Abstract Abstract
Cytogenetic data for the genus Acromyrmex Mayr, 1865 are available, to date, for a few species from Brazil and Uruguay, which have uniform chromosome numbers (2n = 38). The recent cytogenetic data of Acromyrmex striatus (Roger, 1863), including its banding patterns, showed a distinct karyotype (2n = 22), similar to earlier studied Atta Fabricius, 1804 species. Karyological data are still scarce for the leafcutter ants and many gaps are still present for a proper understanding of this group. Therefore, this study aimed at increasing cytogenetic knowledge of the genus through the characterization of other six species: Acromyrmex balzani (Emery, 1890), Acromyrmex coronatus Fabricius, 1804, Acromyrmex disciger (Mayr, 1887), Acromyrmex echinatior (Forel, 1899), Acromyrmex niger (Smith, 1858) and Acromyrmex rugosus (Smith, 1858), all of which were collected in Minas Gerais – Brazil, except for Acromyrmex echinatior which was collected in Barro Colorado – Panama. The number and morphology of the chromosomes were studied and the following banding techniques were applied: C-banding, fluorochromes CMA3 and DAPI, as well as the detection of 45S rDNA using FISH technique. All the six species had the same chromosome number observed for already studied species, i.e. 2n = 38. Acromyrmex balzani had a different karyotype compared with other species mainly due to the first metacentric pair. The heterochromatin distribution also showed interspecific variation. Nevertheless, all the studied species had a pair of bands in the short arm of the first subtelocentric pair. The fluorochrome CMA3 visualized bands in the short arm of the first subtelocentric pair for all the six species, while Acromyrmex rugosus and Acromyrmex niger also demonstrated in the other chromosomes. The AT-rich regions with differential staining using DAPI were not observed. 45S ribosomal genes were identified by FISH in the short arm of the first subtelocentric pair in Acromyrmex coronatus, Acromyrmex disciger and Acromyrmex niger. The uniform chromosome number in the genus Acromyrmex (2n = 38) suggests that Acromyrmex striatus (2n = 22) should be transferred to a new genus. Other aspects of the chromosome evolution in ants are also discussed.
Keywords: Chromosome evolution, karyotype, fungus-growing ants, biodiversity, heterochromatin, FISH
Introduction
Fungus-growing ants belong to the Atta-genus group (Ward et al. 2015) corresponding to the tribe Attini in the traditional sense. Leafcutter ants comprise a particular group of fungus-growing ants which are referred to as dominant herbivores of the Neotropics (Hölldobler and Wilson 1990). They include the genera Acromyrmex Mayr, 1865 and Atta Fabricius, 1804 and are exclusively found in the New World, primarily in the Neotropical region (Mayhé-Nunes and Jaffé 1998) and are considered the most derived group of ants arising about 8-12 million years ago (Schultz and Brady 2008, Mehdiabadi and Schultz 2010).
The genus Acromyrmex contains 33 described species (or more than 60 taxa if all subspecies and variations are included) (Bolton 2014, Rabeling et al. 2015). They are distributed from California (USA) to Patagonia (Argentina), excluding Chile. Most Brazilian species are widely distributed, although some of them have more restricted distribution (Gonçalves 1961, Mayhé-Nunes 1991, Delabie et al. 2011).
The genus Acromyrmex has been subdivided into two subgenera, Acromyrmex and Moellerius Forel, 1893 (Emery 1913), based on morphological traits. A phylogenetic study based on the morphological traits of this genus showed that the two subgenera formed distinct groups, of which Moellerius was considered the most derived (Mayhé-Nunes 1991). However, recent phylogenetic molecular studies of the genus Acromyrmex, including five species of the subgenus Moellerius, subdivided Acromyrmex species into distinct clusters (Cristiano et al. 2013). Only two of them, Acromyrmex balzani (Emery, 1890) and Acromyrmex landolti (Forel, 1885), were placed in the same group, suggesting that Acromyrmex and Moellerius could not be monophyletic (Cristiano et al. 2013); similar results were observed by Sumner et al. (2004). These data suggest that the two subgenera Acromyrmex and Moellerius do not represent natural groups.
