Abstract
Electronic medical records (EMRs) for diabetic patients contain information about heart disease risk factors such as high blood pressure, cholesterol levels, smoking status, etc. Discovering the described risk factors and tracking their progression over time may support medical personnel in making clinical decisions, as well as facilitate data modeling and biomedical research. Such highly patient-specific knowledge is essential to driving the advancement of evidence-based practice, and can also help improve personalized medicine and care. One general approach for tracking the progression of diseases and their risk factors described in EMRs is to first recognize all temporal expressions, and then assign each of them to the nearest target medical concept. However, this method may not always provide the correct associations. In light of this, this work introduces a context-aware approach to assign the time attributes of the recognized risk factors by reconstructing contexts that contain more reliable temporal expressions. The evaluation results on the i2b2 test set demonstrate the efficacy of the proposed approach, which achieved an F-score of 0.897. To boost the approach’s ability to process unstructured clinical text and to allow for the reproduction of the demonstrated results, a set of developed .NET libraries used to develop the system is available at https://sites.google.com/site/hongjiedai/projects/nttmuclinicalnet.
Graphical Abstract
Introduction
Heart disease is the leading cause of death in the United States, and coronary heart disease accounts for more than 60% of all incidents of heart disease. In addition, heart attack events can result in several complications such as heart failure, valvular heart diseases and arrhythmia. To prevent the incidence of new myocardial infarction, reduction of known proper risks related to coronary heart disease including smoking, hypertension, hyperlipidemia, obesity, and diabetes mellitus and tracking their progression over time are of utmost importance. To promote the identification of information relevant to heart disease risks and track their progression as documented in electronic medical records (EMRs), this work developed a system that recognizes mentions of medical concepts including the following items: disease names of diabetes and coronary artery disease (CAD); associated tests such as glycated hemoglobin (HbA1C) and test results; events and symptoms of CAD; and measurements related to diabetes, hyperlipidemia, hypertension, and obesity including blood glucose/pressure, cholesterol level, low-density lipoprotein (LDL) level, body mass index (BMI), and waist circumference.
One general approach for progression tracking is to first recognize all temporal expressions, and then assign each to the nearest target concept. The distance between the concept and a temporal expression could be the number of tokens among them or depend on the syntactic parsing. However, associations resulting from such an approach may not always be correct, especially if the text processed by the natural language processing (NLP) system was incomplete or contains arbitrary line breaks. In light of this, this work proposes a context-aware approach which first reconstructs the context to enrich it with reliable temporal information. The algorithm then assigns the corresponding time attributes for all recognized concepts with respect to the creation time of the medical records (hereinafter referred to as the document creation time or DCT) based on the temporal information of the constructed context. The proposed algorithm and core library used to develop the system are available at https://sites.google.com/site/hongjiedai/projects/nttmuclinicalnet to allow for the enhancement of the proposed method and the reproduction of the demonstrated results.
Materials and Methods
NTTMUNSW System
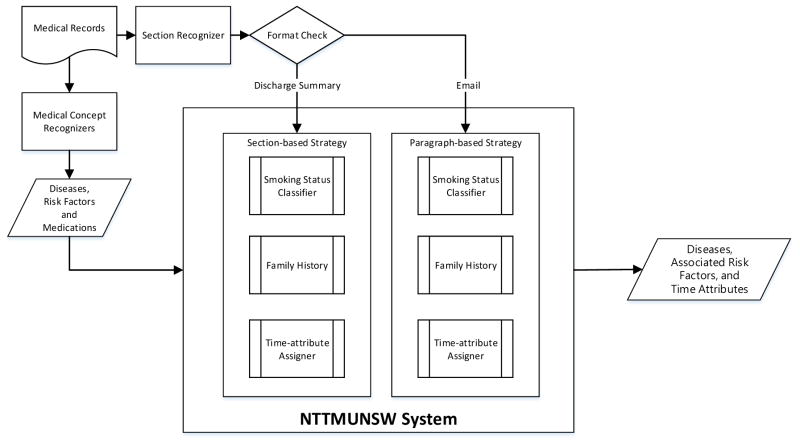
Figure 1 displays a flowchart of the developed NTTMUNSW system —a joint work of National Taitung University (NTTU), Taipei Medical University (TMU), the University of New South Wales (UNSW) and Taiwan’s Academia Sinica. For each EMR, a section recognizer constructed in our previous work [1] is first used to identify section headings. The text between two section headings is considered as the corresponding section content of the preceding section. For example, in Fig. 2, the content of the “Narrative History” section is “55 y/o woman who presents for f/u … Still with hot flashes, wakes her up at night. …”
Figure 1.
