Abstract
Background
Echinocandins are nonribosomal lipopeptides produced by ascommycete fungi. Due to their strong inhibitory effect on fungal cell wall biosynthesis and lack of human toxicity, they have been developed to an important class of antifungal drugs. Since 2012, the biosynthetic gene clusters of most of the main echinocandin variants have been characterized. Especially the comparison of the clusters allows a deeper insight for the biosynthesis of these complex structures.
Results
In the genome of the echinocandin B producer Aspergillus nidulans NRRL 8112 we have identified a gene cluster (Ani) that encodes echinocandin biosynthesis. Sequence analyses showed that Ani is clearly delimited from the genomic context and forms a monophyletic lineage with the other echinocandin gene clusters. Importantly, we found that the disjunct genomic location of the echinocandin B gene cluster in A. pachycristatus NRRL 11440 on two separate subclusters, Ecd and Hty, at two loci was likely an artifact of genome misassembly in the absence of a reference sequence. We show that both sequences can be aligned resulting a single cluster with a gene arrangement collinear compared to other clusters of Aspergillus section Nidulantes. The reassembled gene cluster (Ecd/Hty) is identical to a putative gene cluster (AE) that was previously deposited at the NCBI as a sequence from A. delacroxii NRRL 3860. PCR amplification of a part of the gene cluster resulted a sequence that was very similar (97 % identity), but not identical to that of AE.
Conclusions
The Echinocandin B biosynthetic cluster from A. nidulans NRRL 8112 (Ani) is particularly similar to that of A. pachycristatus NRRL 11440 (Ecd/Hty). Ecd/Hty was originally reported as two disjunct sub-clusters Ecd and Hty, but is in fact a continuous sequence with the same gene order as in Ani. According to sequences of PCR products amplified from genomic DNA, the echinocandin B producer A. delacroxii NRRL 3860 is closely related to A. pachycristatus NRRL 11440. A PCR-product from the gene cluster was very similar, but clearly distinct from the sequence published for A. delacroxii NRRL 3860 at the NCBI (No. AB720074). As the NCBI entry is virtually identical with the re-assembled Ecd/Hty cluster, it is likely that it originates from A. pachycristatus NRRL 11440 rather than A. delacroxii NRRL 3860.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2885-x) contains supplementary material, which is available to authorized users.
Keywords: Nonribosomal peptides, Biosynthesis, Aspergillus section Nidulantes, Fungi, Sequence analysis
Background
Echinocandins are fungal nonribosomal cyclic hexapeptides conjugated with a fatty acid or highly reduced polyketide side chain (Fig. 1).
Fig. 1.
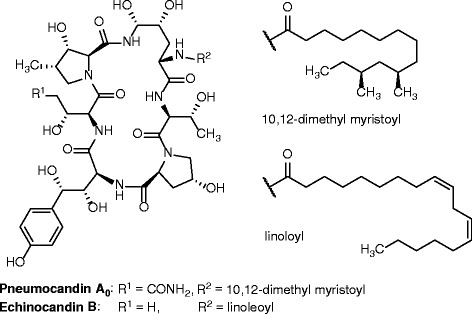
Structures of Pneumocandin A and Echinocandin B, two important members of the echinocandin family
They have a strong antifungal activity because they specifically inhibit β-1,3-glucan synthase, the enzyme responsible for the main polymer in cell wall biosynthesis. Semi-synthetic derivatives have become important antifungal drugs for treatment of invasive mycoses [1–3]. Besides their pharmacological properties, their extraordinary structures offer numerous challenging questions for biosynthetic studies. Depending on the individual echinocandin, four or five of the six amino acids are non-proteinogenic. Most amino acid modifications are due to hydroxylations catalyzed by cytochrome P450 monoxygenases or non-heme dioxygenases. However, in two amino acids, 3-hydroxy-4-methylproline and dihyroxyhomothyrosine, the carbon skeleton is also non-canonical. The peptide ring of echinocandins is closed by an N-acyl-hemiacetal moiety, which is very sensitive to hydrolysis as a rule. Nevertheless, in echinocandins it is conformationally stabilized, so that the cyclopeptides can be handled without protection. The first systematic studies on echinocandin biosynthesis were carried out the early 1990s when researchers from Merck & Co. Research Laboratories identified the biosynthetic building blocks of the pneumocandin through 13C-labeling experiments and developed a comprehensive concept of pneumocandin biosynthesis [4, 5]. However, it was not until two decades later that echinocandin biosynthesis was elucidated at the genetic level [6, 7]. In the genome of Aspergillus pachycristatus NRRL 11440 [8], previously named as Emericella rugulosa, two separate partial clusters were located on different contigs of the assembly and were found to be responsible for encoding the synthesis of echinocandin B. The larger section, Ecd, encoded most of the enzymes required for echinocandin assembly and decoration. This included genes for a nonribosomal peptide synthetase (NRPS) with domains for the assembly of six amino acids and peptide cyclization, an Acyl Amp ligase for acylation with linoleic acid, three oxygenases, and an ABC transporter. The other cluster, Hty, contained the genes for the homotyrosine biosynthetic pathway and two additional oxygenases. The functions of many of the biosynthetic enzymes encoded by Ecd were demonstrated experimentally [6, 7]. In 2014, we described an α-ketoglutarate/Fe2+-dependent proline hydroxylase involved in pneumocandin biosynthesis in Glarea lozoyensis [9]. More recently, the functions of four oxygenases in G. lozoyensis cluster GL were demonstrated by deletion in the host strain [10, 11]. The results were consistent with and complemented the findings for A. pachycristatus.