Leafcutter ants are one of the most studied groups of fungus-growing ants (Mayhé-Nunes 1991), both in terms of biology and geographic distribution. Their status as agricultural pests has contributed to their knowledge, although taxonomic limits of different species are sometimes unclear (Delabie et al. 2011, Bacci et al. 2009). Under these circumstances, the so-called “integrative taxonomy” can produce more consistent results by complementing data obtained by different techniques (Schlick-Steiner et al. 2010). Nowadays, ant cytogenetics is a rapidly developing research field (Delabie et al. 2012). Cytogenetic data on fungus-growing ants with information for at least the chromosome number and morphology are available at present for 38 taxa (reviewed in Barros et al. 2011, Cristiano et al. 2013, Barros et al. 2013, 2014a, 2014b, Cardoso et al. 2014, Barros et al. 2015), corresponding to about 10% of described species (Brandão et al. 2011). In some ant genera, e.g. in Mycetarotes Emery, 1913 and Cyphomyrmex Mayr, 1862, chromosome numbers are variable at the species level (reviewed in Barros et al. 2011). However, species within the genera Atta (Barros et al. 2014a, Fadini and Pompolo 1996, Murakami et al. 1998, Barros et al. 2015) and Acromyrmex (Fadini and Pompolo 1996, Goñi et al. 1983) have the same chromosome numbers, 2n = 22 and 2n = 38, respectively, and similar chromosome morphology.
Cytogenetic data on the leafcutter ants are scarce. Namely, these data are available for five Atta species (Barros et al. 2014a, Fadini and Pompolo 1996, Murakami et al. 1998, Barros et al. 2015). In these species, 2n = 22 and a karyotypic formula of 2n = 18m+2sm+2st were found. Similar banding patterns were also observed in different species (Barros et al. 2014a, Murakami et al. 1998, Barros et al. 2015) which belonged to the three of four species groups defined on the basis of molecular data (Bacci et al. 2009). Cytogenetic data on Acromyrmex are also restricted but available for some taxa collected in Brazil: Acromyrmex (Acromyrmex) crassispinus (Forel, 1909); Acromyrmex (Acromyrmex) subterraneus molestans Santschi, 1925; Acromyrmex (Acromyrmex) subterraneus subterraneus Forel, 1893 (Fadini and Pompolo 1996); and in Uruguay: Acromyrmex (Acromyrmex) ambiguus Emery, 1888; Acromyrmex (Acromyrmex) hispidus Santschi, 1925; and Acromyrmex (Moellerius) heyeri (Forel, 1899) (Goñi et al. 1983). All the species had the same chromosome number, 2n = 38. However, Acromyrmex (Moellerius) striatus (Roger, 1863) has recently shown 2n = 22, with a karyotypic formula of 2n = 20m+2sm (Cristiano et al. 2013), the same chromosome number found in all Atta species studied to date (Barros et al. 2014a, Fadini and Pompolo 1996, Murakami et al. 1998, Barros et al. 2015). Since Acromyrmex striatus belongs to the well-supported clade which is quite distinct from other members of the genus Acromyrmex, it is suggested that this species is a sister group of all other leafcutter ants, which split before the divergence between Acromyrmex and Atta (Cristiano et al. 2013). Despite the same chromosome number, karyotypes of Acromyrmex striatus and Atta species differ in morphology of two chromosome pairs as well as in their banding patterns (Cristiano et al. 2013, Barros et al. 2014a). The aim of the present study is therefore to describe chromosome sets of six species of the genus Acromyrmex to update our knowledge of karyotype evolution of leafcutter ants and Neotropical Formicidae in general.
Material and methods
Six cytogenetically studied Acromyrmex species were collected between August 2008 and March 2010 in the state of Minas Gerais – Brazil, except for Acromyrmex echinatior (Forel, 1899) which was collected in Panama (Table 1). Metaphases were obtained according to Imai et al. (1988) using larval ganglia or testes of freshly defecated larvae. To study chromosome morphology, metaphases were analyzed using conventional 4% Giemsa staining. The karyotypes were composed by arranging chromosomes according to their size and chromosome arm ratio (r) (Levan et al. 1964). Ten best metaphases per species with a similar degree of condensation were measured. Karyotypes were composed using the Corel Photopaint X3® software. The colonies and individuals analyzed are listed in Table 1. For the banding techniques, 4 to 10 individuals per species were used: C-banding for heterochromatin detection was performed according to Sumner (1972) with minor adaptations suggested by Barros et al. (2013); sequential fluorochrome staining with CMA3/DA/DAPI (Schweizer 1980) was done to reveal specific GC- and AT-rich regions. To detect (NORs) in three species, 2 to 4 individuals of each species were studied using (FISH) with the 45S rDNA probe isolated from Arabidopsis thaliana (Moscone et al. 1996). The metaphases were observed and photographed using an Olympus® BX 60 microscope attached to a Q Color 3 Olympus® image capture system. For fluorochrome analysis, filters WB (450-480 nm) and WU (330-385 nm) were used for studying CMA3 and DAPI staining, respectively, as well as Leica microscope DMRA2 filter Y3 (545/30 nm), attached to a D Leica IM50 Version 5 Release 190 software was used for FISH analysis.
Table 1.
