Abstract
Background
Circadian rhythm disruption is a prevalent feature of modern day society that is associated with an increase in pro-inflammatory diseases and there is a clear need for a better understanding of the mechanism(s) underlying this phenomenon. We have previously demonstrated that both environmental and genetic circadian rhythm disruption causes intestinal hyperpermeability and exacerbates alcohol-induced intestinal hyperpermeability and liver pathology. The intestinal microbiota can influence intestinal barrier integrity and impact immune system function; thus, in the current study, we sought to determine if genetic alteration of the core circadian clock gene, Clock, altered the intestinal microbiota community.
Methods
Male ClockΔ19 mutant mice (mice homozygous for a dominant-negative mutant allele) or littermate wild-type mice were fed one of three experimental diets: (1) a standard chow diet, (2) an alcohol-containing diet, or (3) an alcohol-control diet in which the alcohol calories were replaced with dextrose. Stool microbiota was assessed with 16S ribosomal RNA gene amplicon sequencing.
Results
The fecal microbial community of Clock mutant mice had lower taxonomic diversity, relative to wild type mice and the ClockΔ19 mutation was associated with intestinal dysbiosis when mice were fed either the alcohol-containing or the control diet. We found that alcohol consumption significantly altered the intestinal microbiota in both wild type and Clock mutant mice.
Conclusion
Our data support a model by which circadian rhythm disruption by the ClockΔ19 mutation perturbs normal intestinal microbial communities and this trend was exacerbated in the context of a secondary dietary intestinal stressor.
Keywords: clock mutation, circadian rhythm disruption, stool microbiota, dysbiosis, alcohol
Introduction
Circadian rhythms regulate nearly every aspect of our lives including behavior, biological systems, organ function, cellular, and molecular processes. Disruption of these rhythms can negatively impact health and immune system function which may contribute to the development or progression of inflammatory-mediated diseases following circadian rhythm disruption (Castanon-Cervantes et al., 2010; Cermakian et al., 2013). One important mechanism that may contribute to immune system dysfunction following the disruption of normal circadian rhythmicity is through effects on the intestinal microbiota.
The intestinal microbiota regulates physiological processes and it is becoming clear that disruption of the normal intestinal flora (i.e., intestinal dysbiosis) is associated with numerous diseases that are promoted or exacerbated by inflammation (Albenberg and Wu, 2014; Chan et al., 2013; Hollister et al., 2014; Kamada and Nunez, 2014). The intestinal microbiota shares a symbiotic relationship with the host whereby the host supplies food sources for the microbiota and the microbiota produce byproducts that can benefit the host. For example, short chain fatty acids (SCFA) produced by certain bacteria can fortify intestinal barrier integrity (Macia et al., 2012; Viladomiu et al., 2013) and a reduction in these SCFA-producing bacteria can negatively impact barrier integrity, thereby allowing pro-inflammatory bacteria products (e.g., lipopolysaccharide (LPS)) to pass into the systemic circulation where they can cause and promote inflammation-mediated diseases. Recent studies also show that microbial products, especially SCFA, are critical for maintaining normal mucosal immune function (Arpaia et al., 2013; Ivanov and Honda, 2012; Smith et al., 2013).
We have shown that circadian rhythm disruption induced by repeated changes in the 24h light:dark cycle (i.e., environmental disruption) or ClockΔ19 mutation-induced changes in the molecular circadian clock (i.e., genetic disruption) promotes intestinal hyperpermeability and markedly exacerbates alcohol-induced intestinal leakiness, endotoxemia, inflammation, and hepatic pathology (Summa et al., 2013). Although it is known that alcohol changes the intestinal microbiota, the consequence of altered circadian organization on the composition and structure of the intestinal microbiota is relatively unknown. We recently demonstrated that environmental circadian rhythm disruption causes intestinal dysbiosis when combined with a high-fat, high-sugar diet (Voigt et al., 2014) and others have shown that perturbation of normal circadian rhythms in the host can impact the intestinal microbiota (Leone et al., 2015; Thaiss et al., 2014). In the current study, we expanded these findings and investigated the consequence of genetic circadian rhythm disruption, achieved via mutation of the Clock gene, on the intestinal microbiota and also evaluated how circadian rhythm disruption may impact alcohol-induced dysbiosis.
Materials and Methods
Animals and Experimental Protocol
Mice homozygous for the dominant negative Δ19 allele of the Clock gene (ClockΔ19 mutant) were used to study genetic disruption of circadian rhythms (Antoch et al., 1997; King et al., 1997; Vitaterna et al., 1994) and wild-type littermates were used as controls. The mutation was induced in C57BL/6J mice; thus, all mice were coisogenic C57BL/6J. Clock mutant mice have a mutation in the canonical circadian regulatory gene, Clock, which impacts the period, precision, and persistence of circadian rhythms at the molecular, cellular, tissue, and behavioral levels (Herzog et al., 1998; Miller et al., 2007; Vitaterna et al., 2006). This model has been well-validated and widely used to explore the link between circadian rhythms and a variety of biological processes (McClung, 2007; Naylor et al., 2000; Turek et al., 2005).
Briefly, young adult (7–9 week old) male mice of each genotype were individually housed on a 12 hour light:12 hour dark light cycle (i.e., 12:12 LD) and fed a standard rodent chow diet (Harlan Teklad Global; Madison, WI) and subsequently placed in either the alcohol-containing or the alcohol-control experimental group. The alcohol diet is comprised of 29% of total daily calories from alcohol (4.5% v/v) and the control group received an isocaloric, alcohol-free diet with the alcohol calories being replaced with dextrose. This diet is a modification of the Leiber-Dicarli diet that has been widely used to study alcohol-induced effects on the liver. The diet is prepared fresh daily and provided to mice in a specialized feeding tube that allows for daily monitoring of food intake. There is a two-week introduction to alcohol (i.e., an alcohol ramp, 3–29% of daily calories from alcohol), followed by eight weeks on the alcohol diet (29% of daily calories from alcohol, 4.5% v/v). Mice were weighed bi-weekly and food consumption was noted daily. It is well-established that Clock mutant mice have metabolic abnormalities Laposky et al, 2008; Turek et al, 2005) and in our study Clock mutant mice did weigh more (significant effect of genotype, p<0.00) and tended to consume more food on a daily basis (significant effect of genotype, p=0.01) than their wild-type counterparts. However, when food consumption was normalized to body weight there was no effect of genotype on food consumption in control-fed or alcohol-fed mice (p>0.05, Supplemental Fig 1).
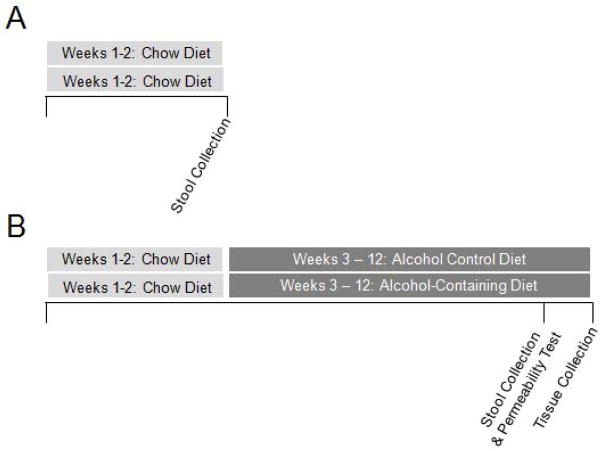
Stool samples were collected after two weeks on the standard rodent chow diet (Fig. 1A) or after ten weeks of an alcohol-containing diet or the alcohol-control diet (Fig. 1B). Stool samples were collected from each mouse over a five hour period (ZT0-ZT5, ZT0=lights on) during a test to assess intestinal barrier integrity (see Methods section below, Correlation Analysis-Intestinal Barrier Integrity). Stool samples were then subsequently frozen and stored at −80°C until use. These samples were then used in sequencing for determination of intestinal microbial community composition.
Figure 1.
