists of 116 exons. SYNE2 belongs to the family of giant spectrin-repeat (nesprins), which plays an important role in linking the nucleus to the cytoskeleton and is a key component of the linker of the nucleoskeleton and cytoskeleton (LINC) complex47,48. Study has reported that SYNE2 depletion may reduce the endothelial cell migration and angiogenic loop formation by regulating the architecture between nucleus and cytoplasm49. Recently, nesprins were identified as candidate cancer genes by high-throughput genome sequencing50,51,52. Strikingly, SYNE2 alterations have also been revealed in many cancers, including breast cancer, head and neck cancer and colorectal cancer50,53,54. Meanwhile, our study showed that the mRNA expression of SYNE2 was significantly down-regulated in HCC (Fig. 4A). These findings imply the significant roles of SYNE2 in cancer biology. Currently, how nesprins regulate and control the cancer development and progression is still poorly understood. Derek et al.55 found that siRNA induced SYNE2 depletion augmented extracellular signal-regulated MAPK1 and 2 (ERK1/2) activity, leading to the increased cell proliferation. p21 has been implicated in cell cycle progression and proliferation via activating ERK1/2 pathway in HCC56. Interestingly, the mRNA expression of SYNE2 and p21 is positively correlated in HBV-related HCC (Fig. 4C) and both are involved in the cell cycle progression and proliferation through activating ERK1/2 pathway. On the basis of above findings and our result that SYNE2 variants were associated with p21 expression, we speculate that there is a close relationship between SYNE2 and p21, even SYNE2 is able to regulate p21 expression, but this is needed to be confirmed in future studies.
Kao et al.28 reported that p21 expression was an independent prognostic factor of favorable survival. Survival analysis in our study showed, although p21 expression was not associated with OS of HBV-related HCC patients, patients positive for p21 expression had higher HR than those negative for p21 expression (HR = 1.13, 95%CI = 0.83–1.54, P = 0.443, Table 3). This was not conflicting with above findings. Remarkably, SNPs (rs4027405 and rs1890908) of SYNE2 gene were not only significantly associated with OS, but with p21 expression in HBV-related HCC. The G allele is a risk allele in rs4027405, and G allele carriers have a higher p21 expression and a worse prognosis. Similar results were observed in rs1890908, largely thanking to the strong linkage disequilibrium between it and rs4027405. Sebastian et al.57 revealed that SYNE2 expression was associated with shorter disease-free survival of gastrointestinal stromal cancer patients. Hence, rs4027405 and rs1890908 are likely to influence the prognosis through affecting the expression or function of SYNE2 and p21. Moreover, univariate and multivariate Cox regression analyses indicated that rs4027405 and rs1890908 were independent predictors of poor OS of HBV-related HCC patients, and it combined with serum AFP can suggest that patients with AFP > 400 ng/ml and higher p21 expression allele have significantly higher risk for death than those with AFP ≤ 400 ng/ml and lower p21 expression allele.
However, our study has several limitations. This was a single center study, and a small fraction of clinical data was missing. The SNPs were not functionally characterized to reveal their effects on the p21 expression and clinical outcome of HBV-related HCC. Therefore, additional studies with larger sample size are needed to further validate the association of these SNPs with p21 expression in HBV-related HCC and with the clinical outcome of HBV-related HCC patients, and in vitro studies are required to clarify the underlying mechanisms.
In conclusion, our results demonstrate that SNPs of EGFR (rs6950826 and rs2227983) and SYNE2 (rs4027405, rs1890908 and rs8010699) are associated with p21 expression in HBV-related HCC. rs4027405 and rs1890908 are potential biomarkers for clinical outcome of HBV-related HCC Chinese patients after hepatectomy in Guangxi. In GWAS, TP53 expression as a covariate, and hence, our findings indicate that variants of EGFR and SYNE2 genes may play important roles in the regulation of p21 expression and affect the outcome of HBV-related HCC via TP53-indenpdent manner.
Methods
Ethic statement
All experimental protocols were approved by the Ethical Review Committee of the First Affiliated Hospital of Guangxi Medical University (Approval Number: 2015 [KY-E-032]). The methods were carried out in accordance with the approved guidelines. Written informed consent was obtained from all subjects before study.