Acromyrmex spp. cytogenetically studied in this paper. Locality, sample size (number of colonies/individuals stained with Giemsa), diploid (2n) and haploid (n) chromosome number and karyotypic formula.
| Acromyrmex species | Locality (coordinates) | Colony – Individuals | 2n (n) | Karyotypic formula |
|---|---|---|---|---|
| Acromyrmex (Moellerius) balzani (Emery, 1890) | Viçosa – MG – Brasil (20°45'S; 42°51'W) | 3 – 12 | 38 | 2n = 12m + 10sm + 14st + 2a |
| Acromyrmex (Moellerius) balzani (Emery, 1890) | Paraopeba – MG – Brazil (19° 17'S; 44° 29'W) | 2 – 15 | 38 | 2n = 12m + 10sm + 14st + 2a |
| Acromyrmex (Acromyrmex) coronatus Fabricius, 1804 | São Tiago – MG – Brazil (20°54'S; 44°30'W) | 1 – 10 | 38 (19) | 2n = 12m + 8sm + 16st + 2a |
| Acromyrmex (Acromyrmex) coronatus Fabricius, 1804 | Paraopeba – MG – Brazil (19°17'S; 44°29'W) | 5 – 20 | 38 | 2n = 12m + 8sm + 16st + 2a |
| Acromyrmex (Acromyrmex) disciger (Mayr, 1887) | Santos Dumont – MG – Brazil (21°27'S; 43°32'W) | 2 – 15 | 38 | 2n = 10m + 12sm + 14st + 2a |
| Acromyrmex (Acromyrmex) niger (Smith, F. 1858) | Viçosa – MG – Brazil (20°45'S; 42°51'W) | 3 – 21 | 38 | 2n = 12m + 14sm + 10st + 2a |
| Acromyrmex (Acromyrmex) rugosus (Smith, F. 1858) | Florestal – MG – Brazil (19°52'S; 44°24'W) | 1 – 6 | 38 | 2n = 16m + 12sm + 8st + 2a |
| Acromyrmex (Acromyrmex) rugosus (Smith, F. 1858) | Paraopeba – MG – Brazil (19° 17'S; 44° 29'W) | 5 – 22 | 38 | 2n = 16m + 12sm + 8st + 2a |
| Acromyrmex (Acromyrmex) echinatior (Forel, 1899) | Barro Colorado – Panama (9°9'N; 79°50'W) | 2 – 10 | 38 | 2n = 8m + 6sm + 14st + 10a |
Adult ant specimens were identified by J.H.C. Delabie and deposited in the ant collection at the (CPDC/Brazil).
Results
All studied species had the same diploid chromosome number, 2n = 38 (Table 1, Fig. 1); males of Acromyrmex coronatus Fabricius, 1804 showed n = 19 (Table 1).
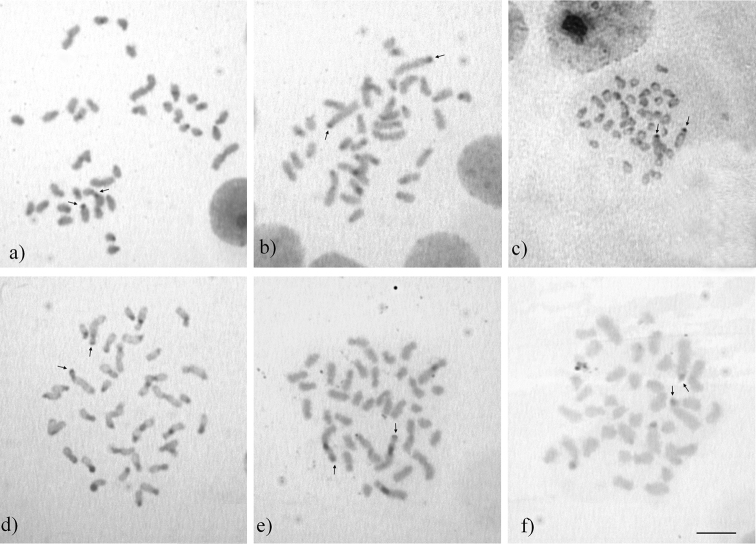
Figure 1.
Karyotype of Acromyrmex species. a Acromyrmex balzani b Acromyrmex coronatus c Acromyrmex disciger d Acromyrmex rugosus e Acromyrmex niger f Acromyrmex echinatior. All species have 2n = 38. Bar = 5 µm.
Chromosome measurements revealed morphological differences between similar karyotypes (Table 1). Several chromosome pairs were easily recognizable among different species, i.e. the first metacentric pair, the two largest subtelocentric pairs and the largest (or unique in some species) acrocentric pair (Fig. 1). In the karyotype of Acromyrmex balzani, the first metacentric pair was larger than in any other studied species (Fig. 1). The size of the first metacentric and the first subtelocentric chromosome pairs were similar in all species (Fig. 1b–f), again except for Acromyrmex balzani (Fig. 1a). No geographical intraspecific differences between the karyotypes were found (Table 1).