Experimental Protocol. Young adult (7–9 week) male ClockΔ19 mutant mice and wild type littermates (C57BL/6J) were housed individually and maintained on a 12 hour light:12 hour dark light cycle (i.e., 12:12 LD). (A) Mice were fed the standard chow (Harlan Teklad Global) and stool was collected two weeks later for analysis. Mice were placed in a metabolic chamber and stool was collected for five hours (ZT0-ZT5). (B) Mice were fed standard rodent chow for the first two weeks of the study then fed either an alcohol-control diet or an alcohol-containing diet (two weeks of a gradual increase in alcohol concentration, followed by eight weeks at the full alcohol dose, 29% of total daily calories from alcohol (4.5% v/v)) for 10 weeks. Mice were placed into a metabolic chamber for five hours (ZT0-ZT5) and urine and stool were collected for analysis of intestinal barrier integrity and stool microbiota analysis, respectively. One day later mice were euthanized at ZT6 and proximal colon tissue was collected for analysis.
Mice were euthanized by conscious decapitation. Serum collected at the time of the tissue harvest evaluated for LPS using a commercially available ELISA kit (MBS-722939; MyBioSource, San Diego, CA) according to the manufacturer’s instructions. Formalin-fixed liver was stained with hematoxylin and eosin (H&E) and a blind assessment of liver steatosis was conducted by a gastrointestinal pathologist. Steatosis severity was scored as percent hepatocyte involvement (0 = <5%, 1 = 5–33%, 2 = 34–66%, 3 = >67%), corresponding to the fraction of lipid-containing hepatocytes.
Analysis of Microbial Community Structure
Genomic DNA was extracted from mouse fecal samples (Fast DNA SPIN Kit for Soil, #116560200, MP Biomedicals; Solon, OH). PCR amplification was performed targeting bacterial small subunit (SSU or 16S) ribosomal RNA (rRNA) genes with primers Gray28F/Gray519R, as described previously (Ishak et al., 2011). Sequencing reactions were performed on a Roche 454 FLX instrument (Roche; Indianapolis, IN) with titanium reagents and procedures, a one-step PCR, and a mixture of Hot Start and Hot Star high-fidelity Taq polymerases.
Raw sequence data were imported into the software package CLC genomics workbench (v8.0; CLC Bio, Qiagen; Boston, MA). Subsequently, merged data were quality trimmed (Q15), and sequences shorter than 350 bases were removed. The remaining sequences were exported as FASTA and processed through the software package QIIME (v1.8.0) (Caporaso et al., 2010). Briefly, sequences were screened for chimeras using the usearch61 algorithm (Edgar, 2010) and putative chimeric sequences were removed from the dataset. Data were pooled, renamed, and clustered into operational taxonomic units (OTU) at 97% similarity using the usearch61 de novo OUT picking algorithm. Representative sequences from each OTU were extracted, and these sequences were classified using the “assign taxonomy” algorithm implementing the uclust algorithm, utilizing the Greengenes reference OTU build (v13_8). A biological observation matrix (BIOM);(McDonald et al., 2012) was generated at taxonomic levels from phylum to genus using the “make OTU table” algorithm. Subsequently, each sample sequence set was sub-sampled to 1,250 sequences to reduce analytical issues associated with variable library size (Gihring et al., 2012). The sub-sampled BIOMs used for statistical analysis and visualization using group-average clustering, non-metric multidimensional scaling (NMDS), analysis of similarity (ANOSIM) and similarity percentages (SIMPER) analyses, as described previously (Voigt et al., 2014). Differences in the relative abundance of individual taxa between a priori defined groups were tested for significance using the “group significance” algorithm, implemented within QIIME. Tests were performed using the non-parametric Kruskal-Wallis test, generating a Benjamini-Hochberg false-discovery rate (FDR) corrected p-value. Taxa with an average abundance of <1% across the entire sample set were removed from such analyses. In silico community functional predications were performed using the software package PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) and significant differences in KEGG ortholog (KO) abundances between groups were identified and analyzed using the KEGG Mapper pathway search function.
Correlation Analysis
We sought to determine if intestinal dysbiosis associated with alcohol or circadian rhythm disruption was associated with changes in gene expression in the proximal colon. Tissue samples were collected one day after the stool collection (Fig. 1B). Mice were euthanized by conscious decapitation and proximal colon samples placed into RNA Later (Qiagen; Valencia, CA), frozen in liquid nitrogen, and stored at −80°C until use. Proximal colon samples were prepared as tissue homogenates and mRNA levels were determined using a Luminex-based custom multiplex bead array. This QuantiGene 2.0 Multiplex Assay (Affymetrix; Santa Clara, CA) was used to assess the level of expression of genes of interest. Gene expression was normalized to the average of two housekeeping genes (Phosphoglycerate kinase 1 (Pgk1) and polymerase polypeptide A (Polr2a)). Thus, mice in the following groups were included in the correlation analysis: wild type alcohol-control diet, wild type alcohol diet, ClockΔ19 mutant alcohol-control diet, and ClockΔ19 mutant alcohol diet.
Relative RNA level for each gene was correlated with bacterial taxa that were greater than 5% of the total microbiota community at the genus level (i.e., Lactobacillus, Clostridium, Allobaculum, Akkermansia). P-values were false discovery rate (FDR) corrected for multiple comparisons. In vivo assessment of intestinal permeability was conducted as described previously (Keshavarzian et al., 2009; Shaikh et al., 2015; Summa et al., 2013). Briefly, mice were fasted for eight hours prior to the test, which was performed at ZT0 (i.e., ZT0=lights on). A 200 μL solution containing lactulose (3.2 mg), sucrose (0.45 mg), sucralose (0.45 mg) and mannitol (0.9 mg) was administered orally (i.e., gavage), and 2 mL 0.9% saline was administered subcutaneously to promote urine production. Urine was collected for five hours and intestinal barrier integrity was determined by measuring urinary sugar concentration using gas chromatography, enabling calculation of the amount of orally administered sugar excreted in the urine over five hours. Intestinal permeability was measured after eight weeks on the full alcohol concentration (i.e., 29% of total daily calories from alcohol). Thus, mice in the following groups were included in the correlation analysis: wild type alcohol-control diet, wild type alcohol diet, ClockΔ19 mutant alcohol-control diet, and ClockΔ19 mutant alcohol diet.
Percent urinary sugar excretion was correlated with bacterial taxa that were greater than 5% of the total microbiota community at the genus level (i.e., Lactobacillus, Clostridium, Allobaculum, Akkermansia). P-values were false discovery rate (FDR) corrected for multiple comparisons.