NTTMUNSW System overview
Figure 2.
Sections recognized by the developed section recognizer are highlighted in bold.
The content of each section is then processed by medical concept recognizers to identify disease mentions, along with their corresponding risk factors and medications. Subsequently, the time-attribute assigner component uses the proposed context-aware algorithm with two different context range parameters to determine the relative time attributes of the medical concepts. The setting of the range parameter determines the maximal expansion of the algorithm. In practice, the parameters were set according to the format of the given EMR. As shown in Fig. 1, if the record is a medical note, such as a discharge summary, the section-based strategy is used to indicate that the range parameter was set to “section”—the text in between two section headings. Otherwise, the record is considered to be a narrative description in a text like a consultation letter, and the paragraph-based method is used. The details of the proposed context-aware algorithm are described in the “Time-attribute Assigner” section. Other steps of the system workflow are elaborated in the following sub-sections.
Development of the Medical Concept Recognizers
Recognition of Mention Concepts
The mention concept herein refers to the descriptions in an EMR that indicate a diagnosis of the two target diseases—diabetes and CAD, as well as hypertension, hyperlipidemia, and obesity. In this work, two methods were used to develop the mention concept recognizer. The first is a dictionary-based method, and the second is based on machine learning.
For the dictionary-based method, we collected a dictionary consisting of 220 terms for all five mention types by manually inspecting all texts tagged as a “mention” concept within the training dataset. In addition, all extracted relevant mentions from the dictionary used in our previous work [2] were merged. For the machine learning-based approach, the training dataset annotated with the occurrence of mention concepts was selected as the training set. The IOB2 tagging scheme was used to formulate the recognition problem as a sequential labeling task. This work then used the conditional random field (CRF) algorithm with the features proposed in our previous works [3, 4] including part-of-speech tags, shallow parser tags, dictionary matching and bag of words to build the model.
Recognition of Other Risk Factors
In addition to the mention concepts described in the previous section, a list of relevant numeric risk factors was defined for each mention type. For instance, blood pressure (BP) with a value over 140/90 mm/hg is considered as a risk factor for hypertension. Table 1 summarizes the medical concepts, their corresponding risk factors, and the standard values at which each factor is deemed to be risky. For each risk factor category, a list of keywords was collected from the training dataset by extracting terms annotated as the category. The list was then used as a dictionary by our system to tag the given medical record. For a candidate risk factor, if its adjacent numeric value (recognized by using regular expression patterns) satisfied the requirement listed in Table 1, the factor was then reserved for the assignment of the time attribute in the later stage. For non-numeric type risk factors, such as the surgery event of CAD, the corresponding keywords were collected from the training set, and the same dictionary-based approach was then used to recognize those factors.
Table 1.
Summary of the targeted diseases and their corresponding risk factor definitions
| Category | Risk Factor | Numeric Value |
|---|---|---|
| Diabetes | High A1C | >=6.5 |
| Diabetes | High glucose | >126 |
| Hyperlipidemia | High cholesterol | >=240 |
| Hyperlipidemia | High LDL | >=100 mg/dL |
| Hypertension | High blood pressure | >=140/90 mm/hg |
| Obesity | BMI | > 30 |
| Obesity | Waist circumference | Men:>= 40 inches; Women: >= 35 inches |
Recognition of Medications
This work also recognizes all medications related to the targeted risk factors within an EMR. For instance, thiazide diuretics, often used to treat hypertension, are a target medication category. Similar to the mention concept recognizer, the same CRF algorithm and feature sets were used to build a model that can recognize target medications. All recognized medications were then matched with a medication name-category mapping file to determine the corresponding medication categories through a normalization step. The mapping file consists of medication terms collected from Wikipedia, including the generic names and classes of all drugs. In addition, as medications are usually presented by brand names in clinical texts, such as “Lozol” for thiazide diuretics, information from the Drugs.com website was therefore consulted for the brand names of all drugs, and the file was further expanded using the vocabularies of Drugbank [5]. Finally, the fourth author, who is a medical doctor, manually verified and finalized the mapping file. The final file contains 21 categories and a total of 474 names. Some recognized medication names cannot be directly mapped to their categories due to typographical errors, so a partial matching algorithm was employed to map the recognized mention with the compiled mapping file.