The C-banding results of Acromyrmex balzani, Acromyrmex coronatus, Acromyrmex disciger (Mayr, 1887) and Acromyrmex rugosus (Smith, 1858) indicated bands in some chromosomes: in the short arms of the submetacentric and subtelocentric and also in the centromeric regions of the metacentric chromosomes (Fig. 2). The largest subtelocentric pair (denominated as ST1) showed bands in all the species. Acromyrmex disciger (Fig. 2c), Acromyrmex rugosus (Fig. 2d) and Acromyrmex coronatus (Fig. 2b) had bands in the telomeric region of the short arm. In Acromyrmex balzani (Fig. 2a) and Acromyrmex echinatior (Fig. 2f) karyotypes, the heterochromatic bands were observed in the short arm of ST1 pair. Acromyrmex niger (Smith, 1858) (Fig. 2e) showed bands in the telomeric region of the short arm of the ST1 pair and also in the long arm. Moreover, Acromyrmex rugosus and Acromyrmex niger had additional bands in the pericentromeric region and in the long arm region of the second subtelocentric pair, respectively.
Figure 2.
C-banded metaphases of Acromyrmex species. a Acromyrmex balzani b Acromyrmex coronatus c Acromyrmex disciger d Acromyrmex rugosus e Acromyrmex niger f Acromyrmex echinatior. Arrows indicate C-bands in the largest subtelocentric chromosome pair. Bar = 5 µm.
The short arms of the ST1 pair revealed differences in the banding patterns among the species with fluorochrome CMA3. Acromyrmex disciger (Fig. 3c) and Acromyrmex coronatus (Fig. 3b) in the telomeric regions. Acromyrmex balzani (Fig; 3a) in the short arms; and Acromyrmex echinatior in the interstitial region (Fig. 3f). However, Acromyrmex niger (Fig. 3e) and Acromyrmex rugosus (Fig. 3d), besides the telomeric regions of the ST1 pair, also showed bands in additional chromosomes. Acromyrmex niger (Fig. 3e) had additional bands in the long arm of the ST1 pair and in the long arm of the second larger subtelocentric pair. Acromyrmex rugosus (Fig. 3d) showed small bands in the telomeric regions of ST1 and in at least three other chromosomes.
Figure 3.
Metaphases of Acromyrmex species stained with CMA3 and DAPI, respectively. a Acromyrmex balzani b Acromyrmex coronatus c Acromyrmex disciger d Acromyrmex rugosus e Acromyrmex niger f Acromyrmex echinatior. Arrows indicate CMA3-positive bands in the largest subtelocentric pair. Arrowheads indicate additional CMA3-positive bands in Acromyrmex niger and Acromyrmex rugosus. Bar = 5 µm.
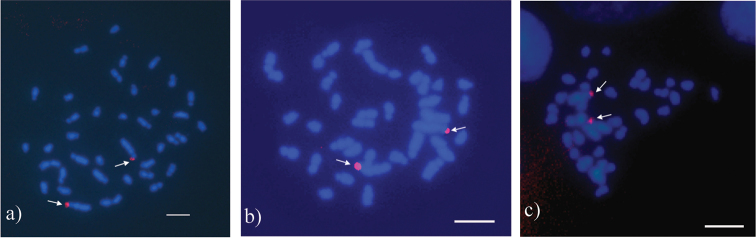
DAPI-positive bright bands which could correspond to AT-rich regions were not revealed (Fig. 3a–f). Instead, DAPI-negative regions which co-localized with CMA3-positive bands were visualized. FISH with the 45S rDNA probe visualized telomeric bands in ST1 pairs of Acromyrmex coronatus, Acromyrmex disciger and Acromyrmex niger (Fig. 4).
Figure 4.
Metaphases of Acromyrmex species. FISH with 45S rDNA probe. a Acromyrmex disciger b Acromyrmex coronatus c Acromyrmex niger. Arrows indicate hybridization signals in the subtelocentric pair. Bar = 5 µm.
Discussion
Karyotypes of Acromyrmex species observed in this study can be distinguished only on the basis of chromosomal measurements. Differential heterochromatin growth is therefore responsible for small but robust differences in chromosomal morphology, and these differences could not be observed using classification proposed by Imai (1991). This classification is based on heterochromatin distribution, and it does not reflect differences in the chromosome size among similar karyotypes. However, some chromosomes had the chromosome arm ratio (r) within the limits of the classification (submetacentric or subtelocentric). In the ants Acromyrmex coronatus and Acromyrmex niger the chromosomes were classified according to the greater ratio (r), i.e., as subtelocentrics.
The largest metacentric pair of Acromyrmex balzani strongly differs in size from that of other species probably due to complex chromosomal rearrangements that need to be further investigated. This can be explained by the fact that Acromyrmex balzani forms a separate clade together with Acromyrmex landolti according to the molecular phylogeny presented by Cristiano et al. (2013).