Results
Impact of the ClockΔ19 Mutation - Chow-fed Mice
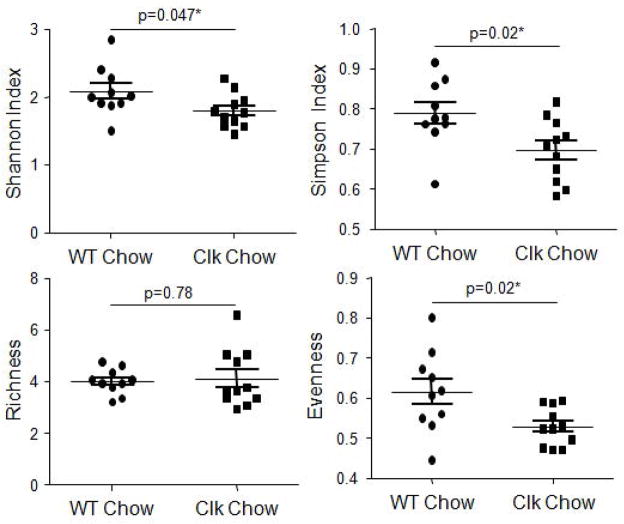
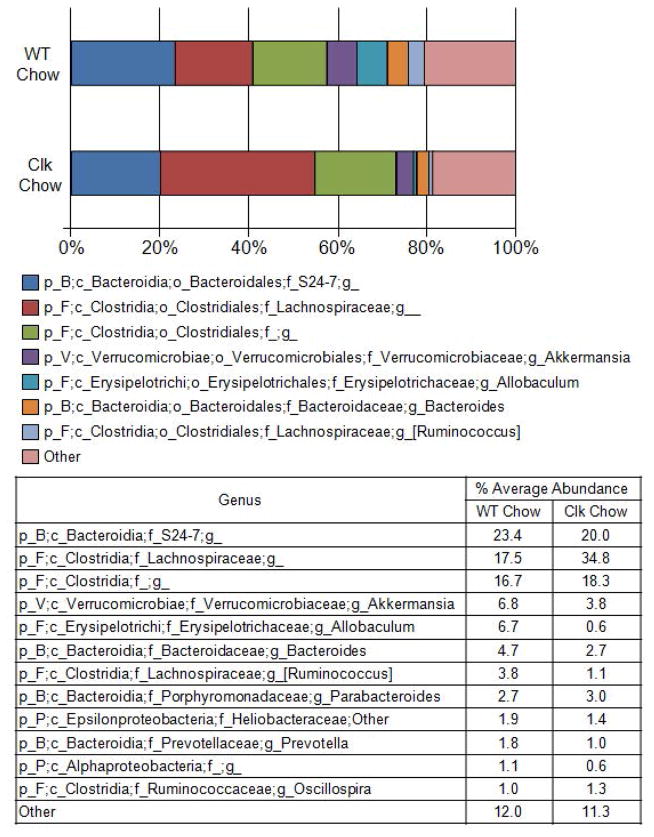
We have shown that environmental disruption of circadian rhythms via changes in the light:dark cycle has no impact on the community structure of the intestinal microbiota when mice are fed a standard chow diet (Voigt et al., 2014). We sought to determine if this effect was specific to environmental disruption of circadian rhythms during adult life (12 weeks) or if it would be also observed in genetically disrupted mice where circadian homeostasis has been disrupted since birth. Following two weeks maintenance on standard chow diet, fecal samples were collected from both wild type and Clock mutant mice, and microbial communities were characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons (Fig. 1A). Analysis of the rRNA gene amplicons demonstrated that the gut microbiota of mice with the ClockΔ19 mutation had lower evenness and diversity (Shannon and Simpson indices) while microbial richness was not significantly different (Fig. 2). The effects on evenness and diversity were observed at the taxonomic levels of genus, family, order, and class (Supplementary Table 1). Similar to the environmental circadian rhythm disruption, no significant differences in community structure were observed between wild type and ClockΔ19 mice fed the standard rodent chow diet using analysis of similarity (ANOSIM) (Table 1) or a predictive assessment of microbial community functional potential (PICRUSt) (Table 2). Although not significant, the average relative abundance of bacteria from the family Lachnospiraceae was higher in mutant mice (17.5–34.8%) (Fig. 3), and this shift contributed to an increase in the Firmicutes/Bacteroidetes ratio in ClockΔ19 mice compared to the wild type mice (Supplementary Fig 2A.).
Figure 2.
The ClockΔ19 mutation reduces bacterial diversity and evenness. Stool was collected from wild type (n=10) and ClockΔ19 mutant mice (n=10) fed a standard rodent diet (Harlan Teklad Global) for two weeks and the stool was characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons. Analysis of the rRNA gene amplicons was used to compare bacterial diversity (Shannon and Simpson indices), richness, and evenness. Genus level comparisons are shown (mean ± standard error of the mean (SEM)) and p-values indicate between group analyses using a Student’s t-test followed by a false discovery rate correction for multiple comparisons. WT, wild type; Clk, ClockΔ19 mutant mice; Chow, standard rodent chow diet.
Table 1.
Mutation of the ClockΔ19 gene impacts stool microbiota community structure when mice were fed an alcohol-control diet (i.e., high-fat, high-sugar) or an alcohol-containing diet. Bray-Curtis metric was calculated in a pair-wise fashion and analysis of similarity (ANOSIM) was performed. Global R is based on ANOSIM analysis. P-values are calculated based on a permutations analysis, employing 999 permutations and p –values were false discovery rate corrected for multiple comparisons. WT, wild type mice, ClockΔ19 mutant mice; Chow, standard rodent chow diet; Control, alcohol-control diet (i.e., high-fat, high-sugar diet); Alcohol, alcohol-containing diet.
| Taxonomic level | Global R | p-value | ||
|---|---|---|---|---|
| Standard Chow Diet | ||||
| WT Chow (n=10) | ClockΔ19 Chow (n=10) | Phylum | 0.04 | 0.24 |
| Class | 0.04 | 0.24 | ||
| Order | 0.03 | 0.26 | ||
| Family | 0.01 | 0.28 | ||
| Genus | 0.01 | 0.30 | ||
| Alcohol Control Diet | ||||
| WT Control (n=8) | ClockΔ19 Control (n=8) | Phylum | 0.10 | 0.13 |
| Class | 0.22 | 0.03* | ||
| Order | 0.22 | 0.03* | ||
| Family | 0.24 | 0.02* | ||
| Genus | 0.21 | 0.02* | ||
| Alcohol Diet | ||||
| WT Alcohol (n=8) | ClockΔ19 Alcohol (n=8) | Phylum | 0.21 | 0.01* |
| Class | 0.22 | 0.01* | ||
| Order | 0.22 | 0.02* | ||
| Family | 0.25 | 0.01* | ||
| Genus | 0.30 | <0.00* | ||
Table 2.
The ClockΔ19 mutation significantly impacts KEGG ortholog abundances in alcohol-fed mice. KEGG ortholog abundance was compared between groups using a Student’s t-test and p-values were false discovery rate corrected for multiple comparisons. The number of significant between group differences was tabulated for each KEGG ortholog category. WT, wild type mice; Clk, ClockΔ19 mutant mice; Chow, standard rodent chow diet; Control, alcohol-control diet (i.e., high-fat, high-sugar diet); Alcohol, alcohol-containing diet.
| WT Chow v Clk Chow | WT Control v Clk Control | WT Alcohol v Clk Alcohol | ||
|---|---|---|---|---|
|
| ||||
| Cellular Processes | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 2 (20) | |
|
| ||||
| Environmental Information Processing | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 2 (15) | |
|
| ||||
| Genetic Information Processing | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 17 (65) | |
|
| ||||
| Human Diseases | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 9 (29) | |
|
| ||||
| Metabolism | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 34 (25) | |
|
| ||||
| Organismal Systems | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 5 (28) | |
|
| ||||
| Unclassified | Pathways Increased (%) | 0 (0) | 0 (0) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | 6 (22) | |
Figure 3.
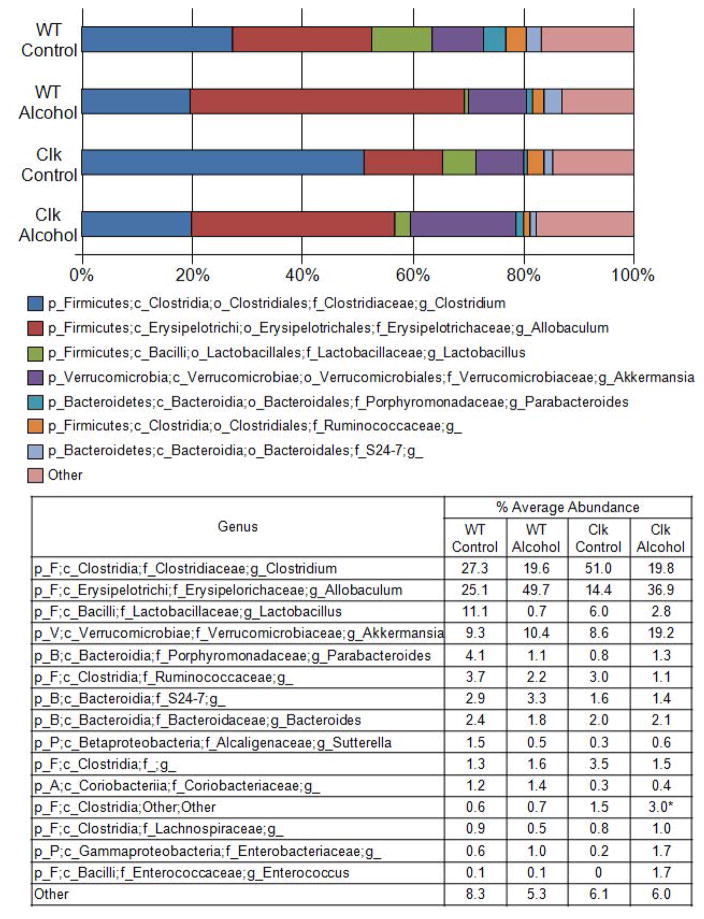
Percent average abundance of the most abundant taxon for wild type and ClockΔ19 mice fed a standard rodent chow diet. Stool was collected from wild type (n=10) and ClockΔ19 mutant mice (n=10) fed a standard rodent diet (Harlan Teklad Global) for two weeks and the stool was characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons. The bar graph graphically represents and the table summarizes the average relative abundance of classified bacterial SSU rRNA gene amplicons belonging to the most abundant taxon at the genus level. WT, wild type mice; Clk, ClockΔ19 mutant mice; Chow, standard rodent chow diet; p_, phylum; c_, class; o_, order; f_, family; g_, genus; B, Bacteroidetes; F, Firmicutes; P, Proteobacteria; V, Verrucomicrobia.