Time-attribute Assigner
The time-attribute assigner determines the time attributes for all recognized medical concepts. Before the execution of the time-attribute assigner, the following NLP steps are applied to the raw text of the given EMR. This work reserved the original line breaks of the raw text, which enables us to easily capture paragraphs in some record layouts. For each text block distinguished by the line breaks, the text was processed by the MedPost tagger[6] and our section classifier [7] to recognize sentences, tokens, and section headings. Note that due to the variety of record layouts, the sentences generated by the above NLP steps may be incomplete as shown in the right column of Fig. 3. The record date of the given EMR was also extracted for later usage. The information can be extracted by checking the content of the “Record date:” section of each record. Subsequently, all recognized medical concept instances along with their corresponding span information were collected as a list. Each instance in the list was then processed by the time-attribute assigner to determine its time attribute.
Figure 3.
An example of text blocks (left column) and sentences generated by the NLP steps (right column).
Context-aware time attribute assignment
The proposed context-aware time-attribute assigner was developed based on the notion that the algorithm for the assignment of a medical concept’s time attribute should both sense and react in a context-dependent manner. The present work defines the context as the narrative containing any information that can be used to characterize the time attribute of a medical concept. If the assigner senses that the current context is insufficient to determine the associated time attribute, the context-aware algorithm reacts by extending the current context to a certain extent and re-checks the context again until it resolves the time attribute or there is no further context to be expanded. More specifically, in this work, the (incomplete) sentence s which contains the target medical concept mc was first checked to determine whether or not it contains a clue that indicates the time attribute of mc. In this case, two types of context were involved, including the mention mc itself and s. The text of mc was considered as the superior context to s, since in some cases the mention itself contains the temporal information. For instance, our risk factor recognizer will recognize the entire statement “HBA1C 05/25/2092 7.30” as a High A1C risk factor (7.30 > 6.5), which indicates that the risk factor was observed on 05/25/2092.
If the assignment failed and s is an incomplete sentence, s is extended to include surrounding tokens to form a complete sentence s′, which is then re-checked by the algorithm. Assuming that the extended context still does not provide any clues, the context is then further extended until either the time attribute is assigned or a predefined maximal extent (defined by the context range parameter) is achieved. Note that this work assumes that the temporal information associated with the target medical concept should be described before or within the current context. Therefore, when starting to expand the context of the complete sentence s′, the algorithm will only consider the content described preceding s′ to generate the current context c. The content following c will be included only when s′ is not the last sentence of a paragraph or the context range parameter was set to use the section-based strategy.
In practice, this work applies two range parameters to implement the proposed context-aware approach for different formats of a given record. The format was determined by judging the numbers of section headings observed by the employed section recognizer and indicator phrases, such as “sincerely” and “with warm regards”. As depicted in Fig. 1, if the given record was a narrative written in a consultation letter, the range parameter was set to “paragraph”, indicating that the paragraph-based strategy was used to determine the time attribute. In this case, the maximal context of a medical concept considered by the context-aware algorithm includes the whole paragraph that contains the medical concept. Take the second to the last line of the right column in Fig. 3 as an example. This line contains the risk factor high BP, which is highlighted in bold. However, the incomplete sentence itself doesn’t provide any clue for time attribute assignment. Therefore, an algorithm was used to further include the three preceding lines to form the complete sentence c. The algorithm then re-checks the sentence and finds that c contains the temporal mention “last January”, which indicates that the event occurred before the DCT. Consider another case in which the above process cannot determine the target concept’s time attribute. The context will be extended to include the entire paragraph, which are all of the lines after the sentence “Dear Dr. Taylor :” as shown in Fig. 3.
On the other hand, for a discharge summary with explicit section headings as shown in Fig. 2, the range parameter was set to “section” and the section-based strategy was used to determine the time attribute. In this case, the maximal context of a medical concept considered by the algorithm can include several paragraphs and even the complete section consisting of several paragraphs that contains the medical concept. Regardless of the format, if two or more temporal mentions are observed in a given context, the temporal mention that is nearest to the target concept will be assigned.