The six studied species showed heterochromatic segments on the short arms of ST1 pair. It was observed that these GC-rich heterochromatic regions correspond to NOR which is, in turn, confirmed by FISH with the 45S rDNA probe in the chromosomes of Acromyrmex coronatus, Acromyrmex disciger and Acromyrmex niger. In the latter species, this technique revealed a single NOR, although additional multiple CMA3-positive bands also were observed. This means that these additional bands are not related to the ribosomal genes. NORs are generally GC-rich and CMA3-positive in different organisms (Reed and Phillips 1995). However, CMA3-positive regions are not always rDNA clusters (Sumner 1990), as was observed in Acromyrmex niger. Multiple CMA3-positive bands and a single NOR revealed by FISH were observed in the fungus-growing ant Mycocepurus goeldii (Forel, 1893) (Barros et al. 2010, 2012); however, ribosomal gene mapping studies of Formicidae of the Neotropical region using FISH are scarce.
The nonspecific banding pattern of DAPI staining revealed in the present work is similar to those observed for other fungus-growing ants such as Mycocepurus goeldii (Barros et al. 2010), Acromyrmex striatus (Cristiano et al. 2013), Trachymyrmex fuscus Emery, 1934 (Barros et al. 2014) and Atta species (Barros et al. 2014, 2015).
Up to now, 12 Acromyrmex species (plus the only subspecies) are cytogenetically studied. All of them show 2n = 38, including both subgenera Acromyrmex and Moellerius (Fadini and Pompolo 1996, Goñi et al. 1983). However, Acromyrmex striatus with 2n = 22 differs from other already known species (Cristiano et al. 2013). The latter chromosome number is also characteristic of all Atta species. Both Acromyrmex and Atta are considered the most derived genera of fungus-growing ants (Schultz and Brady 2008, Mehdiabadi and Schultz 2009). Since differences in chromosomal morphology and banding patterns can be observed within Acromyrmex, it differs in this respect from Atta.
Patterns of heterochromatin distribution on short arms of some submetacentric and subtelocentric chromosomes of Acromyrmex species suggest that centric fissions which contributed to the origin of the derived karyotype with 2n = 38, probably occurred in the karyotype of the most recent common ancestor of this group. Moreover, recent molecular phylogenetic reconstruction by Cristiano et al. (2013) also suggests that Acromyrmex striatus is a sister group to the remaining leafcutter ants. The above-mentioned fissions were followed by heterochromatin growth which played an important role in maintaining telomeric stability according to the minimum interaction theory proposed by Imai et al. (1994). Differential heterochromatin growth in Acromyrmex is responsible for interspecific variation in the size of heterochromatic blocks. However, different species of this genus retain the same chromosome number (2n = 38), except for Acromyrmex striatus with 2n = 22 (Cristiano et al. 2013).
Besides data on the chromosome numbers, multiple GC-rich segments were observed in the fungus-growing ants Mycocepurus goeldii (Barros et al. 2010), Sericomyrmex sp. (Barros et al. unpublished data), Trachymyrmex fuscus (Barros et al. 2014b) and Acromyrmex striatus (Cristiano et al. 2013). Nevertheless, single CMA3-positive bands were found in the short arms of ST1 pairs in all Acromyrmex spp. and in the fourth chromosome pair of Atta species. Multiple GC-rich segments observed in Acromyrmex niger do not represent NORs, and therefore are probably derived.
Cytogenetic data permitted the differentiation among four of the six Acromyrmex species studied. Acromyrmex balzani, included in the Moellerius subgenus, showed the largest metacentric chromosome pair with lower size compared with the other species. Acromyrmex echinatior, besides the higher quantity of acrocentric chromosomes, also had interstitial bands in the ST1 pair for the fluorochrome CMA3, differing from the other species, which suggest the possibility of inversion. Acromyrmex niger showed multiple CMA3-positive bands: in the telomeric regions of the short arms of the ST1 pair, in the pericentromeric regions of the long arm of the ST1 pair and in the second largest subtelocentric pair. Acromyrmex rugosus had a greater proportion of metacentric chromosomes compared with the other Acromyrmex and also showed small bands in the telomeric regions of at least three other chromosomes. Acromyrmex coronatus and Acromyrmex disciger could only be cytogenetically differentiated from the other species by slight differences in the morphology that are probably due to the differential growth of heterochromatin on the short arms of the chromosomes.
