Abstract
The PI3K/AKT/mTOR pathway is activated through multiple mechanisms in colorectal carcinoma. Here, the clinicopathologic and molecular features of AKT1 E17K–mutated colorectal carcinoma in comparison with PIK3CA-mutated colorectal carcinoma are described in detail. Interestingly, in comparison with PIK3CA mutants, AKT1 E17K was significantly associated with mucinous morphology and concurrent BRAF V600E mutation. Among PIK3CA mutants, exon 21 mutations were significantly associated with BRAF V600E mutation, MSI-H status, and poor differentiation, while exon 10 mutations were associated with KRAS/NRAS mutations. Three of four AKT1 mutants with data from both primary and metastatic lesions had concordant AKT1 mutation status in both. Both AKT1-and PIK3CA-mutant colorectal carcinoma demonstrated frequent loss of PTEN expression (38% and 34%, respectively) and similar rates of p-PRAS 40 expression (63% and 50%, respectively). Both patients with AKT1 E17K alone had primary resistance to cetuximab, whereas 7 of 8 patients with PIK3CA mutation alone experienced tumor shrinkage or stability with anti-EGFR therapy. These results demonstrate that AKT1 E17K mutation in advanced colorectal carcinoma is associated with mucinous morphology, PIK3CA wild-type status, and concurrent RAS/RAF mutations with similar pattern to PIK3CA exon 21 mutants. Thus, AKT1 E17K mutations contribute to primary resistance to cetuximab and serve as an actionable alteration.
Introduction
The phosphoinositide 3-kinase/v-akt murine thymoma viral oncogene/mammalian target of the rapamycin (PI3K/AKT/mTOR) pathway is activated in colorectal carcinoma through various mechanisms, including missense mutations in PIK3CA and AKT1 (v-akt murine thymoma viral oncogene homologue 1) and loss of PTEN expression. The PIK3CA gene encodes the p110α catalytic subunit of PI3K. Its activation promotes carcinogenesis through increased cell proliferation and survival (1). Eighty percent of oncogenic mutations in PIK3CA are clustered at hotspots in exon 10 at codons 542 and 545 and exon 21 at codon 1047 (2). Recently, proximal location and mucinous histology have been associated with PIK3CA-mutant colorectal carcinoma (3). Differences between the molecular profiles of PIK3CA exon 10– and 21–mutant colorectal carcinoma include a higher prevalence of concurrent KRAS mutation with PIK3CA exon 10 mutation and higher prevalence of BRAF co-mutation and microsatellite instability (MSI) with PIK3CA exon 21 mutation (4).
In 2007, Carpten and colleagues (5) first described the pleckstrin homology domain missense mutation AKT1 E17K in various types of human and mouse tumors, including breast, colorectal, ovarian, and myeloid malignancies. Mutant Akt (E17K) is more readily ubiquitinated and phosphorylated than wild-type Akt. The ubiquitinated-phosphorylated-Akt (E17K) translocates more efficiently to the nucleus than wild-type Akt, which may contribute to the transforming potential of E17K-Akt6 (6). Kumar and Purohit (7) have shown that the oncogenic effects of AKT1 E17K mutation may be due to rapid conformational changes affecting activation status. Activation of the protein encoded by the gene occurs via membrane localization, which is mediated by the pleckstrin homology domain, and followed by the subsequent phosphorylation of AKT1 at S473 and T308 positions (8).
Unlike PIK3CA and PTEN alterations, AKT1 mutations have not been thoroughly studied in colorectal carcinoma due to low numbers, although past studies have identified rare cases. While previous studies have identified rare cases, analysis of characteristics, including mutation prevalence, histopathologic features, concurrent driver mutations, and MSI status, has not been performed. In this study, we aimed to characterize the clinicopathologic, molecular, and pathway activation status of the AKT1 pleckstrin homology domain hotspot mutation E17K and to compare these cases with PIK3CA mutants in a large cohort of colorectal carcinoma.
Patients and Methods
Patients
After approval from the local Institutional Review Board, data were reviewed for a total of 2,631 colorectal carcinoma cases consecutively submitted for clinical molecular testing at MSKCC from 2009 to 2013. Testing was primarily to assess eligibility for anti-EGFR antibody therapy. Therefore, the vast majority of cases had distant metastases. An additional 491 cases with mutational analysis were identified through the cBio portal from The Cancer Genome Atlas (TCGA; ref. 9). All 141 colorectal carcinoma cases with AKT1 p. E17K, PIK3CA p.R88Q, p.E542K, p.E545K/G/D, or PIK3CA p. H1047L/R were included for further histopathologic, molecular (including RAS/RAF status), IHC, and clinical review.