Contextual clues for time-attribute assignment
The time-attribute assigner relies on several contextual clues to determine the time attribute. Along with the updated context c, a similar method was used to re-check the temporal information until either the time attribute of mc is updated or no new context is sensed by the context-aware algorithm. A temporal expression extraction system based on our previous work [8] was executed on a given text to extract temporal information. Table 2 shows examples of extracted temporal expressions and their distributions in the training set. The system uses a pattern-based matching method to extract several types of temporal expressions described in a variety of ways, such as “in month|year”, “from year to year”, “this year”, and “6/12”, and normalizes them to their corresponding numeric values by using a mapping table. In addition, several post-processing rules were implemented to remove false positive cases, such as 150/70, which is not a temporal expression. Finally, non-numeric temporal expressions matched with keywords such as “today”, “last” and “recent” were assigned with their predefined time attributes. The numeric temporal expression is compared with the record date to determine the time attributes if the expression includes the year information. Otherwise, the context surrounding the expression is considered to determine the time attribute. For example, the time attribute for the expression “past 10 years” is “Before DCT”.
Table 2.
Summary of the distribution of each temporal expression in a single document.
| Numeric temporal expressions | Non-numeric temporal expressions | |
|---|---|---|
|
| ||
| Training set | 16.09 | 2.53 |
|
| ||
| Testing set | 15.60 | 2.37 |
|
| ||
| Examples | … start : 6 / 12 / 2070 end : 01 / 13 / 2072 – inactivated | … performed some pacing maneuvers today in clinic … |
| … from a study of august 06, 2086. | … she presented with prior to this year. | |
| … your blood sugar over the past 3 months | … last used several months ago as … | |
In addition, certain types of medical concepts, such as the “mention” indicator shown in Table 3, may not have explicit temporal expressions. Subsequently, the algorithm by default assigns the most common time attribute for such concept types based on the statistical results if there is no available temporal information. However, if the associated time attribute values have a (nearly) uniform distribution in the dataset used by this work, the algorithm will generate all of the three DCT as its output.
Table 3.
Summary of the distribution of each indicator. (The numerator represents the number of various time attributes in the indicator; the denominator represents the total amount in the indicator.)
| Risk factors | Indicators | Time attributes | Amount of Training | Amount of testing |
|---|---|---|---|---|
| CAD | Test | Before DCT | 266/311 (86%) | 49/53 (92%) |
| During DCT | 45/311 (14%) | 4/53 (8%) | ||
| After DCT | 0/311 (0%) | 0/53 (0%) | ||
| Mention | Before DCT | 1276/3773 (34%) | 166/504 (33%) | |
| During DCT | 1255/3773 (33%) | 171/504 (34%) | ||
| After DCT | 1242/3773 (33%) | 167/504 (33%) | ||
| Event | Before DCT | 1143/1233 (93%) | 126/135 (93%) | |
| During DCT | 84/1233 (7%) | 8/135 (6%) | ||
| After DCT | 6/1233 (0%) | 1/135 (1%) | ||
| Symptom | Before DCT | 192/281 (68%) | 39/67 (58%) | |
| During DCT | 79/281 (28%) | 25/67 (37%) | ||
| After DCT | 10/281 (4%) | 3/67 (4%) | ||
| Hypertension | Mention | Before DCT | 2453/7326 (33%) | 357/1074 (33%) |
| During DCT | 2444/7326 (33%) | 359/1074 (33%) | ||
| After DCT | 2430/7326 (33%) | 358/1074 (33%) | ||
| High bp | Before DCT | 168/1524 (11%) | 19/191 (10%) | |
| During DCT | 1356/1524 (89%) | 172/191 (90%) | ||
| After DCT | 0/1524 (0%) | 0/191 (0%) | ||
| Hyperlipidemia | Mention | Before DCT | 1459/4381 (33%) | 232/696 (33%) |
| During DCT | 1461/4381 (33%) | 232/696 (33%) | ||
| After DCT | 1461/4381 (33%) | 232/696 (33%) | ||
| High LDL | Before DCT | 79/114 (69%) | 25/28 (89%) | |
| During DCT | 35/114 (31%) | 3/28(11%) | ||
| After DCT | 0/114 (0%) | 0/28 (0%) | ||
| High chol. | Before DCT | 26/29 (90%) | 8/10 (80%) | |
| During DCT | 3/29 (10%) | 2/10 (20%) | ||
| After DCT | 0/29 (0%) | 0/10 (0%) | ||
| Diabetes | Mention | Before DCT | 2558/7699 (33%) | 346/1041 (33%) |
| During DCT | 2583/7699 (34%) | 348/1041 (33%) | ||
| After DCT | 2558/7699 (33%) | 347/1041 (33%) | ||
| A1C | Before DCT | 381/451 (84%) | 69/80 (86%) | |
| During DCT | 70/451 (16%) | 11/80 (14%) | ||
| After DCT | 0/451 (0%) | 0/80 (0%) | ||
| Glucose | Before DCT | 68/96 (71%) | 17/32 (53%) | |
| During DCT | 28/96 (29%) | 15/32 (47%) | ||
| After DCT | 0/96 (0%) | 0/32 (0%) | ||
| Obese | Mention | Before DCT | 569/1776 (32%) | 78/242 (32%) |
| During DCT | 641/1776 (36%) | 86/242 (36%) | ||
| After DCT | 566/1776 (32%) | 78/242 (32%) | ||
| BMI | Before DCT | 10/77 (13%) | 2/17 (12%) | |
| During DCT | 61/77 (79%) | 13/17 (76%) | ||
| After DCT | 6/77 (8%) | 2/17 (12%) | ||
| Medication | N/A | Before DCT | 12716/37646 (34%) | 1869/5534 (34%) |