Taking all these results together, the nearly complete echinocandin biosynthesis can be modelled, though the precise order of some of the oxidation steps remain to be confirmed.
In the meantime, the sequences of additional echinocandin biosynthetic clusters were deposited at the NCBI by researchers from Toyama Prefectural University (Table 1). These sequences now allow phylogenetic reconstructions and more sound investigations of sequence-structure relationships and [12, 13].
Table 1.
| Cluster acronym | Organism (synonyms) | Echinocandin Product | NCBI-ID |
|---|---|---|---|
| AA | Aspergillus aculeatus ATCC 16872 | aculeacin [27] | GOLD Project ID: Gp0010055 |
| AM | Aspergillus mulundensis Y-30462 = DSMZ 5745 (= Aspergillus sydowii var. mulundensis) | mulundocandin [28] | KP742486 |
| Ani | Aspergillus nidulans NRRL 8112 (= Emericella nidulans DSM 946) | echinocandin B | KT806042 |
| CC | Coleophoma crateriformis | antibiotic WF738A | AB720076 |
| CE_1 | Coleophoma empetri F-11899 | antibiotic WF11899A-C | AB723722 |
| CE_2 | Coleophoma empetri No. 14573 | antibiotic WF14573A,B | AB720725 |
| Ecd/Hty (identical with AE) | Ecd/Hty: Aspergillus pachycristatus NRRL 11440 (= Emericella rugulosa, Aspergillus nidulans var. roseus ATCC 58397) AE: stated strain: Aspergillus delacroxii NRRL 3860 (= Emericella nidulans var. echinulatus | echinocandin B |
Ecd/Hty: assembly of Ecd JX421684 and Hty JX421685 AE: AB720074 |
| GL, Glo | GL: Glarea lozoyensis wildtype strain ATCC 20868 Glo: G. lozoyensis mutant strain ATCC 74030 |
pneumocandin | NW_007360987, AGUE01000179 |
| PH | Phialophora cf. hyalina No. 16616 (=Tolypocladium parasiticum) | antibiotic WF16616 | AB720726 |
Here we present the echinocandin B gene cluster from A. nidulans NRRL 8112 (Ani) identified by whole genome sequencing and sequence analysis. The Ani gene cluster is compared with the known echinocandin biosynthesis clusters from Aspergillus section Nidulantes. We also found that the two separated semi-clusters Ecd and Hty encoding echinocandin B biosynthesis in A. pachycristatus can be aligned into a continuous sequence (Ecd/Hty), which is virtually identical with the echinocandin B biosynthesis cluster AE deposited at the NCBI as a sequence from A. delacroxii NRRL 3860 (= E. nidulans var. echinulata).