Study population
A total of 426 patients with pathologically proven HBV-related HCC were enrolled from the First Affiliated Hospital of Guangxi Medical University between 2005 and 2014. HCC tissues were collected during surgery and immediately stored at −80 °C until the DNA extraction. Total DNA was extracted using the TIANamp Genomic DNA Kit (TIANGEN BIOTECH [BEIJING] CO, LTD). DNA concentration and purity were measured with the NanoDrop2000 system (Thermo Fisher Scientific, Waktham, MA, USA). Immunohistochemical staining of p21 and TP53 in HCC tissues was done in the Department of Pathology at the First Affiliated Hospital of Guangxi Medical University. Finally, 114 patients were found to be positive for p21 expression and 312 negative for p21 expression. Patients with p21 positive expression were included in positive group and those negative for p21 expression of patients in negative group.
According to previous findings30, the TP53 249Ser mutation was evidence of AFB1 exposure and detected by sanger sequencing after amplification with polymerase chain reaction (PCR) (Sequencing primers in Supplementary Table 1).
The clinicopathological characteristics of included patients were obtained from medical records and pathological reports, including age, gender, smoking status, drinking status, pathological grade, biobehaviors of the cancer, serum AFP level, hepatic cirrhosis, radical resection and use of transcatheter hepatic arterial chemoembolization (TACE). Tumor status was classified according to the Barcelona Clinic Liver Cancer (BCLC) staging system58. Child-pugh class was defined as previously published59. Portal vein tumor thrombus (PVTT) was determined according to previous classifications60: vp1 = PVTT in distal to second order portal branches; vp2 = PVTT in second order portal branches; vp3 = PVTT in first order branches; vp4 = PVTT in main trunk. Smoking status, drinking status and radical resection were defined as previously reported61.
Immunohistochemistry
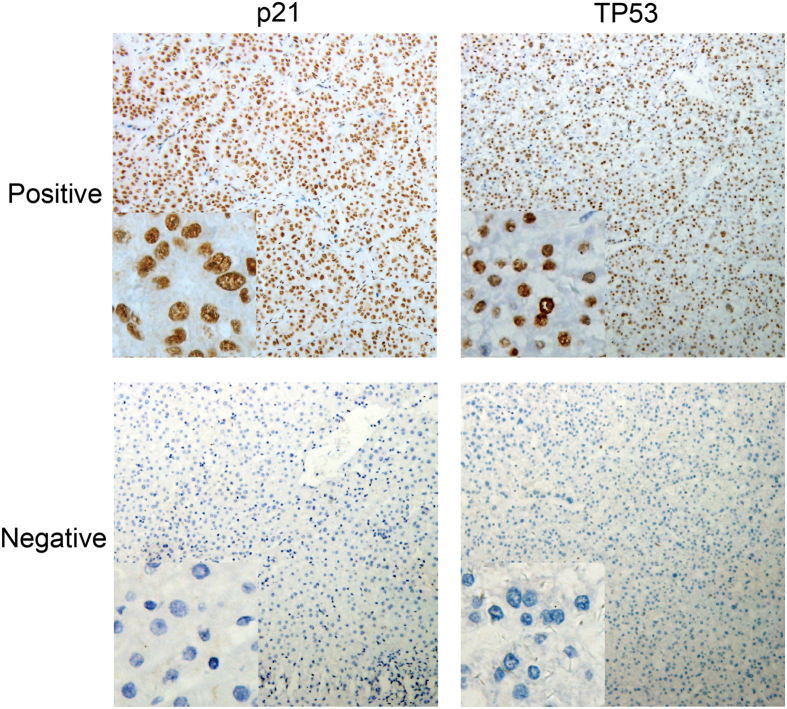
Paraffin embedded HCC tissues were used in immunohistochemistry for p21 and TP53 with general two-step method. Briefly, the antigen retrieval was done with EDTA Tris at a high temperature for 2.5 min. Then, sections were treated with 3% hydrogen peroxide for 10 min to inactivate endogenous peroxidase. After washing thrice with PBS, the mouse against human p21 (1:150) (ZSGB-BIO ORIGENE, Beijing, China) and mouse against human TP53 (1:150) (ZSGB-BIO ORIGENE, Beijing, China) antibodies were independently added, followed by incubation overnight at 4 °C. The sections were incubated with secondary antibody (Dako Cytomation, Glostrup, Denmark) at 37 °C for 30 min. Finally, these sections were visualized with DAB (Dako Cytomation) and counterstained with hematoxylin. Positive and negative controls were included in each immunohistochemical reaction. All the sections were reviewed and scored by two pathologists independently who were blind to the clinical characteristics of patients. At least 10 fields were randomly selected at a high-power (x400 magnifications) at the regions distant from necrotic areas, and the proportion of positive cells was calculated with the following formula: (number of positive cells/total number of the cells ×100%). Positive cells had brown granules in the nucleus. According to previous criteria, positive p21 expression was defined as the presence of ≥5% positive cancer cells28,62, and positive staining of TP53 as ≥10% positive cancer cells28,63,64 (Fig. 7).