| During DCT | 12559/37646 (33%) | 1832/5534 (33%) | ||
| After DCT | 12371/37646 (33%) | 1833/5534 (33%) |
Family History Status Classifier
As defined in the annotation guidelines [9] of the i2b2 track 2 dataset, the family history status possesses an attribute with only the value "present" or "not present". This tag should only be marked as present if the patient has a first-degree relative (i.e., parents, siblings, or children) who was diagnosed with premature CAD (younger than 55 for male relatives, younger than 65 for female relatives). As a result, the classification of the family history status is divided into two steps. First, a dictionary consisting of 12 first-degree relatives was assembled to recognize keywords indicating first-degree relatives in the record. For instance, “father”, “mother”, “dad”, and “mom” were all included in the dictionary. This work also analyzes family history status from the semantic perspective by generating dictionaries of negation and CAD-related terms, and establishing two criteria to determine the attribute value: (1) The context contains a male/female first-degree relative word, along with specific age-related information (over 55 years old for male and 65 years old for female); (2) the context contains CAD-related terms and even numbers of negation terms. When both criteria are met, the indicator attribute is “present”. Otherwise, it is “not present”.
Smoking Status Classifier
For the detection of smoking status, a list of smoking-related keywords, such as “smoking” and “cigarette”, was compiled. The keyword list was then used by the Smoking Status Classifier as a dictionary to tag the given EMR. Afterwards, the text containing the listed terms was regarded as the context of the smoking status, and several weighted rules developed for different smoking statuses were applied to determine the patient’s smoking status, which follows the same rationale of our context-aware algorithm. If the context did not provide sufficient information to determine the smoking status, a similar context-aware approach was used to expand the context and re-apply the developed rules until either the status was determined or no further updated context was available.
Dataset
The 2014 i2b2 NLP shared task [9] released a dataset consisting of medical documents annotated with information related to the progression of heart disease in diabetic patients for the identification of risk factors of heart disease over time. Annotations of the dataset include the presence and progression of diseases (diabetes, CAD), and associated risk factors (hypertension, hyperlipidemia, smoking status, obesity status, and family history), and the time they were present in the patient’s medical history. We used the dataset in our work to evaluate the proposed medical concept progression tracking method. Following the same manner of the i2b2 risk factor track, the two training sets released by the track were used to develop rules. For the machine learning-based system, a tenfold cross validation on the same dataset was applied to select efficient features for mention concept and medication recognition.
Results and Discussion
Pipeline-based Baseline System
This work implements a baseline system to explore the performance of existing systems in tracking the progression of heart disease risk factors. Apache clinical Text Analysis and Knowledge Extraction System (cTAKES) v3.1.1 with UMLS 2014AA as the underlying dictionary was selected as the baseline system to recognize medications and the mention concepts. A post-processing component was developed and used to filter out medical concepts which are not related to heart disease risk factors. The post-processing component also maps the recognized medications to their medication categories through the same normalization steps described in the "Recognition of Medications” section. For laboratory values of risk factors that cannot be captured by cTAKES, such as BP and HbA1C, the Apache Ruta language was used to define patterns that can recognize the values as illustrated in Table 1. The output of the post-processing component was merged with the output from the cTAKES system to generate the final recognition results. Since cTAKES is not capable of assigning time attributes to the recognized concepts, this work develops a time attribute assignment component based on the naïve Bayes classifier to assign the time attribute shown in Table 3. All occurrences of the medical concepts in the training set are treated as training instances, and their associated time attributes serve as the corresponding class labels. The features used include the word tokens of the target medical concept, its surrounding word tokens, the section information, and the type of the concept (e.g. mention, event, or symptom). The classifier was tuned by employing tenfold cross validation on the training set. The best performing model was used on the test set. All custom developed components are integrated into cTAKES via the Unstructured Information Management Architecture framework [10].