Results and discussion
Echinocandin biosynthesis cluster from A. nidulans NRRL 8112
A. nidulans strain NRRL 8112 was originally isolated from India and was described as a weak producer of echinocandin B in a patent from Eli Lilly in 1976 (3.5 g/200 L culture broth) [14]. As several other A. nidulans strains are well characterized and often used for molecular genetic research, we thought that echinocandin B biosynthesis by this strain would be of particular interest. To identify the corresponding cluster from A. nidulans, genomic DNA of NRRL 8112 was sequenced using the Illumina technique. Originally a reference assembly with the genome of A. nidulans FGSC A4 (assembly ASM14920v1) was attempted, however, more than one million reads remained unmapped. As an alternative, the reads were assembled de novo resulting a genome of 31.6 Mbp partitioned in 1,572 contigs. By BLASTn search using genes of the Ecd biosynthetic cluster as query sequences we localized an echinocandin biosynthesis cluster of about 51 kb (Ani) in a relatively small contig (56 kbp). The set of genes in Ani is equivalent to that of echinocandin B biosynthesis cluster AE (≙ Ecd/Hty) and the mulundocandin biosynthesis cluster. The gene orders are fully collinear with those from Aspergilli depicted previously [12].
Determination of terminal regions by comparison with AE from A. delacroxii
A comparison with the currently known echinocandin biosynthetic clusters (Table 1) revealed a highly conserved “core” set of genes, most of them encoding enzymes required for biosynthesis. Though evidence was rather vague, possible genes in the periphery of the core cluster were considered to be linked to echinocandin biosynthesis [7, 15]. However, similar genomic context in the flanking regions of the clusters indicating a function for echinocandin biosynthesis was only found in two closely related clusters from Coleophoma empetri strains (CE_1 and CE_2 in Table 1) [12].
The gene order in Ani was identical to that in the AE-cluster, which was deposited at the NCBI as a sequence from A. delacroxii NRRL 3860, previously named as E. nidulans var. echinulata (Table 1) [16–18]. This allowed to align the DNA-sequences of the clusters over the whole lengths including the right- and left-flanking regions (Fig. 2). For the region considered as the “core” cluster, the average nucleotide sequence identity is about 89 %. At both ends of this region it drops abruptly to about 30 %. A BLAST analysis revealed that the left- and right-flanking regions comprised a genomic context orthologous to sections in the genome of A. nidulans FGSC A4 (ATCC 38163), a strain that lacks the echinocandin cluster. Thus, even in closely related species, echinocandin gene clusters appear as discrete genomic units and lack similarity in the near genomic neighborhoods [12]. This observation strongly suggests that genes in close proximity to the “core” cluster are not involved in echinocandin biosynthesis. As the available DNA-segments encoding echinocandin B biosynthesis were relatively short, an analysis of more distant regions was not possible.
Fig. 2.
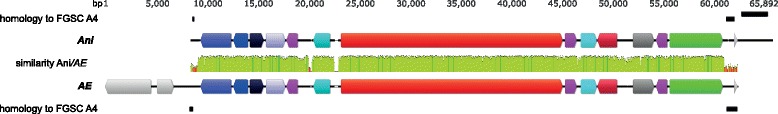
Schematic representation of a DNA sequence alignment of the echinocandin B biosynthesis clusters and flanking regions of the echinocandin gene clusters Ani and AE. Pairwise sequence identities are depicted as columns showing the overall identity about 50 bp. The following color code is used for sequence identity: dark green: 100 %; bright-green: 30 % – <100 % and red: <30 %. The black bars indicate segments with homology to sections in the genome of A. nidulans FGSC A4 determined by Discontiguous Megablast (e-values <10−130)
Comparison of cluster AE with Ecd and Hty