Impact of the ClockΔ19 Mutation – Alcohol-Control Diet-Fed Mice
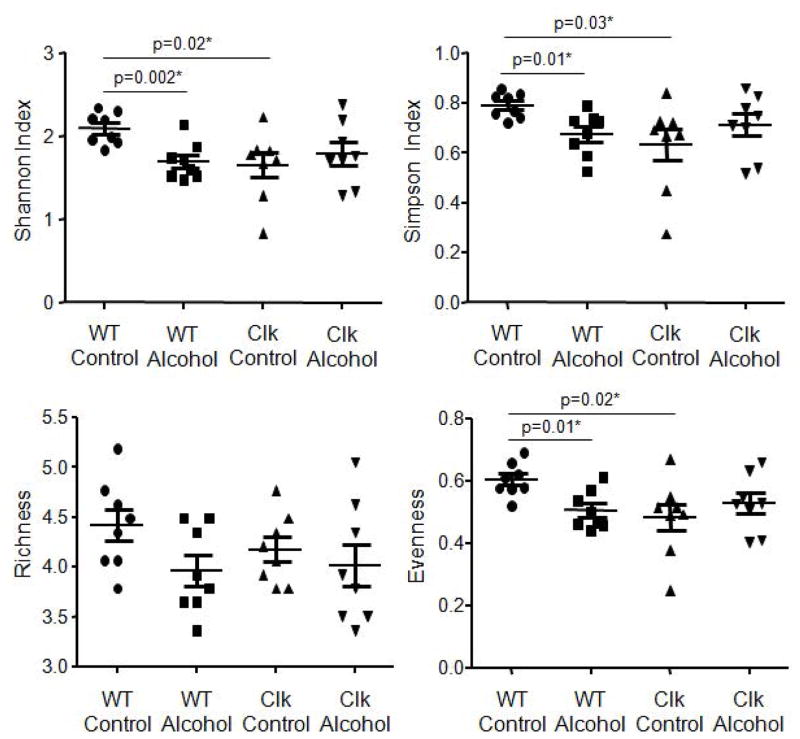
Our group has demonstrated that environmental circadian rhythm disruption significantly impacts stool microbiota composition when mice are fed an alcohol-control diet (i.e., high-fat, high-sugar diet, 36% protein, 29% carbohydrate (dextrose), and 35% fat)) (Voigt et al., 2014) and in the current study we investigated if the same was true in ClockΔ19 mutant mice. Microbial community analyses were performed on stool samples collected following 10 weeks of alcohol-control diet feeding (Fig. 1B). Microbial community diversity (Shannon and Simpson indices) and evenness were significantly lower in ClockΔ19 mice fed the alcohol-control diet relative to wild type mice fed the same diet (Fig. 4, Supplementary Table 1). Similarly, microbial community structure was significantly different between alcohol-control diet-fed ClockΔ19 mice compared to wild type mice fed the alcohol-control diet, and these differences were observed at all taxonomic levels except phylum (Table 1). Although substantial shifts in individual taxa were observed between wild type and Clock mutant mice, these shifts were not significant when adjusted with the false-discovery rate correction nor were any differences revealed using a predicted functional analysis (PICRUSt) (Table 2). Nonetheless, alcohol-control diet-fed ClockΔ19 mice had communities dominated by bacteria from the genus Clostridum, and relatively low abundance of bacteria from the genus Allobaculum (class Erysipelotrichi) compared to their wild type counterparts (Fig. 5). The overall changes in the community structure contributed to a slight, but not significant, increase in the Firmicutes/Bacteroidetes ratio in ClockΔ19 mice fed the alcohol-control diet compared to wild type mice (Supplementary Fig 2B.).
Figure 4.
The ClockΔ19 mutation and alcohol consumption reduce bacterial diversity and evenness. Stool was collected from wild type and ClockΔ19 mutant mice fed a standard rodent diet (Harlan Teklad Global) for two weeks followed by 10 weeks on an alcohol-control (wild type n=8, ClockΔ19 mutant n=8) or an alcohol-containing (wild type n=8, ClockΔ19 mutant n=8) experimental diet and the stool was characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons. Analysis of the rRNA gene amplicons was used to compare bacterial diversity (Shannon and Simpson indices), richness, and evenness. Genus level comparisons are shown (mean ± standard error of the mean (SEM)) and p-values indicate between group analyses using a Student’s t-test followed by a false discovery rate correction for multiple comparisons. WT, wild type; Clk, ClockΔ19 mutant mice; Chow, standard rodent diet. WT, wild type mice; Clk, ClockΔ19 mutant mice; Control, alcohol-control diet; Alcohol, alcohol-containing diet.
Figure 5.
Percent average abundance of the most abundant taxon for wild type and ClockΔ19 mice fed either an alcohol-control diet or an alcohol-containing diet. Stool was collected from wild type and ClockΔ19 mutant mice fed a standard rodent diet (Harlan Teklad Global) for two weeks followed by 10 weeks on an alcohol-control (wild type n=8, ClockΔ19 mutant n=8) or an alcohol-containing (wild type n=8, ClockΔ19 mutant n=8) experimental diet. Stool was characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons. The bar graph graphically represents and the table summarizes the average relative abundance of classified bacterial SSU rRNA gene amplicons belonging to the most abundant taxon at the genus level. WT, wild type mice; Clk, ClockΔ19 mutant mice; Control, alcohol-control diet; Alcohol, alcohol-containing diet; p_, phylum, c_, class; o_, order; f_, family, g_genus; A, Actinobacteria; B, Bacteroidetes; F, Firmicutes; P, Proteobacteria; V, Verrucomicrobia.
Impact of the ClockΔ19 Mutation - Alcohol-fed Mice
Chronic alcohol feeding significantly decreased microbiota community evenness and diversity in wild type mice but the ClockΔ19 mutation did not further impact microbial community diversity, richness, or evenness (Fig. 4, Supplementary Table 1). However, community structure was significantly different between ClockΔ19 mice compared to wild type mice fed the alcohol-containing diet, and these differences were observed at all taxonomic levels (Table 1). Only a single taxon was significantly altered by the ClockΔ19 mutation in alcohol-fed mice, which was an increase in an unidentified genus within the order Clostridiales (Firmicutes, class Clostridia, p<0.04) (Fig. 5).
Predictive assessment of the microbial community functional potential (PICRUSt) was performed and revealed many significant differences in the predicted function of the microbial population between alcohol-fed wild type and alcohol-fed ClockΔ19 mutant mice. These changes were dominated by a decrease in pathways including cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, organismal systems, as well as additional unclassified pathways (Table 2, Supplementary Table 2). ClockΔ19 mice are reported to have metabolic abnormalities (Turek et al., 2005) and indeed we identified 34 metabolic pathways that were significantly decreased by the ClockΔ19 mutation in alcohol-fed mice. Specifically, we identified a decrease in energy metabolism, metabolism of amino acids, metabolism of terpenoids and polyketides, metabolism of carbohydrates, metabolism of cofactors and vitamins, metabolism of nucleotides, and lipid metabolism. In addition, biosynthesis of other secondary metabolites, xenobiotics degradation and metabolism, glycan biosynthesis and metabolism, and enzyme families were significantly different between wild type and ClockΔ19 mice fed an alcohol-containing diet, with the majority of these pathways exhibiting a significant decrease in the ClockΔ19 mice compared to their wild type counterparts (Supplementary Table 2).