Overall results
Table 4 presents the performance of our three submitted runs during the participation of the track and the aggregated statistics of the 49 submissions from 20 teams in this track. The best result of our NTTMUNSW system, which ranked 7 out of all 20 teams, achieved an overall micro F-score (F) of 0.8973 on the test set, outperforming the median and mean F-scores by 0.025 and 0.082, respectively.
Table 4.
Performance of each submitted run of the NTTMUNSW system and the aggregated results of all runs.
| Baseline | NTTMUNSW Run 1 (Rank 7) | NTTMUNSW Run 2 | Mean | Median | |
|---|---|---|---|---|---|
| Micro P | 0.6779 | 0.8594 | 0.8384 | 0.808 | 0.852 |
| Micro R | 0.7566 | 0.9387 | 0.9404 | 0.835 | 0.908 |
| Micro F | 0.7151 | 0.8973 | 0.8865 | 0.815 | 0.872 |
Both Runs 1 and 2 used the machine learning-based medication recognizer and the proposed context-aware approach for the assignment of time attributes. The only difference between Runs 1 and 2 is that the former replaced the machine learning-based recognizer with the dictionary-based recognizer for the recognition of mention concepts. As shown in Table 4, the precision (P) of the dictionary-based hybrid system (Run 1) is higher than the pure machine learning-based hybrid system (Run 2), while the recall (R) of the machine learning-based system (Run 2) is higher than that of the dictionary-based system. The results are in line with the common understanding of the distinct advantages of the two approaches, although they may not differ significantly. A close examination of the dataset revealed that the target medical concepts share limited and fixed terms. Therefore, the thoroughly selected collection of these terms achieved a satisfying recall. Finally, according to Table 4, the NTTMUNSW system significantly outperforms the baseline system.
Time Attribute Assignment Result Comparison
Table 5 compares the performance between three time-attribute assignment approaches regarding the five main medical concepts. The first is the result of the naïve Bayes method used by the pipeline-based baseline system. The other two are results based on Run 1 with or without the proposed context-aware algorithm. Configuration w/o the context-aware method was implemented by selecting the temporal information nearest to the target concept as its associated temporal expression and comparing the description with the DCT. If no temporal information is observed, the distribution shown in Table 3 is used to assign the time attribute. For example, the time attributes of “CAD-Test” are assigned the “before DCT” attribute since the probability of 86% is larger than the sum of the other two DCTs.
Table 5.
Performance comparison of the five main medical concepts between w/ and w/o context-aware approaches.
| naïve Bayes (Macro/Micro) | w/o context-aware (Macro/Micro) | w/context-aware (Macro/Micro) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Concept Type | P | R | F | P | R | F | P | R | F |
| Hypertension | 0.343/ 0.558 | 0.483/ 0.615 | 0.401/ 0.585 | 0.683/ 0.83 | 0.736/ 0.959 | 0.708/ 0.890 | 0.734/ 0.933 | 0.751/ 0.981 | 0.744/ 0.957 |
| Hyperlipidemia | 0.447/ 0.843 | 0.441/ 0.925 | 0.444/ 0.883 | 0.442/ 0.827 | 0.454/ 0.952 | 0.447/ 0.885 | 0.449/ 0.859 | 0.437/ 0.923 | 0.443/ 0.890 |
| Diabetes | 0.658/ 0.869 | 0.675/ 0.952 | 0.666/ 0.909 | 0.650/ 0.864 | 0.672/ 0.95 | 0.661/ 0.905 | 0.687/ 0.928 | 0.676/ 0.953 | 0.681/ 0.941 |
| OBESE | 0.140/ 0.845 | 0.142/ 0.851 | 0.141/ 0.848 | 0.159/ 0.770 | 0.168/ 0.981 | 0.163/ 0.862 | 0.159/ 0.771 | 0.167/ 0.977 | 0.163/ 0.862 |
| CAD | 0.214/ 0.665 | 0.231/ 0.560 | 0.222/ 0.608 | 0.275/ 0.432 | 0.415/ 0.943 | 0.331/ 0.593 | 0.357/ 0.74 | 0.326/ 0.781 | 0.341/ 0.760 |
| Average | 0.360/ 0.756 | 0.394/ 0.781 | 0.375/ 0.767 | 0.441/ 0.745 | 0.489/ 0.957 | 0.462/ 0.827 | 0.477/ 0.846 | 0.472/ 0.923 | 0.475/ 0.882 |
The baseline system based on the naïve Bayes algorithm didn’t fully benefit from the annotated corpus, which may result from simplified and noisy feature sets. Compared with the w/o context-aware approach, the context-aware approach achieved better precision scores among all concept types, especially the CAD concept, and resulted in a better overall F-score (0.882 .vs. 0.827). The results demonstrated the strength of the proposed context-aware approach for assigning time attributes in improving the precision of the temporal information assignment. As shown in Fig. 3, the i2b2 2014 dataset contains incomplete sentences or arbitrary line breaks. Therefore, the temporal expression for a target medical concept may not be the nearest temporal information observed in the context. For example, a temporal expression mentioned