In contrast to all other clusters, echinocandin B biosynthesis by A. pachycristatus was reported to be split in two separate clusters, Ecd and Hty, which were found on different contigs of the genome assembly [7]. A minimal chromosomal distance of at least 42.5 kb was estimated. When we compared genes of the AE-cluster with those from Ecd and Hty, we found strikingly high sequence identities (>95 %). As the gene orders in Ecd and Hty are collinear with that of AE, both half-clusters could be aligned to AE (Fig. 3). Notably, there was a coinciding section of about 400 bp between Ecd and Hty (96.0 % sequence identity) (Fig. 4). This overlap was used to assemble both partial clusters into a continuous sequence (Ecd/Hty). To confirm the overlap of Ecd and Hty, this area and the flanking regions were amplified by PCR from A. pachycristatus genomic DNA (Fig. 3b and Additional file 1: Figures S1–S3 and Table S1). The PCR-product of the 5′-terminus proved to be identical with Hty and contained a part of the cytochrome-P450 oxygenase gene HtyF. At the 3′-terminus, a section of Ecd up to the NRPS-gene (EcdA) was found. In contrast, PCR experiments that aimed at the amplification of Hty and Ecd as separate clusters, were unsuccessful. The gene arrangement in the fused cluster Ecd/Hty was identical with that in AE, Ani and AM (cf. Table 1). Notably, the sequence identity between Ecd/Hty and AE was 99.9 % over the full lengths of the alignment. Furthermore, most of the non-matching sites were due to gaps and ambiguities in one of the sequences. Only three base pairs in the 53.9 kbp alignment were genuinely inconsistent (99.99 % sequence identity). The near complete sequence identity strongly suggests that the same strain has been used in both sequencing projects. However, according to the literature, the strains originated from unrelated sources. A. delacroxii (A-32204, NRRL 3860) was first described by researchers from the Ciba-Geigy AG, Switzerland [16, 17] as A. nidulans var. echinulatus. It was originally isolated from beech leaves at a non-specified place. A. pachycristatus NRRL 11440 (formerly E. rugulosa, ATCC 58397) was isolated by researchers of Eli Lilly from a soil sample from Greenfield, Indiana. It was originally classified as A. nidulans var. roseus [19]. Comprehensive experimental studies reported in the literature and our PCR-control of genomic DNA confirmed that strain ATCC 58397 actually has a cluster of the reported sequence. To find out more about the relationship of the strains, we first amplified and sequenced the internal transcribed spacers (ITS) region of both strains [20]. The resulting sequences differed only in one basepair. A large number of ITS sequences with 99 % identity was found at the NCBI Nucleotide database. Most of them were from A. nidulans strains, but some also from A. pachychristatus (E. rugulosa) and A. delacroxii (E. echinulata) (Additional file 1: Table S2 and Figure S5). As the ITS DNA marker was not sufficient to differentiate the strains, a part of the calmoludin gene was analyzed [8, 21]. In an alignment of calmodulin gene sequences from A. delacroxii and A. pachycristatus strains deposited at the NCBI Nucleotide database more than 30 species-conserved differences were found (Additional file 1: Figure S6), which allows a clear differentiation of both. This was also visualized in a phylogenetic tree based on the calmodulin sequences of about 120 Aspergillus (Emericella) strains, in which the A. delacroxii and A. pachycristaus strains were located in distinct subclades (Additional file 1: Figure S7).
Fig. 3.
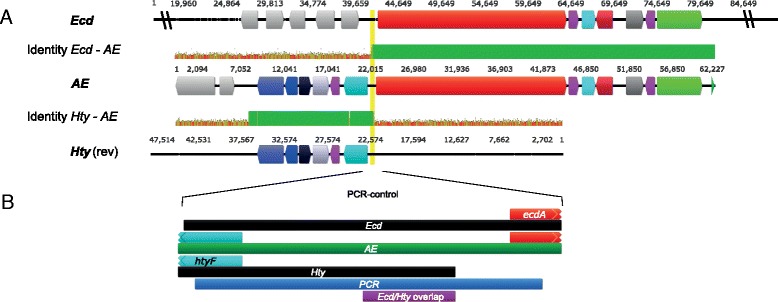
a Schematic representation of a reference alignment of Ecd and Hty to AE. Pairwise sequence identities are depicted as bars between the sequences. One bar is define by about 50 bp. The following color code is used for sequence identity: green = 100 %; greeny-brown 30 % – <100 % and red <30 %. The yellow mark indicates a region of approximately 400 bp in which Ecd and Hty are highly similar (96 % identity). b Excerpt showing the overlapping region (purple) and the section verified by sequencing of PCR product (blue)
Fig. 4.

Overlapping region of Ecd and Hty aligned with the PCR product amplified from genomic A. pachycristatus DNA. (An alignment with the entire PCR-product as outlined in 2b is shown in Additional file 1: Figure S3)
Surprisingly, the calmodulin sequence of A. delacroxii NRRL 3860 (Additional file 1: Figure S8) differed in only two basepairs from that of A. pachycristatus NRRL 11440. Both sequences could be clearly assigned to the A. rugulosa/A. pachychristatus group rather than to the E. delacroxii (E. echinulata) strains (Additional file 1: Figure S7).