Impact of Alcohol
Alcohol significantly reduced evenness and diversity in wild type mice; however, this effect was not observed in ClockΔ19 mutant mice (Fig. 4, Supplementary Table 3). Microbiota community structure was altered by alcohol in both wild type and ClockΔ19 mice (Table 3); but, the specific changes induced by alcohol were different in each genotype (Table 4). The microbiome of alcohol-fed wild type mice had significantly higher relative abundance of bacteria from the genus Allobaculum and lower relative abundance of bacteria from the genus Lactobacillus compared to wild type mice fed the alcohol-control diet. ClockΔ19 mutant mice fed the alcohol-containing diet demonstrated a significant reduction in bacteria in the class Clostridia compared ClockΔ19 mutant mice fed the alcohol-control diet (Fig. 5, Table 4).
Table 3.
Alcohol impacts stool microbiota community structure in both wild type and ClockΔ19 mutant mice. Bray-Curtis metric was calculated in a pair-wise fashion and analysis of similarity (ANOSIM) was performed. Global R is based on ANOSIM analysis. P-values are calculated based on a permutations analysis, employing 999 permutations and p-values were false discovery rate corrected for multiple comparisons. WT, wild type mice, ClockΔ19 mutant mice; Control, alcohol-control diet (i.e., high-fat, high-sugar diet); Alcohol, alcohol-containing diet.
| Taxonomic level | Global R | p-value | ||
|---|---|---|---|---|
| Wild Type Mice | ||||
| WT Control (n=8) | WT Alcohol (n=8) | Phylum | 0.01 | 0.44 |
| Class | 0.46 | <0.00* | ||
| Order | 0.45 | 0.01* | ||
| Family | 0.41 | <0.00* | ||
| Genus | 0.363 | <0.00* | ||
| ClockΔ19 Mutant Mice | ||||
| ClockΔ19 Control (n=8) | ClockΔ19 Alcohol (n=8) | Phylum | 0.19 | 0.02* |
| Class | 0.32 | <0.00* | ||
| Order | 0.32 | <0.00* | ||
| Family | 0.40 | <0.00* | ||
| Genus | 0.379 | <0.00* | ||
Table 4.
Alcohol significantly impacts stool microbiota community structure in both wild type and ClockΔ19 mutant mice. Abundance of each taxon was compared between a priori defined comparisons using a Student’s t-test and p-values were false discovery rate corrected for multiple comparisons. WT, wild type mice; Clk, ClockΔ19 mutant mice; Control, alcohol-control diet; Alcohol, alcohol-containing diet; NC, no change; p_, phylum; c_, class; o_, order; f_, family; g_, genus; B, Bacteroidetes; F, Firmicutes; P, Proteobacteria; V, Verrucomicrobia..
| WT Control v WT Alcohol | Clk Control v Clk Alcohol | |
|---|---|---|
| Class | ||
| p_F, c_Clostridia | NC | ↓ (p=0.03*) |
| p_F, c_Erysipelotrichi | ↑ (p=0.02*) | NC |
| p_F, c_Bacilli | ↓ (p=0.01*) | NC |
| Order | ||
| P_F, c_Clostridia, o_Clostridiales | NC | ↓ (p=0.03*) |
| p_F, c_Erysipelotrichi, o_Erysipelotrichales | ↑ (p=0.02*) | NC |
| p_F, c_Bacilli, o_Lactobacillales | ↓ (p=0.01*) | NC |
| Family | ||
| p_F, c_Clostridia, o_Clostridiales, f_Clostridiaceae | NC | ↓ (p=0.04*) |
| p_F, c_Erysipelotrichi, o_Erysipelotrichales, f_Erysipelotrichaceae | ↑ (p=0.03*) | NC |
| p_F, c_Bacilli, o_Lactobacillales, f_Lactobacillaceae | ↓ (p=0.01*) | NC |
| p_F, c_Clostridia, o_Clostridiales, f_Ruminococcaceae | NC | ↓ (p=0.02*) |
| p_F, c_Clostridia, o_Clostridiales, f_Lachnospiraceae | ↓ (p=0.02*) | NC |
| Genus | ||
| p_F, c_Erysipelotrichi, o_Erysipelotrichales, f_Erysipelotrichaceae, g_Allobaculum | ↑ (p=0.048*) | NC |
| p_F, c_Bacilli, o_Lactobacillales, f_Lactobacillaceae, g_Lactobacillus | ↓ (p=0.01*) | NC |
Predictive assessment of the microbial community functional potential (PICRUSt) was performed and while numerous pathways were altered by alcohol in wild type mice no pathways were significantly altered by alcohol in ClockΔ19 mice (Table 2). Alcohol-induced changes in wild type mice were characterized by an increase in pathways including cellular processes, environmental information processing, genetic information processing, human diseases, organismal systems, metabolism, as well as additional unclassified pathways (Table 5, Supplementary Table 4). To further characterize these changes, we used PICRUSt to evaluate bacterial secretion systems and lipopolysaccharide (LPS) biosynthesis as these are factors that can influence intestinal barrier integrity and inflammation. Seven bacterial secretion systems in the general secretory pathway (i.e., sec-SRP) were more abundant in wild type alcohol-fed mice and one type IV secretion system (i.e., VirD4) was also significantly increased (Table 6). In addition, two of three pathways in lipopolysaccharide (LPS) biosynthesis were significantly upregulated in alcohol-fed mice (Table 6). Evaluation of serum LPS levels indicated there was not a significant difference between control- and alcohol-fed wild type mice (Student’s t-test, p>0.05, data not shown). We have previously demonstrated that serum LPS rhythms exhibit a robust circadian pattern (Summa et al, 2013); thus, analysis of serum LPS at a single time point may have obscured between group differences.
Table 5.
Alcohol significantly impacts KEGG ortholog abundances in wild type mice. KEGG ortholog abundance was compared between groups using a Student’s t-test and p-values were false discovery rate corrected for multiple comparisons. The number of significant between group differences was tabulated for each KEGG ortholog category. WT, wild type mice; Clk, ClockΔ19 mutant mice; Chow, standard rodent chow diet; Control, alcohol-control diet (i.e., high-fat, high-sugar diet); Alcohol, alcohol-containing diet.
| WT Control v WT Alcohol | Clk Control v Clk Alcohol | ||
|---|---|---|---|
|
| |||
| Environmental Information Processing | Pathways Increased (%) | 10 (77) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | |
|
| |||
| Cellular Processes | Pathways Increased (%) | 3 (30) | 0 (0) |
| Pathways Decreased (%) | 1 (10) | 0 (0) | |
|
| |||
| Genetic Information Processing | Pathways Increased (%) | 23 (88) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | |
|
| |||
| Human Diseases | Pathways Increased (%) | 10 (32) | 0 (0) |
| Pathways Decreased (%) | 1 (3) | 0 (0) | |
|
| |||
| Metabolism | Pathways Increased (%) | 88 (65) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | |
|
| |||
| Organismal Systems | Pathways Increased (%) | 7 (39) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | |
|
| |||
| Unclassified | Pathways Increased (%) | 20 (74) | 0 (0) |
| Pathways Decreased (%) | 0 (0) | 0 (0) | |
Table 6.