Figure 7. Expression of p21 and TP53 in four hepatocellular carcinoma tissues (Immunohistochemistry).
(background: 100×; left down corner: 400×) Fig. 1.
Genotyping
All samples were genotyped using the Illumina HumanExome BeadChip-12-1_A, which includes 242,901 markers on the protein-altering variants. These markers include non-synonymous single nucleotide polymorphisms (SNPs), SNPs in splice sites, stop variants, SNPs in promoter regions, SNPs in extended MHC region, GWAS tag markers, HLA tags, etc. Details about SNPs can be found at the exome array design webpage. Samples were processed according to the manufacturer’s instructions. Genotype calling was carried out using the Genotyping Module v1.0 in GenomeStudio version 2011.1, and average call rate was 99.84%. A total of 50 samples (>10%) were randomly selected and sequenced for the candidate loci using the ABI Prism 3100 (Applied Biosystems, Shanghai Sangon Biological Engineering Technology & Services Co, Ltd, Shanghai, China). The candidate loci and primers used for sequencing are shown in Supplementary Table 1. The results of sequencing are 100% concordant with the results of genotyping by BeadChip-12-1A.
Follow up
All patients were followed up via telephone or hospital visit until death or the last time follow-up was done in September 2014. The median follow-up period in 426 patients was 41 months (ranging: 3–123 months), and the median survival time (MST) was 47 months.
Statistical analysis
Quality Control
A quality control (QC) procedure was used before association analysis as follows: 1. The sample was removed if they (i) had an overall genotyping rate of <95%; (ii) had ambiguous gender; (iii) had genome-wide identity-by-descent (IBD) >0.1875; (iv) were outliers in principal components analysis (PCA) for ancestry and population stratification. 2. The SNP was excluded if they had (i) a call rate of <95%; (ii) a P value in Hardy-Weinberg equilibrium (HWE) <1 × 10−6; (iii) a Minor allele frequencies (MAF) <0.05. 3. PCA: Guangxi in Southern China is a multi-ethnic region. An analysis of population stratification by PCA65 was performed to eliminate the multi-ethnic interference using the EIGENSOFT package. Genomic inflation factors (GIF)66 was used to investigate the residual population stratification, which was calculated with MATLAB 7.0 (see URLs). The procedure was performed with the Plink version 1.07, R 3.0.1 and EIGENSOFT package67.
Association Analysis
The association analysis was evaluated using Single Variant Test68 (chose Logistic Score Test) with EPACTS package version 3.2.6 (see URLs), after adjustment for age, gender, BCLC stage, TP53 expression status, TACE status before hepatectomy, pathological grade and hepatic cirrhosis. The Chi-squared test and logistic regression model were independently used to analyze the association of genetic model of SYNE2 and EGFR with p21 expression. Local linkage disequilibrium (LD) and recombination patterns nearby SYNE2 and EGFR were analyzed using the LocusZoom69 (see URLs).