In addition to the common genomic marker sequences, a region between the NRPS and the adjacent cytochrome p450 oxygenase gene of the echinocandin biosynthesis cluster was analyzed. It was the same region which had already been sequenced from A. pachycristatus to confirm the connectivity of Ecd and Hty. As for the calmodulin fragments, the sequences of A. delacroxii and A. pachychrytatus (and the AE cluster) were very similar, but clearly distinct (97 % identity, see Additional file 1: Figure S10). From these results, we concluded that the AE cluster most likely originates from A. pachycristatus NRRL 11440 and not from A. delacroxii NRRL 3860.
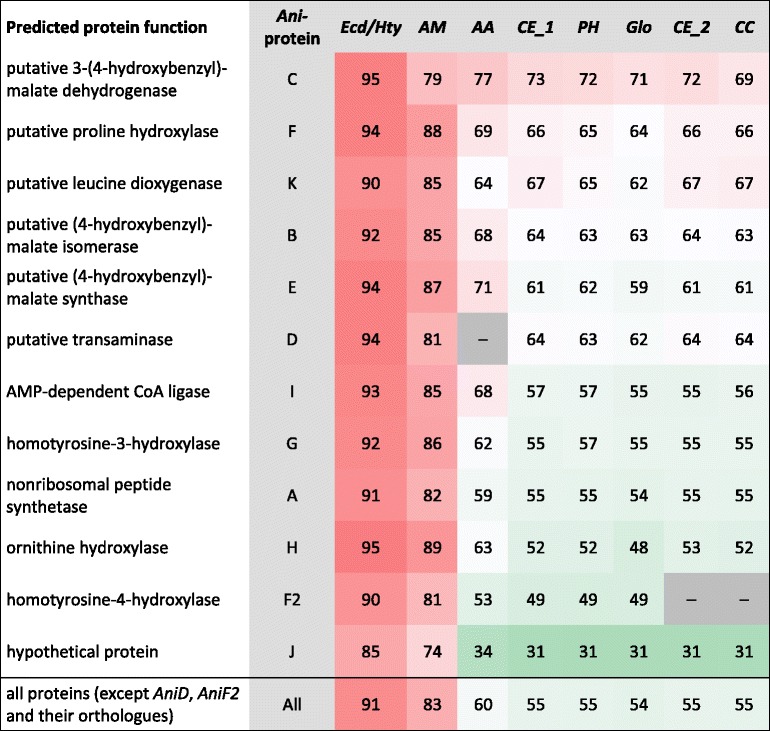
So far, echinocandin biosynthesis has been found only in relatively narrow phylogenetic regions of two related lineages of fungi, the Aspergillacea (Eurotiomycetes) and Helotiales (Leotiomycetes) [12]. Although these clades diverged an estimated 290 – 390 million years ago [22], echinocandin biosynthesis clusters share a remarkable degree of similarity. The sequence identities of Ani proteins with their orthologues range from 48 to 94 %. The only exception is AniJ, a putative protein of unknown function (31–85 %), whose orthologs are generally more diverse. Recently, Bills, An and coworkers have published a detailed phylogenetic analysis of the currently known echinocandin biosynthetic clusters [12]. The phylogenetic trees of individual pathway proteins were monophyletic. The degree of sequence divergence in the echinocandin pathway genes was similar to that in genes involved in primary carbohydrate metabolism, thus strongly suggesting a monophyletic lineage of echinocandin biosynthesis clusters originating from a common ancestor of the Aspergillaceae and Helotiales. No evidence was found for horizontal gene transfer at later evolutionary stages. Table 2 shows the sequence identities of Ani proteins with their orthologues in the other clusters. It reveals consistent trends depending on the individual cluster and the type of protein. No significant deviations were found in Ani proteins. Thus, Ani conforms well to the concept of a monophyletic lineage for echinocandin biosynthesis. The closest clusters to Ani were Ecd/Hty from A. pachycristatus and AM from A. mulundensis with 91 % and 83 % overall identity, respectively.
Table 2.