Alcohol significantly impacts the KEGG ortholog abundances of bacterial secretion systems and lipopolysaccharide biosynthesis in wild type mice. KEGG ortholog abundance was compared between groups using a Student’s t-test and p-values were false discovery rate corrected for multiple comparisons. The number of significant between group differences was tabulated for each KEGG ortholog category. Values are mean ± standard error of the mean. WT, wild type mice; Clk, ClockΔ19 mutant mice; Chow, standard rodent chow diet; Control, alcohol-control diet (i.e., high-fat, high-sugar diet); Alcohol, alcohol-containing diet.
| KEGG Ortholog | WT Control (n=8) | WT Alcohol (n=8) | p-Value | |
|---|---|---|---|---|
| Bacterial Secretion System | ||||
| K03070 | preprotein translocase subunit SecA (sec-SRP) | 1701±252 | 3325±302 | 0.01 |
| K03073 | preprotein translocase subunit SecE (sec-SRP) | 938±95 | 1621±146 | 0.01 |
| K03075 | preprotein translocase subunit SecG (sec-SRP) | 1111±130 | 1931±162 | 0.01 |
| K03076 | preprotein translocase subunit SecY (sec-SRP) | 1497±210 | 2979±275 | 0.01 |
| K03106 | signal recognition particle subunit SRP54 (sec-SRP) | 1132±132 | 1948±162 | 0.01 |
| K03110 | fused signal recognition particle receptor (sec-SRP) | 1080±132 | 1947±162 | 0.01 |
| K03205 | type IV secretion system protein VirD4 (type IV) | 968±111 | 1735±156 | 0.01 |
| K03217 | preprotein translocase subunit YidC (sec-SRP) | 1203±138 | 1952±162 | 0.02 |
| Lipopolysaccharide Biosythesis | ||||
| K02840 | UDP-D-galactose:(glucosyl) LPS alpha-1,6-D-galactosyltransferase [EC:2.4.1.-] | 25±6 | 6±2 | 0.045 |
| K03271 | phosphoheptose isomerase [EC:5.-.-.-] | 642±136 | 1464±144 | 0.01 |
| K07031 | None | 394±91 | 1068±124 | 0.01 |
Functional Assessment of Intestinal Microbiota on the Host
Liver tissue was collected one day following stool collection (Fig. 1). Evaluation of the liver revealed that steatosis (the fraction of lipid containing hepatocytes) was significantly increased in groups demonstrating intestinal dysbiosis (Fig. 6). Both genotype (F(1,26)=8.21, p=0.01) and diet (F(1,26)=19.30, p<0.00) had a significant effect on the liver pathology suggesting that intestinal dysbiosis may contribute to fatty liver associated with alcohol consumption and circadian rhythm disruption.
Figure 6.
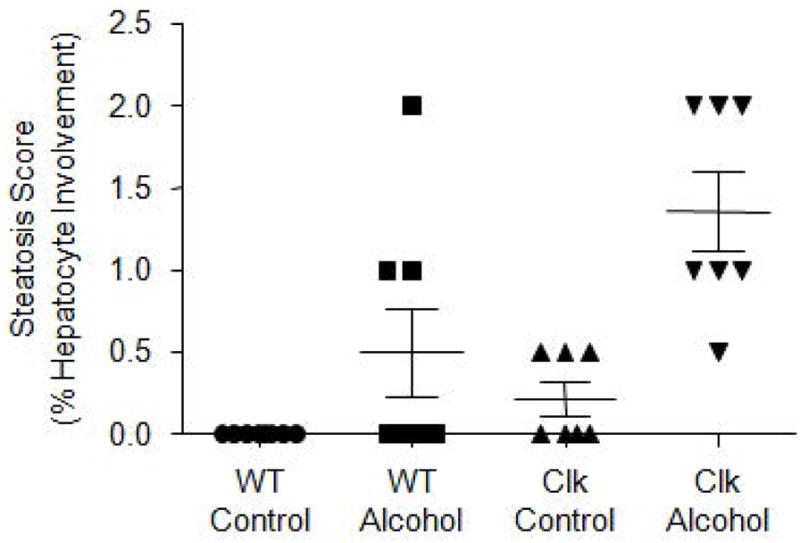
Liver steatosis is observed in groups demonstrating intestinal dysbiosis. Formalin-fixed liver was stained with hematoxylin and eosin (H&E) steatosis severity was scored as percent hepatocyte involvement (0 = <5%, 1 = 5–33%, 2 = 34–66%, 3 = >67%), corresponding to the fraction of lipid-containing hepatocytes. Data were analyzed via two-way ANOVA and shown as mean±standard error of the mean. WT, wild type mice; Clk, ClockΔ19 mutant mice; Control, alcohol-control diet; Alcohol, alcohol-containing diet.
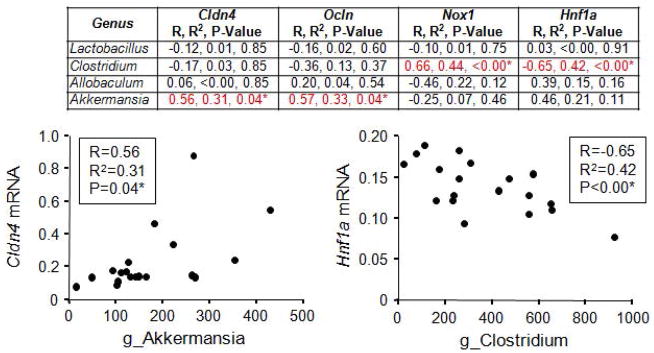
There is a high degree of crosstalk between the intestinal microbiota and the intestinal epithelium; thus, we sought to determine if gene expression in the proximal colon correlated with stool bacteria. Proximal colon tissue was collected one day following collection of stool samples (Fig. 1). Again, we selected those bacterial genera that were greater than 5% of the total microbiota community on the genus level (i.e., Lactobacillus, Clostridium, Allobaculum, Akkermansia) and correlated them with genes of interest involved in intestinal barrier integrity, oxidative stress, and inflammation in tissue collected from the proximal colon. Specifically, these included tight junction genes claudin 4 (Cldn4) and occludin (Ocln) as well as oxidative stress production gene nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 1 (Nox1) and hepatocyte nuclear factor 1a (Hnf1a). Clostridium negatively correlated with inflammation marker Hnf1a and positively correlated with oxidative stress marker Nox1 while Akkermansia positively correlated with Cldn4 and Ocln (Fig. 7). No correlations were observed between Lactobacillus or Allobaculum and any gene of interest (Fig. 7).
Figure 7.
Clostridium and Akkermansia correlate with proximal colon markers of intestinal barrier integrity and inflammation. Stool and proximal colon tissue were collected from wild type and ClockΔ19 mutant mice fed a standard rodent diet (Harlan Teklad Global) for two weeks followed by 10 weeks on an alcohol-control (wild type n=8, ClockΔ19 mutant n=8) or an alcohol-containing (wild type n=8, ClockΔ19 mutant n=8) experimental diet. Stool microbial communities were characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons and proximal colon tissue was analyzed for mRNA expression of genes of interest using an Affymetrix luminex based gene array. Genes included: Cldn4, claudin 4 (tight junction); Ocln, occludin (tight junction); Nox1, nicotinamide adenine dinucleotide phosphase (NAPDPH) oxidase (oxidative stress); Hnf1a, hypoxia inducible factor 1a (inflammation). Bacterial genera that were greater than 5% of the total microbiota community were correlated with mRNA expression (Pearson correlation) and p-values were false discovery rate corrected for multiple comparisons. g_, genus.
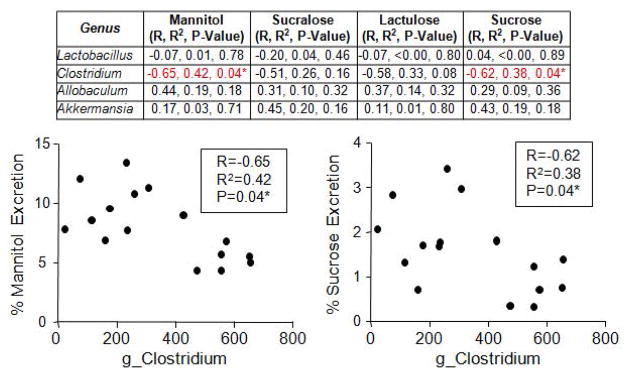
The intestinal microbiota composition can influence intestinal barrier integrity which can elicit mucosal and systemic inflammation and promote/exacerbate a number of inflammation-mediated disorders; thus, we sought to determine if intestinal leakiness associated with the Clock mutation and/or alcohol consumption (Summa et al. 2013) correlated with any bacterial taxon. Stool was collected during the same period as a test for intestinal barrier integrity was administered (Fig. 1B). We selected those bacterial genera that were greater than 5% of the total microbiota community (i.e., Lactobacillus, Clostridium, Allobaculum, Akkermansia) and correlated each taxon with percent urinary sugar excretion (greater percentage of urinary sugar content indicates greater intestinal barrier dysfunction). We found that Clostridium negatively correlated with both mannitol (thought to be an indication of proximal/mid small intestine permeability) and sucrose excretion (thought to be a marker of stomach and duodenum permeability) but not sucralose (primarily small intestine) or lactulose (total gut but primarily large intestine) (Fig. 8). Lactobacillus, Allobaculum, and Akkermansia did not correlate with any serum marker of intestinal barrier integrity (Fig. 8).