at the beginning of a paragraph may be closer to the medical concepts located at the end of the statement preceding that paragraph. In fact, the correct temporal information of the medical concepts was found at the very beginning of the statement or even the paragraph or section. Incomplete sentences containing medical concepts may also post similar problems when more than two temporal expressions exist. The closer or syntactically temporal expression may be mentioned in the next statement, but the current line contains another temporal expression. A comparison of the results between the w/o and w/ context-aware approaches demonstrates that our algorithm can construct a more reliable context for time attribute assignment.
nttmuClinical.NET Library
All of the NLP components cited in this work, including the proposed context-aware time attribute assigners and resources used, such as the dictionary file for our smoking status classifier, can be downloaded from https://sites.google.com/site/hongjiedai/projects/nttmuclinicalnet or http://btm.tmu.edu.tw/nttmuClinicalNET/. The core library was implemented in the Microsoft .NET environment, which can be easily installed through the NuGet package manager of the Microsoft development platform. The website also provides the application programming interface documentation (refer to http://btm.tmu.edu.tw/nttmuClinicalNET/api) for references and runnable NUnit sample code that can be used to test the functionality of the installed library.
Related Work
The task of tracking progression of medical concepts requires recognizing risk factors for heart disease and determines the time attribute associated with the recognized concepts. For the recognition of risk factors, both dictionary-based and machine-learning-based approaches were used. For example, J Urbain [11] used CRF model to recognize risk factor instances. A Khalifa and SM Meystre [12] collected risk factor terms from a training dataset and adapted a dictionary-based lookup component based on existing NLP tools such as cTAKES to recognize medical concepts. For the time attribute assignment issue, certain types of medical concepts have a uniform distribution for the assignment of DCT in the i2b2 dataset. There are 23,363 before DCT attributes, 22,704 during DCT attributes and 20,650 after DCT attributes in the whole training set. Some participants [11–13] employed statistical information to assign the time attributes for recognized concepts, or tracked the progression of these medical concepts by developing heuristic rules [14], a combination of both [15, 16], or machine learning-based approaches [17, 18]. In comparison with the aforementioned related works, the main contribution of this work was the proposed context-aware algorithm that can sense the context of a target medical concept and react accordingly if the context does not provide sufficient information for determining its time attribute. Our experimental results have demonstrated the ability of the proposed approach in improving the precision of the time attribute assignment. We believe that the algorithm can be integrated into other related works to boost performance, and we plan to address this topic in future work.
Conclusion
This work proposes a context-aware approach to track the progression of medical concepts through the determination of their time attributes. The approach recognizes the concepts based on both dictionary and machine learning-based techniques. Subsequently, the context-aware algorithm iteratively refines the context for the recognized concepts until the associated temporal information is resolved. The non-context-aware method, which uses the overall probability of the time attribute from the training set without considering the temporal information from the text, exhibited an acceptable F-score of 0.827 for the five main medical concepts. Nevertheless, the proposed method outperformed the non-context-aware method in terms of precision, and resulted in an improvement of 0.055 in F-score. The advantage of the proposed context-aware algorithm is that it intelligently collects temporal information from the text, includes information concealed across different sentences. Consequently, this method has a better chance of determining the correct time attributes. By contrast, the assignment of time attribute based on the nearest temporal information or the probability distribution calculated from the training sample may not be competent when applied to a dataset with different characteristics. With the proposed context-aware approach, the system based on the nttmuClinical.NET library achieved an overall micro F-score of 0.8973 on the i2b2 2014 test set.