Sequence identity of Ani-proteins to their orthologs in other echinocandin biosynthetic clusters. Color shading was used to visualize sequence identities. Color code: red: 100 %, white: 50 % and green 0 %. Protein sets: Ani: A. nidulans NRRL 8112, AA: A. aculeatus ATCC 16872, CC: C. crateriformis, CE_1: C. empetri F-11899, CE_2: C. empetri, PH: T. parasiticum No. 16616, Ecd/Hty: A. pachycristatus NRRL 11440, formerly E. rugulosa, AM: A. muludensis DSMZ 5745, GL: G. lozoyensi ATCC 20868, Glo: G. lozoyensis mutant strain ATCC 74030 (GL and Glo are identical except from two residues in protein GloC ≙ GLOXY4)
Conclusions
In summary, we have identified the echinocandin B biosynthesis cluster Ani in the genome of the echinocandin B producer strain A. nidulans NRRL 8112. The gene order in Ani is collinear with that in the other known clusters of Aspergilli section Nidulantes (AE ≙ Ecd/Hty and AM). Sequence comparison of the individual proteins revealed that Ani forms a strictly monophyletic clade with the other echinocandin gene clusters. In the course of sequence comparisons, we found that the seemingly distant subclusters in A. pachychristatus NRRL 11440, Ecd and Hty, can be assembled into a single sequence (Ecd/Hty). The overlap of both sequences was confirmed by PCR. Moreover, Ecd/Hty was found to be virtually identical to echinocandin B biosynthesis-cluster AE which was deposited at the NCBI as a sequence from A. delacroxii NRRL 3860. To scrutinize this unexpected finding, genomic DNA from this strain and A. pachychristatus NRRL 11440 was amplified by PCR and sequenced. The calmodulin marker sequences of both strains were very similar, but not entirely identical (2 different bp). BLAST search and phylogenetic analysis showed that both of them were most similar to calmodulin sequences from other A. pachycristatus strains (99–100 % identity), and they clearly differed from those of A. delacroxii strains (92–94 % identity). The sequence of a PCR product from a section of the echinocandin gene cluster in A. delacroxii was very similar (97 % identity), but clearly distinct from the corresponding sequence of A. pachycristatus (≙ AE). Therefore, we suppose that AE originates from A. pachycristatus NRRL 11440 and not A. delacroxii NRRL 3860. Moreover, the partial sequence of the calmodulin gene and the remarkably high similarity of the gene cluster fragments strongly suggest that strain NRRL 3860 belongs to the A. pachychristatus/A. rugulosa group rather than to A. delacroxii. An in-depth examination of this strain will be necessary to clarify the taxonomic status.
Methods
Aspergillus nidulans NRRL 8112 (= ATCC 58396) and A. pachycristatus NRRL 11440 (= A. nidulans var. roseus ATCC 58397) were purchased from LGC Standards/ATCC (UK). A. delacroxii NRRL 3860 (= E. nidulans var. echinulata) was obtained from the Agricultural Research Service Collection (ARS) of the United States Department of Agriculture (USDA), Peoria IL (USA).
The genome of A. nidulans NRRL 8112 was sequenced and assembled by the De Novo Sequencing Service of BaseClear (Netherlands) using Illumina technique. (Assembly statistics: N50: 65,796 bp, L50: 138, largest contig: 462,827 bp). The paired-end library was deposited at the NCBI in Bioproject PRJNA89151 SRA experiment SRX128799. The genome was annotated with the automatic gene prediction tool AUGUSTUS [23] and the “Antibiotics and Secondary Metabolite Analysis Shell” (antiSMASH) [24].
For PCR-experiments genomic DNA from A. pachycristatus NRRL 11440 and A. delacroxii NRRL 3860 was prepared as follows: A culture was grown from a spore suspension in potato dextrose medium (250 mL) at 24 °C and 160 rpm for 14 days. The mycelium was harvested, frozen in liquid nitrogen and ground to a fine powder. The DNA was then extracted as per GenElute™ Plant Genomic DNA Miniprep Kit (Qiagen, Germany).
PCR experiments were conducted as per NEB protocol using Phusion™ Master Mix. The following profile was used for amplification: 96 °C for 10 s (hot start), pause and addition of polymerase; 96 °C for 5 min; then 30 cycles at 96 °C for 30 s; 57 °C (TM-2) for 20 s; 72 °C for 2 min; and a final extension at 72 °C for 10 min. For primer pair WH2_fw and WH2_rv a melting temperature of TM = 53 °C was used. Primer sequences are given in Table S1 in Additional file 1.
DNA and protein sequences were edited, compared and visualized with the Geneious 8.1 software platform [25]. Sequence identities were determined by aligning each set of protein sequences with MUSCLE [26] implemented in Geneious 8.1. with the default settings. The global sequence identity of the clusters was determined through a concatenation of the alignments of all proteins which occur in all clusters.
Abbreviations
The acronyms for the biosynthetic clusters are explained in Table 1.