Figure 8.
Clostridium correlates with markers of intestinal hyperpermeability. Stool and urine were collected from wild type and ClockΔ19 mutant mice fed a standard rodent diet (Harlan Teklad Global) for two weeks followed by 10 weeks on an alcohol-control (wild type n=8, ClockΔ19 mutant n=8) or an alcohol-containing (wild type n=8, ClockΔ19 mutant n=8) experimental diet. Stool microbial communities were characterized using Roche 454 pyrosequencing of bacterial rRNA gene amplicons and urine was analyzed for urinary sugar content following a gavage of non-digestible, non-absorbable sugars (mannitol, sucralose, lactulose, sucrose). Bacterial genera that were greater than 5% of the total microbiota community were correlated with percent urinary urinary sugar excretion (Pearson correlation) and p-values were false discovery rate corrected for multiple comparisons. g_, genus.
Discussion
In the current study, the ClockΔ19 mutation was associated with intestinal dysbiosis. Specifically, (1) ClockΔ19 chow-fed mice had a significant reduction in bacterial evenness and diversity compared to WT mice fed the same diet, (2) ClockΔ19 mutant mice fed the alcohol-control diet (i.e., a high-fat, high-sugar diet) had a significant reduciton in evenness and diversity as well as a significant change in community structure compared to WT control-fed mice, and (3) alcohol-fed ClockΔ19 mice demonstrated a signficant alteration in community structure compared to WT mice fed alcohol. These changes may have an important funcitonal relevance as we found that liver pathology was found in all groups that exhibited intestinal dysbiosis and that specific bacterial taxon significantly correlated with markers of intestinal barrier integrity and gene expression in the proximal colon.
A reduction in bacterial diversity is a feature associated with numerous pathological conditions (Mondot et al., 2013; Round and Mazmanian, 2009). Thus, it is perhaps not surprising that we observed a reduction in bacterial diversity in ClockΔ19 mice that have a variety of physiological abnormalities including metabolic syndrome (Turek et al., 2005). It is interesting that diversity was not further impacted by alcohol consumption in ClockΔ19 mice; however, this may represent a floor effect as both alcohol and the ClockΔ19 mutation impact bacterial diversity. We observed signficant alterations in community structure when ClockΔ19 mice were fed the alcohol-control or alcohol-containing diet but not when mice were fed the standard rodent chow diet. This finding is in agreement with our previous report in which we proposed a “two hit” hypothesis whereby circadian rhythm disruption by itself may prime the microbiota community (manifested as low bacterial diversity) but may not be sufficient to promote pathology while overt effects are only observed when circadian disruption is combined with a second insult (e.g., alcohol or a high-fat, high-sugar diet). Despite the significant differences in overall community structure observed in ClockΔ19 mice, only a single bacterial taxon was signficantly altered by the mutation and this was observed in the ClockΔ19 alcohol-fed mice; an increase in an unidentified member of the order Clostridiales (Firmicutes, class Clostridia). The fact that this is an unidentified member makes it hard to judge how this may be influencing the mouse or why the change has taken place and further investigation will be necessary to more fully understand this alteration. Evaluation of the overall community structure using PICRUSt revealed that microbial changes in stool collected from ClockΔ19 mice were characterized by a reduction in the number of metabolic pathways. This change is highly relevant as numerous metabolic abnormalities are a feature associated with the ClockΔ19 mutation (Turek et al., 2005). It is not clear if this change in bacterial metabolism is a cause or a result of changes in metabolism in the host as recent publications indicate that the host can influence the microbiota and vice versa (Leone et al., 2015; Thaiss et al., 2014). It is possible that the metabolic syndrome assoicated with the ClockΔ19 mutation may be, in part, a consequence of changes in the intestinal microbiota.
Alcohol consumption has a profound and long-lasting effect on the intestinal microbiota (Bode et al., 1984; Bull-Otterson et al., 2013; Mutlu et al., 2009; Mutlu et al., 2012; Queipo-Ortuno et al., 2012; Yan et al., 2011). The specific changes vary depending on species (mouse, rat, human), sample collection site (feces, tissue), as well as alcohol consumption protocol/history. Our results in wild type mice recapitulate some previous findings in mice: a significant decrease in Lactobacillus (although Lactobacillus has also been reported to be increased in fecal samples from alcohol-fed mice), a significant decrease in Lachnospiraceae (family), and a significant increase in Allobaculum (Bull-Otterson et al., 2013; Yan et al., 2011). In the current study, alcohol reduced bacterial diversity in wild type mice which is in agreement with our findings in human alcoholics and alcohol-fed rats (Mutlu et al., 2009; Mutlu et al., 2012). While alcohol significantly impacted the bacterial communities in both wild type and ClockΔ19 mice it is interesting to note that the alcohol-induced changes in wild type and ClockΔ19 mice were overlapping but not identical indicating that alcohol-induced effects are, at least in part, dependent on the underlying genetic background of the host. In the current study, Clostridia (class) were primarily impacted in ClockΔ19 mutant mice while wild type mice had changes that were dominated by bacteria in the classes Erysipelotrichi and Bacilli. Evaluation of predicted microbial community function using PICRUSt revealed an increase in bacterial secretion systems and LPS biosynthesis suggesting that pathological changes associated with chronic alcohol consumption (e.g., inflammation and liver pathology) may be, in part, a consequence of changes in the intestinal microbiome. Despite the predicted upregulation of LPS biosynthesis we did not observe a significant increase in serum LPS levels. This finding may be due to a number of factors incluing the time of sample collection. We previously demonstrated that serum LPS levels exhibit a robust circadian pattern (Summa et al., 2013) which may the consequence of the time of LPS biosynthesis in the bacteria, the intestinal barrier integrity of the host, or the ability of the host immune system to respond to and eliminate serum LPS. Future studies will be necessary to evaluate these hypotheses. Alternatively, the predicted increase in LPS biosynthesis identified via PICRUSt may not have been sufficient to result in a statistically different serum LPS level.
Little is known about the relationship between circadian rhythms and the microbiota, although recent studies have demonstrated that interactions between commensal bacteria and circadian Clock genes help maintain intestinal homeostasis (Mukherji et al., 2013) and that circadian rhythmicity of intestinal bacteria is regulated by the host (Leone et al., 2015; Thaiss et al., 2014). We have previously demonstrated that the ClockΔ19 mutation causes intestinal barrier dysfunction and this effect is exacerbated by alcohol consumption (Voigt et al., 2014). Intestinal barrier integrity can be influenced by the intestinal microbiota (Sharma et al., 2010); thus, we evaluated if particular bacterial taxon were influencing intestinal barrier integtrity. Correlation analysis of predominant bacterial species in the stool (>5% of the total population) with serum markers of intestinal leakiness as well as the expression of genes that can influence barrier integrity revealed two bacterial taxa that may be contributing, Akkermansia and Clostridium. Akkermansia, a mucolytic bacterium, positively correlated with the tight junction genes Cldn4 and Ocln and indeed this finding is in agreement with previous data demonstrating that Akkermansia mucinphila is anti-inflammatory and beneficial for intestinal barrier integrity (Everard et al., 2013). Clostridium negatively correlated with markers of barrier integrity suggesting that it may exert beneficial effects on the host, although poteintially negative effects were observed including a negative correlation with Hnf1a gene expression (generally considered to be beneficial) and a positive correlation with Nox1 mRNA (encodes pro-inflammatory NADPH oxidase). We have shown that alcohol consumption disrupts intestinal barrier function (Keshavarzian et al., 1999; Keshavarzian et al., 2009; Summa et al., 2013) and in the current study Clostridium was markedly decreased by alcohol consumption in both wild type and ClockΔ19 mice. Furthrmore, the family Clostridiaceae (contains the genus Clostridium) was significantly reduced in ClockΔ19 mice; supporting the hypothesis that Clostridium plays a beneficial role in promoting intestinal barrier integrity. Bacterial fermentation by Bacteroidetes and Clostridium can produce short chain fatty acids (SCFA) (Cummings et al., 1987) and SCFA, especially butyrate, can influence intestinal barrier integrity (Hamer et al., 2008; Kinoshita et al., 2002; Peng et al., 2009). Thus, SCFA production is one mechanism by which Clostridium could have positively impacted intestinal barrier integrity, but further studies will be required to test this hypothesis.