Five of the six Acromyrmex species studied in this paper were collected in a particular area in the South East of South America. However, Acromyrmex echinatior was collected in Central America, which is more than 5,000 km from the main study area. Moreover, another three species analyzed by Goñi et al. (1983) were collected in Uruguay, expanding the knowledge of South American Acromyrmex. The chromosome number is uniform among different species of Acromyrmex (sensu stricto), although chromosome morphology and banding patterns of these ants allow the identification of some species via their karyotypes. However, karyotype structure of Acromyrmex striatus suggests that it belongs to a different lineage and therefore, according to sequences obtained from nuclear genes, this species does not belong to the “true” Acromyrmex lineage but to the sister group to the remaining leafcutter ants (Cristiano et al. 2013). A new genus therefore could be erected due to the karyotypic features of Acromyrmex which are further supported by combining the already published cytogenetic and molecular data (Cristiano et al. 2013) together with the additional karyological information. In this case, cytogenetics shows its importance as an additional tool in integrative taxonomy.
Conserved chromosome numbers were found in certain ant genera, as in Pogonomyrmex Mayr, 1868 in which 13 of 15 studied species had the same chromosome number. The two other species were transferred to another subgenus Ephebomyrmex Wheeler, 1902 (Taber et al. 1988). Camponotus (Myrmothrix) spp. also presented uniform chromosome number (Mariano et al. 2003), as did the other members of the genera Pheidole Westwood, 1839, Lasius Fabricius, 1804 and Iridomyrmex Mayr, 1862 (reviewed in Lorite and Palomeque 2010). Other animal groups, such as most birds and different genera of insects of the order Lepidoptera also demonstrated conservatism in respect to the chromosome number (White 1973), as did some bee genera, such as Melipona Illiger, 1806 (reviewed in Rocha et al. 2007) and Partamona Schwarz, 1939 (reviewed in Martins et al. 2009).
Our data confirmed uniformity of the chromosome number (2n = 38) in the studied Acromyrmex species. However, chromosomal rearrangements such as heterochromatin growth are likely to be responsible for karyotypic differentiation in this ant group. Location of rDNA clusters of other leafcutter ants (especially Acromyrmex striatus) also needs to be determined using molecular cytogenetic techniques (FISH). Moreover, cytogenetic studies of other members of fungus-growing ants, e.g. of the genus Trachymyrmex Forel, 1893 which represents the sister group to leafcutter ants, will be important for better understanding of chromosomal evolution of this group and Neotropical Formicidae in general.
Acknowledgments
We are grateful to Terezinha M.C. Della Lucia and Danival de Souza for providing larvae of some species and for helpful suggestions on a previous version of the manuscript; to Pedro Lorite and Vladimir Gokhman for valuable suggestions, to Manoel José de Souza for assistance in the field work, to V & M Florestal for their support in the field work at Paraopeba, and to Global Edico for language reviewing. We are grateful to CAPES and CNPq for the scholarships granted to LACB, as well as to CNPq for the research grant to JHCD and MAC. This research was supported by FAPEMIG and FAPESB.
Citation
Barros LAC, de Aguiar HJAC, Mariano CSF, Andrade-Souza V, Costa MA, Delabie JHC, Pompolo SG (2016) Cytogenetic data on six leafcutter ants of the genus Acromyrmex Mayr, 1865 (Hymenoptera, Formicidae, Myrmicinae): insights into chromosome evolution and taxonomic implications. Comparative Cytogenetics 10(2): 229–243. doi: 10.3897/CompCytogen.v10i2.7612
References
- Bacci Jr M, Solomon SE, Mueller UG, Martins VG, Carvalho AO, Vieira LG, Silva-Pinhati ACO. (2009) Phylogeny of leafcutter ants in the genus Atta Fabricius (Formicidae: Attini) based on mitochondrial and nuclear DNA sequences. Molecular Phylogenetics and Evolution 51(3): 427–437. doi: 10.1016/j.ympev.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Barros LAC, Aguiar HJAC, Mariano CSF, Delabie JHC, Pompolo SG. (2010) Cytogenetic characterization of the lower-Attine Mycocepurus goeldii (Formicidae: Myrmicinae: Attini). Sociobiology 56: 57–66. [Google Scholar]