Highlights.
A context-aware approach is proposed to track the progression of medical concepts through the determination of their time attributes.
The context-aware approach reconstructs the context to enrich it with more reliable temporal expressions.
A .NET library, nttmuClinical.NET, is released for processing unstructured electronic medical records.
Acknowledgments
This work was supported by the Ministry of Science and Technology (MOST-103-2221-E-038-019, MOST-104-2319-B-400-002, MOST-103-2319-B-010-002 and MOST-103-3111-Y-001-027). We are grateful to Dr. Amber Stubbs, an assistant professor at Simmons College, School of Library and Information Science, and Dr. Ozlem Uzuner, an associate professor at the State University of New York for their valuable help in the i2b2 shared task.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen C-W, Chang N-W, Chang Y-C, Dai H-J. Section Heading Recognition in Electronic Health Records Using Conditional Random Fields. In: Cheng S-M, Day M-Y, editors. Technologies and Applications of Artificial Intelligence. Vol. 8916. Springer International Publishing; 2014. pp. 47–55. [Google Scholar]
- 2.Jonnagaddala J, Liaw S-T, Rayb P, Kumarc M, Dai H-J. TMUNSW. Identification of disorders and normalization to SNOMED-CT terminology in unstructured clinical notes. 9th International Workshop on Semantic Evaluations; 2015. [Google Scholar]
- 3.Tsai RT, Sung CL, Dai HJ, Hung HC, Sung TY, Hsu WL. NERBio: using selected word conjunctions, term normalization, and global patterns to improve biomedical named entity recognition. Bmc Bioinformatics. 2006;7(Suppl 5):S11. doi: 10.1186/1471-2105-7-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai HJ, Lai PT, Chang YC, Tsai RT. Enhancing of chemical compound and drug name recognition using representative tag scheme and fine-grained tokenization. Journal of Cheminformatics. 2015;7(Suppl 1):S14. doi: 10.1186/1758-2946-7-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. DrugBank 4. 0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(Database issue):D1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics. 2004;20(14):2320–2321. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- 7.Dai H-J, Chen C-W, Wu C-C, Syed-Abdul S. Recognition and Evaluation of Clinical Section Headings in Clinical Documents Using Token-based Formulation with Conditional Random Fields. BIoMed Research International. 2015 doi: 10.1155/2015/873012. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang YC, Dai HJ, Wu JC, Chen JM, Tsai RT, Hsu WL. TEMPTING system: a hybrid method of rule and machine learning for temporal relation extraction in patient discharge summaries. J Biomed Inform. 2013;46(Suppl):S54–62. doi: 10.1016/j.jbi.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Stubbs A, Uzuner O. Annotating Risk Factors for Heart Disease in Clinical Narratives for Diabetic Patients. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DF, AL Building an example application with the unstructured information management architecture. IBM Systems Journal. 2004;43(3):455–475. [Google Scholar]
- 11.Urbain J. Identifying risk factors for heart disease in diabetic patients over time from electronic medical record text: i2b2 2014 NLP Challenge. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 12.Khalifa A, Meystre SM. Identification of Risk Factors for Heart Disease in Electronic Health Records of Diabetic Patients. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 13.Karystianis G, Dehghan A, Kovačević A, Keane JA, Nenadic G. Using Local Lexicalized Rules for Identification of Heart Disease Risk Factors in Free-text Clinical Notes. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 14.Yang H, Garibaldi J. Automatic Extraction of Risk Factors for Heart Disease in Clinical Texts. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 15.Ju M, Ge C, Jia Z, Li H. Building NLP Systems based on Annotated Corpus for Identifying Risk Factors for Heart Disease Over Time. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 16.Cormack J, Nath C, Milward D, Raja K, Jonnalagadda S. Agile Text Mining for the i2b2 2014 Cardiac Risk Factors Challenge. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 17.Roberts K, Shooshan SE, Rodriguez L, Abhyankar S, Kilicoglu H, Demner-Fushman DNLM. Machine Learning Methods for Detecting Risk Factors for Heart Disease in EHRs. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]
- 18.Chen Q, Li H, Tang B, Liu X, Liu Z, Liu S, Wang W. Identifying risk factors for heart disease over time HITSZ’s system for track 2 of the 2014 i2b2 NLP challenge. Proceeding of the seventh i2b2 shared task and workshop challenges in natural language processing for clinical data; 2014. [Google Scholar]