Survival Analysis
Overall survival (OS) was defined as the time from hepatetomy to death or the last follow-up. MST was calculated for all variables, univariate and multivariate survival analysis were done using the Cox proportional hazard regression model. Hazard ratios (HR) and their 95% confidence interval (CI) were calculated from the Cox proportional hazards regression model with adjustment for age, gender, race, body mass index (BMI), smoking status, drinking status, TACE status, serum AFP level, BCLC stage, child-pugh class, radical resection, pathological grade, hepatic cirrhosis and PVTT. A value of P < 0.05 was considered statistically significant. Statistical analysis was performed with SPSS version 18.0 (SPSS, Inc., Chicago, IL, US).
Additional Information
How to cite this article: Han, C. et al. EGFR and SYNE2 are associated with p21 expression and SYNE2 variants predict post-operative clinical outcomes in HBV-related hepatocellular carcinoma. Sci. Rep. 6, 31237; doi: 10.1038/srep31237 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by the National Nature Science Foundation of China (No: 81560535, 81072321, 30760243, 30460143 and 30560133), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Nature Sciences Foundation (No: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific-Research-Grant Z201018). The authors thank Prof. Yanling Hu for her contribution to data analysis guidance, and Prof. Minhao Peng, Kaiyin Xiao, Xigang Chen, Zhixiong Su, Ming Su, Bin Chen and Xinping Ye for their contribution to sample collection.
Footnotes
Author Contributions C.Y.H., X.W.L. and T.P. designed this manuscript. C.Y.H., X.W.L., W.Q., L.Y., X.G.L., Z.T.L., S.C.L., Z.W.C., H.S., G.Z.Z., Z.L.L., G.C., Z.M.L., X.Q., Y.G., Z.N.M., L.Q.L. and T.P. conducted the study, collected the data, and analyzed the data. C.Y.H. wrote this manuscript.
References
- Torre L. A. et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians 65, 87–108, doi: 10.3322/caac.21262 (2015). [DOI] [PubMed] [Google Scholar]
- El-Serag H. B. & Rudolph K. L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576, doi: 10.1053/j.gastro.2007.04.061 (2007). [DOI] [PubMed] [Google Scholar]
- Mittal S. & El-Serag H. B. Epidemiology of hepatocellular carcinoma: consider the population. Journal of clinical gastroenterology 47 Suppl, S2–6, doi: 10.1097/MCG.0b013e3182872f29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag H. B. Hepatocellular carcinoma. The New England journal of medicine 365, 1118–1127, doi: 10.1056/NEJMra1001683 (2011). [DOI] [PubMed] [Google Scholar]
- Liu Z. M. et al. Hepatitis B virus infection contributes to oxidative stress in a population exposed to aflatoxin B1 and high-risk for hepatocellular carcinoma. Cancer letters 263, 212–222, doi: 10.1016/j.canlet.2008.01.006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner A., Llovet J. M. & Bruix J. Hepatocellular carcinoma. Lancet 379, 1245–1255, doi: 10.1016/S0140-6736(11)61347-0 (2012). [DOI] [PubMed] [Google Scholar]
- Farazi P. A. & DePinho R. A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nature reviews. Cancer 6, 674–687, doi: 10.1038/nrc1934 (2006). [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993). [DOI] [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K. & Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993). [DOI] [PubMed] [Google Scholar]
- Gartel A. L. & Tyner A. L. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Molecular cancer therapeutics 1, 639–649 (2002). [PubMed] [Google Scholar]
- Winters Z. E. et al. Subcellular localisation of cyclin B, Cdc2 and p21(WAF1/CIP1) in breast cancer. association with prognosis. European journal of cancer 37, 2405–2412 (2001). [DOI] [PubMed] [Google Scholar]