BLAST(n), basic local alignment search tool (nucleotide); FGSC, Fungal Genetics Stock Center; NCBI, National Center for Biotechnology Information; NRPS, nonribosomal peptide synthase; NRRL, Northern Regional Research Laboratory; PCR, polymerase chain reaction
Acknowledgements
We thank Prof. Michael Müller for his generous support and James Swezey (NRRL) for assistance in obtaining strain NRRL 3860.
Funding
L.Y. was supported by an Alexander von Humboldt Research Fellowship. The article processing charge was funded by the German Research Foundation (DFG) and the Albert Ludwigs University Freiburg in the funding program Open Access Publishing.
Availability of data and material
The data sets supporting the results for this article are included within the article and Additional file 1.
The annotated sequence of the Ani-cluster is available from the NCBI nucleotide database under accession number KT806042. The partial calmodulin gene sequence of A. delacroxii NRRL 3860 was deposited under accession number KX159480.
Authors’ contributions
WH conceived and supervised the study and drafted the manuscript. KGH performed the PCR-experiments and worked on the manuscript. LY prepared genomic DNA from A. nidulans NRRL 8112, coordinated the genome sequencing and performed initial cluster analyses. B.A.G and S.G. analyzed and annotated the genome. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional file
PCR experiments and sequence alignments. This file containes further information on the PCR-reactions, a sequence alignment of the gene clusters Ecd, Hty and AE with the PCR product from genomic A. pachycristatus DNA, sequences of amplified ITS and calmodulin regions, their alignment with published sequences and a comparison of PCR-product sequences from the clusters of E. pachychristatus and E. delacroxii. (DOCX 8989 kb)
Contributor Information
Wolfgang Hüttel, Phone: (+)49 761 203 67398, Email: wolfgang.huettel@pharmazie.uni-freiburg.de.
Loubna Youssar, Email: lyoussar@gmail.com.
Björn A. Grüning, Email: gruening@informatik.uni-freiburg.de
Stefan Günther, Email: stefan.guenther@pharmazie.uni-freiburg.de.
Katharina G. Hugentobler, Phone: (+)49 761 203 67398, Email: katharina.hugentobler@pharmazie.uni-freiburg.de
References
- 1.Balkovec JM, Hughes DL, Masurekar PS, Sable CA, Schwartz RE, Singh SB. Discovery and development of first in class antifungal caspofungin (CANCIDAS®) – A case study. Nat Prod Rep. 2014;31(1):15–34. doi: 10.1039/C3NP70070D. [DOI] [PubMed] [Google Scholar]
- 2.Stan CD, Tuchilus C, Stan CI. Echinocandins – new antifungal agents. Rev Med Chir Soc Med Nat Iasi. 2014;118(2):528–36. [PubMed] [Google Scholar]
- 3.Emri T, Majoros L, Tóth V, Pócsi I. Echinocandins: production and applications. Appl Microbiol Biotechnol. 2013;97(8):3267–84. doi: 10.1007/s00253-013-4761-9. [DOI] [PubMed] [Google Scholar]
- 4.Adefarati AA, Giacobbe RA, Hensens OD, Tkacz JS. Biosynthesis of L-671,329, an echinocandin-type antibiotic produced by Zalerion arboricola – origins of some of the unusual amino-acids and the dimethylmyristic acid side-chain. J Am Chem Soc. 1991;113(9):3542–5. doi: 10.1021/ja00009a048. [DOI] [Google Scholar]
- 5.Adefarati AA, Hensens OD, Jones ETT, Tkacz JS. Pneumocandins from Zalerion arboricola V. Glutamic acid-derived and leucine-derived amino-acids in pneumocandin A0 (L-671,329) and distinct origins of the substituted proline residues in pneumocandins A0 and B0. J Antibiot. 1992;45(12):1953–7. doi: 10.7164/antibiotics.45.1953. [DOI] [PubMed] [Google Scholar]
- 6.Jiang W, Cacho RA, Chiou G, Garg NK, Tang Y, Walsh CT. EcdGHK are three tailoring iron oxygenases for amino acid building blocks of the echinocandin scaffold. J Am Chem Soc. 2013;135(11):4457–66. doi: 10.1021/ja312572v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cacho RA, Jiang W, Chooi Y-H, Walsh CT, Tang Y. Identification and characterization of the eechinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440. J Am Chem Soc. 2012;134(40):16781–90. doi: 10.1021/ja307220z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuzawa T, Tanaka R, Horie Y, Hui Y, Abliz P, Yaguchi T. The correlation among molecular phylogenetics, morphological data, and growth temperature of the genus Emericella, and a new species. Mycoscience. 2012;53(6):433–45. doi: 10.1007/S10267-012-0188-X. [DOI] [Google Scholar]