Circadian rhythm disruption is a prevalent feature in modern day society (Golombek et al., 2013; Reddy and O’Neill, 2010) that promotes a number of inflammation-mediated diseases. Interestingly, mice harboring a mutation in the ClockΔ19 gene had features that overlapped with mice subjected to envrionmental circadian rhythm disruption (i.e., changes in the light:dark cycle) (Voigt et al., 2014), suggesting that circadian rhythm disruption may be the common feature driving the dysbiotic changes in the intestinal microbiota promoting inflammation and inflammation-mediated diseases.
Our data show for the first time that the ClockΔ19 mutation, which promotes circadian rhythm disruption, has a significant effect on the intestinal microbiota. These findings provide experimental evidence to support the hypothesis that circadian rhythms in the host organism may impact the intestinal microbial community; as alcohol-induced effects were different in wild type and ClockΔ19 mutant mice. Incorporating this new information about effects of alcohol and circadian disruption on the intestinal microbiota opens new avenues in our thinking of alcohol-related diseases, and may have implications more broadly for diseases characterized by circadian disruption and associated changes in the microbiota. Such information is poised to play a critical role in future medical care, when personalized pictures of health and disease risk are generated and assessed in the context of individual patients. This work contributes to a growing foundation based on mechanistic understanding upon which such a future can be constructed. The functional consequences of alcohol-induced intestinal dysbiosis need to be further explored including metabolomics and measurements of short-chain fatty acids to determine how dysbiosis may impact intestinal immune homeostasis, inflammation, and intestinal barrier function.
Supplementary Material
Acknowledgments
Work was supported by the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under award number AA020216 to AK and FWT.
References
- Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146:1564–1572. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Vekken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode JC, Bode C, Heidelbach R, Durr HK, Martini GA. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology. 1984;31:30–34. [PubMed] [Google Scholar]
- Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, Kong M, Barker D, McClain C, Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, Besing RC, Menaker M, Gewirtz AT, Davidson AJ. Dysregulation of inflammatory responses by chronic circadian disruption. J Immunol. 2010;185:5796–5805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Lange T, Golombek D, Sarkar D, Nakao A, Shibata S, Mazzoccoli G. Crosstalk between the circadian clock circuitry and the immune system. Chronobiol Int. 2013;30:870–888. doi: 10.3109/07420528.2013.782315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YK, Estaki M, Gibson DL. Clinical consequences of diet-induced dysbiosis. Ann Nutr Metab. 2013;63(Suppl 2):28–40. doi: 10.1159/000354902. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gihring TM, Green SJ, Schadt CW. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol. 2012;14:285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- Golombek DA, Casiraghi LP, Agostino PV, Paladino N, Duhart JM, Plano SA, Chiesa JJ. The times they’re a-changing: effects of circadian desynchronization on physiology and disease. J Physiol Paris. 2013;107:310–322. doi: 10.1016/j.jphysparis.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146:1449–1458. doi: 10.1053/j.gastro.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol. 2011;61:821–831. doi: 10.1007/s00248-010-9793-4. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology. 2014;146:1477–1488. doi: 10.1053/j.gastro.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzian A, Farhadi A, Forsyth CB, Rangan J, Jakate S, Shaikh M, Banan A, Fields JZ. Evidence that chronic alcohol exposure promotes intestinal oxidative stress, intestinal hyperpermeability and endotoxemia prior to development of alcoholic steatohepatitis in rats. J Hepatol. 2009;50:538–547. doi: 10.1016/j.jhep.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzian A, Holmes EW, Patel M, Iber F, Fields JZ, Pethkar S. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol. 1999;94:200–207. doi: 10.1111/j.1572-0241.1999.00797.x. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Suzuki Y, Saito Y. Butyrate reduces colonic paracellular permeability by enhancing PPARgamma activation. Biochem Biophys Res Commun. 2002;293:827–831. doi: 10.1016/S0006-291X(02)00294-2. [DOI] [PubMed] [Google Scholar]
- Laposky A, Bass J, Kohsaka A, Turek FW. Sleep and circadian rhythms: key components in the regulation of energy metabolism. FEBS Lett. 2008;582:142–151. doi: 10.1016/j.febslet.2007.06.079. [DOI] [PubMed] [Google Scholar]
- Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. 2015 doi: 10.1073/pnas.1501305112. Epub before print: pii:201501305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L, Thorburn AN, Binge LC, Marino E, Rogers KE, Maslowski KM, Vieira AT, Kranich J, Mackay CR. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev. 2012;245:164–176. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- McClung CA. Role for the Clock gene in bipolar disorder. Cold Spring Harb Symp Quant Biol. 2007;72:637–644. doi: 10.1101/sqb.2007.72.031. [DOI] [PubMed] [Google Scholar]
- McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, Wilke A, Huse S, Hufnagle J, Meyer F, Knight R, Caporaso JG. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012;1:7. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S, de Wouters T, Dore J, Lepage P. The human gut microbiome and its dysfunctions. Dig Dis. 2013;31:278–285. doi: 10.1159/000354678. [DOI] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Mutlu E, Keshavarzian A, Engen P, Forsyth CB, Sikaroodi M, Gillevet P. Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats. Alcohol Clin Exp Res. 2009;33:1836–1846. doi: 10.1111/j.1530-0277.2009.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queipo-Ortuno MI, Boto-Ordonez M, Murri M, Gomez-Zumaquero JM, Clemente-Postigo M, Estruch R, Cardona DF, Andres-Lacueva C, Tinahones FJ. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am J Clin Nutr. 2012;95:1323–1334. doi: 10.3945/ajcn.111.027847. [DOI] [PubMed] [Google Scholar]
- Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh M, Rajan K, Forsyth CB, Voigt RM, Keshavarzian A. Simultaneous gas-chromatographic urinary measurement of sugar probes to assess intestinal permeability: use of time course analysis to optimize its use to assess regional gut permeability. Clin Chim Acta. 2015;442:24–32. doi: 10.1016/j.cca.2014.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Young C, Neu J. Molecular modulation of intestinal epithelial barrier: contribution of microbiota. J Biomed Biotechnol. 2010;2010:305879. doi: 10.1155/2010/305879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly Y, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa KC, Voigt RM, Forsyth CB, Shaikh M, Cavanaugh K, Tang Y, Vitaterna MH, Song S, Turek FW, Keshavarzian A. Disruption of the Circadian Clock in Mice Increases Intestinal Permeability and Promotes Alcohol-Induced Hepatic Pathology and Inflammation. PLOS One. 2013;8:e67102. doi: 10.1371/journal.pone.0067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladomiu M, Hontecillas R, Yuan L, Lu P, Bassaganya-Riera J. Nutritional protective mechanisms against gut inflammation. J Nutr Biochem. 2013;24:929–939. doi: 10.1016/j.jnutbio.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci USA. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. Circadian disorganization alters intestinal microbiota. PLOS One. 2014;9:e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan AW, Fouts DE, Brandl J, Starkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.