- Westfall M. D., Mays D. J., Sniezek J. C. & Pietenpol J. A. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Molecular and cellular biology 23, 2264–2276 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller H. A. et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proceedings of the National Academy of Sciences of the United States of America 97, 3260–3265 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S., Ignatova A., Park S. T. & Sun X. H. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Molecular and cellular biology 17, 5888–5896 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H., Jung J. K., Lim J. S., Tiwari I. & Jang K. L. Hepatitis B virus X protein overcomes all-trans retinoic acid-induced cellular senescence by downregulating levels of p16 and p21 via DNA methylation. The Journal of general virology 92, 1309–1317, doi: 10.1099/vir.0.029512-0 (2011). [DOI] [PubMed] [Google Scholar]
- Ahn J. Y., Chung E. Y., Kwun H. J. & Jang K. L. Transcriptional repression of p21(waf1) promoter by hepatitis B virus X protein via a p53-independent pathway. Gene 275, 163–168 (2001). [DOI] [PubMed] [Google Scholar]
- Park U. S., Park S. K., Lee Y. I., Park J. G. & Lee Y. I. Hepatitis B virus-X protein upregulates the expression of p21waf1/cip1 and prolongs G1– >S transition via a p53-independent pathway in human hepatoma cells. Oncogene 19, 3384–3394, doi: 10.1038/sj.onc.1203674 (2000). [DOI] [PubMed] [Google Scholar]
- Qiao L. et al. Hepatitis B virus X protein increases expression of p21(Cip-1/WAF1/MDA6) and p27(Kip-1) in primary mouse hepatocytes, leading to reduced cell cycle progression. Hepatology 34, 906–917, doi: 10.1053/jhep.2001.28886 (2001). [DOI] [PubMed] [Google Scholar]
- Martin-Caballero J., Flores J. M., Garcia-Palencia P. & Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer research 61, 6234–6238 (2001). [PubMed] [Google Scholar]
- Hui A. M., Kanai Y., Sakamoto M., Tsuda H. & Hirohashi S. Reduced p21(WAF1/CIP1) expression and p53 mutation in hepatocellular carcinomas. Hepatology 25, 575–579, doi: 10.1002/hep.510250314 (1997). [DOI] [PubMed] [Google Scholar]
- Kobayashi S. et al. P21WAF1/CIP1 messenger RNA expression in hepatitis B, C virus-infected human hepatocellular carcinoma tissues. Cancer 91, 2096–2103 (2001). [DOI] [PubMed] [Google Scholar]
- Shi Y. Z. et al. Reduced p21(WAF1/CIP1) protein expression is predominantly related to altered p53 in hepatocellular carcinomas. British journal of cancer 83, 50–55, doi: 10.1054/bjoc.2000.1310 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cueva E. et al. Tumorigenic activity of p21Waf1/Cip1 in thymic lymphoma. Oncogene 25, 4128–4132, doi: 10.1038/sj.onc.1209432 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Y. A., Elson A. & Leder P. Loss of p21 increases sensitivity to ionizing radiation and delays the onset of lymphoma in atm-deficient mice. Proceedings of the National Academy of Sciences of the United States of America 94, 14590–14595 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Toki T., Konishi I., Nikaido T. & Fujii S. Expression of p21WAF1/CIP1 in adenocarcinoma of the uterine cervix: a possible immunohistochemical marker of a favorable prognosis. Cancer 82, 2409–2417 (1998). [PubMed] [Google Scholar]
- Baretton G. B., Klenk U., Diebold J., Schmeller N. & Lohrs U. Proliferation- and apoptosis-associated factors in advanced prostatic carcinomas before and after androgen deprivation therapy: prognostic significance of p21/WAF1/CIP1 expression. British journal of cancer 80, 546–555, doi: 10.1038/sj.bjc.6690390 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbia M. et al. Expression of p21WAF1 predicts outcome of esophageal cancer patients treated by surgery alone or by combined therapy modalities. Clinical cancer research: an official journal of the American Association for Cancer Research 4, 2615–2623 (1998). [PubMed] [Google Scholar]
- Kao J. T. et al. P21/WAF1 is an independent survival prognostic factor for patients with hepatocellular carcinoma after resection. Liver international: official journal of the International Association for the Study of the Liver 27, 772–781, doi: 10.1111/j.1478-3231.2007.01499.x (2007). [DOI] [PubMed] [Google Scholar]
- Zhang M. F., Zhang Z. Y., Fu J., Yang Y. F. & Yun J. P. Correlation between expression of p53, p21/WAF1, and MDM2 proteins and their prognostic significance in primary hepatocellular carcinoma. Journal of translational medicine 7, 110, doi: 10.1186/1479-5876-7-110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Qi X., Jia J., Yang M. & Han G. Hepatocellular carcinoma. Lancet 380, 469; author reply 470–461, doi: 10.1016/S0140-6736(12)61283-5 (2012). [DOI] [PubMed] [Google Scholar]