- 9.Houwaart S, Youssar L, Hüttel W. Pneumocandin biosynthesis: involvement of a trans-selective proline hydroxylase. ChemBioChem. 2014;15(16):2365–9. doi: 10.1002/cbic.201402175. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Chen L, Yue Q, Liu X, An Z, Bills GF. Genetic manipulation of the pneumocandin biosynthetic pathway for generation of analogues and evaluation of their antifungal activity. ACS Chem Biol. 2015;10:1702–10. doi: 10.1021/acschembio.5b00013. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Yue Q, Li Y, Niu X, Xiang M, Wang W, Bills GF, Liu X, An Z. Engineering of Glarea lozoyensis for exclusive production of the Pneumocandin B0 precursor of the antifungal drug caspofungin acetate. Appl Environ Microbiol. 2015;81(5):1550–8. doi: 10.1128/AEM.03256-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue Q, Chen L, Zhang X, Li K, Sun J, Liu X, An Z, Bills GF. Evolution of chemical diversity in echinocandin lipopeptide antifungal metabolites. Eukaryot Cell. 2015;14(7):698–718. doi: 10.1128/EC.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bills GF, Li Y, Chen L, Yue Q, Niu X-M, An Z. New insights into the echinocandins and other fungal non-ribosomal peptides and peptaibiotics. Nat Prod Rep. 2014;31(10):1348–75. doi: 10.1039/C4NP00046C. [DOI] [PubMed] [Google Scholar]
- 14.Higgens CE, Michel KH. Antibiotic A-22082 and process for production thereof, US patent 4024246. 1977. [Google Scholar]
- 15.Chen L, Yue Q, Zhang X, Xiang M, Wang C, Li S, Che Y, Ortiz-López F, Bills GF, Liu X, et al. Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics. 2013;14(1):339. doi: 10.1186/1471-2164-14-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benz F, Knüsel F, Nüesch J, Treichler H, Voser W, Nyfeler R, Keller-Schierlein W. Stoffwechselprodukte von Mikroorganismen 143. Mitteilung. Echinocandin B, ein neuartiges Polypeptid-Antibioticum aus Aspergillus nidulans var. echinulatus: Isolierung und Bausteine. Helv Chim Acta. 1974;57(8):2459–77. doi: 10.1002/hlca.19740570818. [DOI] [PubMed] [Google Scholar]
- 17.CIBA GEIGY AG. Antifungal antibiotic A32204 production by aerobic cultivation of Aspergillus nidulans var. echinulatus A32204. Swiss patent 568386. 1975
- 18.Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CHW, Perrone G, Seifert KA, Susca A, Tanney JB, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–73. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boeck LVD, Kastner RE. Method of producing the A-30912 antibiotics, US Patent 4288549. 1981. [Google Scholar]
- 20.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 2012;109(16):6241–6. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong S-B, Go S-J, Shin H-D, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97(6):1316–29. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- 22.Gueidan C, Ruibal C, de Hoog GS, Schneider H. Rock-inhabiting fungi originated during periods of dry climate in the late devonian and middle triassic. Fungal Biol. 2011;115(10):987–96. doi: 10.1016/j.funbio.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Hoff KJ, Stanke M. WebAUGUSTUS – a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013;41(Web Server issue):W123–8. doi: 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, et al. antiSMASH 3.0 – a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hino M, Fujie A, Iwamoto T, Hori Y, Hashimoto M, Tsurumi Y, Sakamoto K, Takase S, Hashimoto S. Chemical diversity in lipopeptide antifungal antibiotics. J Ind Microbiol Biotechnol. 2001;27(3):157–62. doi: 10.1038/sj.jim.7000091. [DOI] [PubMed] [Google Scholar]
- 28.Bills GF, Yue Q, Chen L, Li Y, An Z, Frisvad JC. Aspergillus mulundensis sp. nov., a new species for the fungus producing the antifungal echinocandin lipopeptides, mulundocandins. J Antibiot. 2016;69(3):141–8. doi: 10.1038/ja.2015.105. [DOI] [PubMed] [Google Scholar]