- Barros LAC, Mariano CSF, Pompolo SG, Delabie JHC. (2011) Citogenética de Attini. In: Della Lucia TMC. (Ed.) Formigas-cortadeiras da biotecnologia ao manejo. 1ª. Editora da Universidade Federal de Viçosa, Viçosa, Brazil, 68–79. [In Portuguese] [Google Scholar]
- Barros LAC, Aguiar HJAC, Andrade-Souza V, Mariano CSF, Delabie JHC, Pompolo SG. (2012) Occurrence of pre-nucleolar bodies and 45S rDNA location on the chromosomes of the ant Mycocepurus goeldii (Forel) (Formicidae, Myrmicinae, Attini). Hereditas 149: 50–54. doi: 10.1111/j.1601-5223.2011.02237.x [DOI] [PubMed] [Google Scholar]
- Barros LAC, Mariano CSF, Pompolo SG. (2013) Cytogenetic studies of five taxa of the tribe Attini (Formicidae: Myrmicinae). Caryologia 66(1): 59–64. doi: 10.1080/00087114.2013.780443 [Google Scholar]
- Barros LAC, Teixeira GA, Aguiar HJAC, Mariano CSF, Delabie JHC, Pompolo SG. (2014a) Banding patterns of three leafcutter ant species of the genus Atta (Formicidae: Myrmicinae) and chromosomal inferences. Florida Entomologist 97: 1694–1701. doi: 10.1653/024.097.0444 [Google Scholar]
- Barros LAC, Aguiar HJAC, Mariano CSF, Delabie JHC, Pompolo SG. (2014b) Cytogenetic characterization of the ant Trachymyrmex fuscus Emery, 1934 (Formicidae: Myrmicinae: Attini) with the description of a chromosomal polymorphism. Annales de la Société Entomologique de France 50(1): 1–7. doi: 10.1080/00379271.2013.856201 [Google Scholar]
- Barros LAC, Aguiar HJAC, Teixeira GA, Mariano CSF, Teixeira MC, Delabie JHC, Pompolo SG. (2015) Cytogenetic data on the threatened leafcutter ant Atta robusta Borgmeier, 1939 (Formicidae: Myrmicinae: Attini). Comptes Rendus Biologies 338: 660–665. doi: 10.1016/j.crvi.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Bolton B. (2014) An online catalog of the ants of the world. http://antcat.org [accessed 19 February 2015]
- Brandão CRF, Mayhé-Nunes AJ, Sanhudo CED. (2011) Taxonomia e filogenia das cortadeiras. In: Della Lucia TMC. (Ed.) Formigas-cortadeiras da biotecnologia ao manejo. 1ª. Editora da Universidade Federal de Viçosa, Viçosa, 27–47. [Google Scholar]
- Cardoso DC, Pompolo SG, Cristiano MP, Tavares MG. (2014) The role of fusion in ant chromosome evolution: insights from cytogenetic analysis using a molecular phylogenetic approach in the genus Mycetophylax. PLoS ONE 9: . doi: 10.1371/journal.pone.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano MP, Cardoso DC, Fernandes-Salomão TM. (2013) Cytogenetic and molecular analyses reveal a divergence between Acromyrmex striatus (Roger, 1863) and other congeneric species: taxonomic implications. PLoS ONE 8(3): . doi: 10.1371/journal.pone.0059784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabie JHC, Alves HSR, Strenzel GMR, Carmo AFR, Nascimento IC. (2011) Distribuição das formigas cortadeiras dos gêneros Acromyrmex e Atta no Novo Mundo. In: Della-Lucia TMC. (Ed.) Formigas cortadeiras: da bioecologia ao manejo. 1ª. Editora da Universidade Federal de Viçosa, Viçosa, 80–101. [In Portuguese] [Google Scholar]
- Delabie JHC, Fernandez F, Majer JD. (2012) Editorial Advances in Neotropical Myrmecology. Psyche 2012: 1–3. doi: 10.1155/2012/286273 [Google Scholar]
- Emery C. (1913) Études sur les Myrmicinae. Annales de la Société Entomologique de Belgique 57: 250–262. [Google Scholar]
- Fadini MAM, Pompolo SG. (1996) Cytogenetics of some ant species of the tribe Attini (Hymenoptera, Formicidae) from the region of Viçosa, MG. Revista Brasileira de Genetica 19: 53–55. [Google Scholar]
- Gonçalves CR. (1961) O gênero Acromyrmex no Brasil (Hymenoptera, Formicidae). Studia Entomologica 4: 113–180. [Google Scholar]
- Goñi G, Zolessi LC, Imai HT. (1983) Karyotype of thirteen ant species from Uruguay (Hymenoptera - Formicidae). Caryologia 36: 363–371. doi: 10.1080/00087114.1983.10797677 [Google Scholar]
- Hölldobler B, Wilson EO. (1990) The Ants. Harvard University Press, Harvard, 732 pp. [Google Scholar]
- Imai HT. (1991) Mutability of constitutive heterochromatin (C-bands) during eukaryotic chromosomal evolution and their cytological meaning. Japanese Journal of Genetics 66: 635–661. doi: 10.1266/jjg.66.635 [DOI] [PubMed] [Google Scholar]
- Imai HT, Taylor RW, Crosland MW, Crozier RH. (1988) Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the Minimum Interaction Hypothesis. Japanese Journal of Genetics 63: 159–185. doi: 10.1266/jjg.63.159 [DOI] [PubMed] [Google Scholar]