- Warde-Farley D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research 38, W214–220, doi: 10.1093/nar/gkq537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagayama H. et al. High expression of p21WAF1/CIP1 is correlated with human hepatocellular carcinoma in patients with hepatitis C virus-associated chronic liver diseases. Human pathology 33, 429–434 (2002). [DOI] [PubMed] [Google Scholar]
- Shiraki K. & Wagayama H. Cytoplasmic p21(WAF1/CIP1) expression in human hepatocellular carcinomas. Liver international: official journal of the International Association for the Study of the Liver 26, 1018–1019, doi: 10.1111/j.1478-3231.2006.01320.x (2006). [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110, 669–672 (2002). [DOI] [PubMed] [Google Scholar]
- Schneider M. R. & Wolf E. The epidermal growth factor receptor ligands at a glance. Journal of cellular physiology 218, 460–466, doi: 10.1002/jcp.21635 (2009). [DOI] [PubMed] [Google Scholar]
- Zandi R., Larsen A. B., Andersen P., Stockhausen M. T. & Poulsen H. S. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cellular signalling 19, 2013–2023, doi: 10.1016/j.cellsig.2007.06.023 (2007). [DOI] [PubMed] [Google Scholar]
- Bassullu N. et al. The Predictive and Prognostic Significance of c-erb-B2, EGFR, PTEN, mTOR, PI3K, p27, and ERCC1 Expression in Hepatocellular Carcinoma. Hepatitis monthly 12, e7492, doi: 10.5812/hepatmon.7492 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. C. et al. Association between Osteopontin and EGFR Expression with Clinicopathological Parameters in Hepatocellular Carcinoma. The Chinese journal of physiology 55, 412–420, doi: 10.4077/CJP.2012.BAA082 (2012). [DOI] [PubMed] [Google Scholar]
- Borlak J., Meier T., Halter R., Spanel R. & Spanel-Borowski K. Epidermal growth factor-induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene 24, 1809–1819, doi: 10.1038/sj.onc.1208196 (2005). [DOI] [PubMed] [Google Scholar]
- Webber E. M., Wu J. C., Wang L., Merlino G. & Fausto N. Overexpression of transforming growth factor-alpha causes liver enlargement and increased hepatocyte proliferation in transgenic mice. The American journal of pathology 145, 398–408 (1994). [PMC free article] [PubMed] [Google Scholar]
- Russell W. E., Kaufmann W. K., Sitaric S., Luetteke N. C. & Lee D. C. Liver regeneration and hepatocarcinogenesis in transforming growth factor-alpha-targeted mice. Molecular carcinogenesis 15, 183–189, doi: (1996). [DOI] [PubMed] [Google Scholar]
- Wang Q., Lin Z. Y. & Feng X. L. Alterations in metastatic properties of hepatocellular carcinoma cell following H-ras oncogene transfection. World journal of gastroenterology 7, 335–339 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerger A. et al. Pharmacogenetic angiogenesis profiling for first-line Bevacizumab plus oxaliplatin-based chemotherapy in patients with metastatic colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 17, 5783–5792, doi: 10.1158/1078-0432.CCR-11-1115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriai T., Kobrin M. S., Hope C., Speck L. & Korc M. A variant epidermal growth factor receptor exhibits altered type alpha transforming growth factor binding and transmembrane signaling. Proceedings of the National Academy of Sciences of the United States of America 91, 10217–10221 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Epidermal growth factor receptor (EGFR) pathway genes and interstitial lung disease: an association study. Scientific reports 4, 4893, doi: 10.1038/srep04893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P. W. et al. Genetic variants of EGF and VEGF predict prognosis of patients with advanced esophageal squamous cell carcinoma. PloS one 9, e100326, doi: 10.1371/journal.pone.0100326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi M. L. et al. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. The Journal of biological chemistry 286, 26743–26753, doi: 10.1074/jbc.M111.233700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz M. J. et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Human molecular genetics 18, 607–620, doi: 10.1093/hmg/ddn386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J. et al. Nesprin-1 and nesprin-2 regulate endothelial cell shape and migration. Cytoskeleton 71, 423–434, doi: 10.1002/cm.21182 (2014). [DOI] [PubMed] [Google Scholar]