- Imai HT, Taylor RW, Crozier RH. (1994) Experimental bases for the minimum interaction theory. Chromosome evolution in ants of the Myrmecia pilosula species complex (Hymenoptera: Formicidae: Myrmeciinae). Japanese Journal of Genetics, 137–182. doi: 10.1266/ggs.69.137
- Levan A, Fredga K, Sandberg A. (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52: 201–220. doi: 10.1111/j.1601-5223.1964.tb01953.x [Google Scholar]
- Lorite P, Palomeque T. (2010) Karyotype evolution in ants (Hymenoptera: Formicidae), with a review of the known ant chromosome numbers. Myrmecological News 13: 89–102. [Google Scholar]
- Mariano CSF, Delabie JHC, Campos LAO, Pompolo SG. (2003) Trends in karyotype evolution in the ant genus Camponotus Mayr (Insecta: Hymenoptera: Formicidae). Sociobiology 42: 831–839. [Google Scholar]
- Martins CCC, Duarte OMP, Waldschmidt AM, Alves RMO, Costa MA. (2009) New occurrence of B chromosomes in Partamona helleri (Friese, 1900) (Hymenoptera, Meliponini). Genetics and Molecular Biology 32: 782–785. doi: 10.1590/S1415-47572009005000065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhé-Nunes AJ. (1991) Estudo de Acromyrmex (Hymenoptera, Formicidae) com ocorrência constatada no Brasil: subsídios para uma análise filogenética. Dissertation (Mestrado em Entomologia Agrícola), Universidade Federal de Viçosa, Viçosa Minas Gerais, 122 pp [In Portuguese] [Google Scholar]
- Mayhé-Nunes AJ, Jaffé K. (1998) On the Biogeography of Attini (Hymenoptera: Formicidae). Ecotrópicos 1998: 45–54. [Google Scholar]
- Mehdiabadi NJ, Schultz TR. (2010) Natural history and phylogeny of the fungus-farming ants (Hymenoptera: Formicidae). Myrmecological News 13: 37–55. [Google Scholar]
- Moscone EA, Matzke MA, Matzke AJM. (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105: 231–236. doi: 10.1007/BF02528771 [PubMed] [Google Scholar]
- Murakami T, Fujiwara A, Yoshida MC. (1998) Cytogenetics of ten ant species of the tribe Attini (Hymenoptera, Formicidae) in Barro Colorado Island, Panama. Chromosome Science 2: 135–139. [Google Scholar]
- Rabeling C, Schultz TR, Bacci Jr M, Bollazzi M. (2015) Acromyrmex charruanus: a new inquiline social parasite species of leaf-cutting ants. Insectes Sociaux 62: 335–349. doi: 10.1007/s00040-015-0406-6 [Google Scholar]
- Reed KM, Phillips RB. (1995) Molecular cytogenetic analysis of the double-CMA3 chromosome of lake trout, Salvelinus namaycush. Cytogenetics and Cell Genetics 40: 104–107. doi: 10.1159/000134002 [DOI] [PubMed] [Google Scholar]
- Rocha MP, Pompolo SG, Fernandes A, Campos LAO. (2007) Melipona - Seis décadas de citogenética. Bioscience Journal 23: 111–117. [In Portuguese] [Google Scholar]
- Schweizer D. (1980) Simultaneous fluorescent staining of R bands and specific heterochromatic regions (DA/DAPI-bands) in human chromosomes. Cytogenetics and Cell Genetics 27: 190–193. doi: 10.1159/000131482 [DOI] [PubMed] [Google Scholar]
- Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH. (2010) Integrative taxonomy: a multisource approach to exploring biodiversity. Annual Review of Entomology 55: 421–438. doi: 10.1146/annurev-ento-112408-085432 [DOI] [PubMed] [Google Scholar]
- Schultz TR, Brady SG. (2008) Major evolutionary transitions in ant agriculture. Proceedings of the National Academy of Sciences of the USA 105(14): 5435–5440. doi: 10.1073/pnas.0711024105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner AT. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 83: 304–306. doi: 10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Sumner AT. (1990) Chromosome Banding, Uniwin Hyman, London, 434 pp. [Google Scholar]
- Sumner S, Aanen DK, Delabie JHC, Boomsma JJ. (2004) The evolution of social parasitism in Acromyrmex leaf-cutting ants: a test of Emery´s rule. Insectes Sociaux 51: 37–42. doi: 10.1007/s00040-003-0723-z [Google Scholar]
- Taber SW, Cokendolpher JC, Francke OF. (1988) Karyological study of North American Pogonomyrmex (Hymenoptera: Formicidae). Insectes Sociaux 35: 47–60. doi: 10.5281/zenodo.24794 [Google Scholar]
- Ward PS, Brady SG, Fisher BL, Schultz TR. (2015) The evolution of Myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Systematic Entomology 40: 61–81. doi: 10.1111/syen.12090 [Google Scholar]
- White MJD. (1973) Animal cytology and evolution. Cambridge, 961 pp. [Google Scholar]