- Sjoblom T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274, doi: 10.1126/science.1133427 (2006). [DOI] [PubMed] [Google Scholar]
- Chittenden T. W. et al. Functional classification analysis of somatically mutated genes in human breast and colorectal cancers. Genomics 91, 508–511, doi: 10.1016/j.ygeno.2008.03.002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117, doi: 10.1038/nature09515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160, doi: 10.1126/science.1208130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399, doi: 10.1038/nature10933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren D. T. et al. Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. The Journal of biological chemistry 285, 1311–1320, doi: 10.1074/jbc.M109.032557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Hippocalcin-like 1 suppresses hepatocellular carcinoma progression by promoting p21 stabilization via activating ERK1/2-MAPK pathway. Hepatology, doi: 10.1002/hep.28395 (2015). [DOI] [PubMed] [Google Scholar]
- Schoppmann S. F. et al. Novel clinically relevant genes in gastrointestinal stromal tumors identified by exome sequencing. Clinical cancer research: an official journal of the American Association for Cancer Research 19, 5329–5339, doi: 10.1158/1078-0432.CCR-12-3863 (2013). [DOI] [PubMed] [Google Scholar]
- Bruix J. & Sherman M. & Practice Guidelines Committee, A. A. f. t. S. o. L. D. Management of hepatocellular carcinoma. Hepatology 42, 1208–1236, doi: 10.1002/hep.20933 (2005). [DOI] [PubMed] [Google Scholar]
- Pugh R. N., Murray-Lyon I. M., Dawson J. L., Pietroni M. C. & Williams R. Transection of the oesophagus for bleeding oesophageal varices. The British journal of surgery 60, 646–649 (1973). [DOI] [PubMed] [Google Scholar]
- Kondo K. et al. Surgical strategy for hepatocellular carcinoma patients with portal vein tumor thrombus based on prognostic factors. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract 13, 1078–1083, doi: 10.1007/s11605-009-0854-2 (2009). [DOI] [PubMed] [Google Scholar]
- Yu L. et al. The XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of HBV-related hepatocellular carcinoma after hepatectomy: A cohort study for 10 years follow-up. Hepatology research: the official journal of the Japan Society of Hepatology, doi: 10.1111/hepr.12611 (2015). [DOI] [PubMed] [Google Scholar]
- Ogawa M., Maeda K., Onoda N., Chung Y. S. & Sowa M. Loss of p21WAF1/CIP1 expression correlates with disease progression in gastric carcinoma. British journal of cancer 75, 1617–1620 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote R. J. et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer research 58, 1090–1094 (1998). [PubMed] [Google Scholar]
- Esrig D. et al. Accumulation of nuclear p53 and tumor progression in bladder cancer. The New England journal of medicine 331, 1259–1264, doi: 10.1056/nejm199411103311903 (1994). [DOI] [PubMed] [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics 38, 904–909, doi: 10.1038/ng1847 (2006). [DOI] [PubMed] [Google Scholar]
- Devlin B. & Roeder K. Genomic control for association studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- Patterson N., Price A. L. & Reich D. Population structure and eigenanalysis. PLoS genetics 2, e190, doi: 10.1371/journal.pgen.0020190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. Y. & Tang Z. Z. A general framework for detecting disease associations with rare variants in sequencing studies. American journal of human genetics 89, 354–367, doi: 10.1016/j.ajhg.2011.07.015. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337, doi: 10.1093/bioinformatics/btq419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.