Histopathology review
World Health Organization definitions were used to classify histopathology: mucinous histology was defined as >50% of tumor volume is composed of extracellular mucin. Moderately differentiated colorectal carcinoma was defined as 50% to 95% of tumor volume comprised of glands. Poorly differentiated colorectal carcinoma was defined as <50% of tumor volume forming glands (10).
Mutation detection
Genomic DNA was extracted from formalin-fixed paraffin-embedded tumor tissue after macrodissection using the DNeasy Tissue kit (Qiagen), following the manufacturer's standard protocol. All cases were analyzed with the MassARRAY system (Sequenom) with primers as previously described (11). Missense mutations in hotspots, including KRAS c.34, 35, 37 38, 181, 182, 183, 351, and 437; NRAS c. 34, 35, 37, 38, 181, 182, and 183; BRAF c. 1781, 1798, and 1799; PIK3CA c. 263, 1624, 1633, and 3140; and AKT1 c.49 were recorded.
MSI assays and mismatch repair protein IHC
For colorectal carcinomas harboring AKT1 E17K, PIK3CA E542K, PIK3CA E545K/G/D, and PIK3CA H1047R/L/Y mutations, MSI was assessed either by IHC for mismatch repair proteins MLH1, PMS2, MSH2, and MSH6, as previously described (12), at the time of surgery or by PCR using previously extracted DNA, permitting sufficient remaining DNA. MSI analysis was performed using the MSI analysis system (Promega) according to the manufacturer's instructions. Briefly, fluorescently labeled primers for BAT-25, BAT-26, NR-21, NR-24, and MONO-27 were used for DNA amplification. The PCR products were then separated by capillary electrophoresis. The presence of additional peaks forming a second Gaussian distribution in ≥2 of the 5 loci was interpreted as MSI-high (MSI-H).
PTEN and p-PRAS40 IHC
IHC staining was performed for PTEN (1:100; Cell Signaling Technology) and phospho-PRAS40 (1:40; Cell Signaling Technology), as previously described (13), on cases with AKT1 E17K, PIK3CA exon 10 mutations, or PIK3CA exon 21 mutations for which formalin-fixed paraffin-embedded tissue was available.
Statistical analysis
Statistical analyses were performed with a χ2 test for 2 × 2 categorical analyses. Two-tailed P values between 0.01 and 0.05, and those less than 0.01, were reported separately to account for multiple hypothesis testing.
Results
Mutation frequency and pathway activation
Out of 2,631 cases of colorectal carcinoma genotyped between 2009 and 2013, 18 (0.7%) cases were positive for AKT1 p. E17K (c. 49G>A) mutation, 9 (0.3%) cases were positive for PIK3CA exon 2 R88Q mutations, 82 (3.1%) cases were positive for PIK3CA exon 10 mutations, and 29 (1.1%) cases were positive for PIK3CA exon 21 mutations. Approximately one-third (33%–38%) and one half (47%–63%) of AKT1 E17K, PIK3CA exon 10, and PIK3CA exon 21 mutants lost PTEN expression and were positive for p-PRAS40 expression, respectively. The clinicopathologic and molecular profiles of these cases are summarized in Table 1 and illustrated in Figs. 1 and 2. Outcomes of KRAS/NRAS/BRAF wild-type patients who received anti-EGFR therapy are summarized in Table 2.
Table 1.
Clinicopathologic and molecular characteristics of colorectal carcinomas harboring AKT1 or PIK3CA mutations
| Clinicopathologic parameter | AKT1 E17K | PIK3CA E542K, E545K/G/D | PIK3CA H1047R/L/Y | PIK3CA R88Q | All PIK3CA |
|---|---|---|---|---|---|
| Age (range, mean, median) | 40–76, 57, 57 | 29–90, 58, 56 | 35–85, 62, 66 | 36–68, 58, 59 | 29–90, 59, 58 |
| Sex (M:F) | 10:8 | 39:43 | 12:17 | 3:6 | 54:66 |
| Proximal location | 10/18 (56%) | 32/82 (39%) | 14/29 (48%) | 6/9 (67%) | 52/120 (43%) |
| Hepatic metastases | 14/18 (78%) | 61/82 (74%) | 10/29 (35%)a,b | 4/9 (44%) | 75/120 (63%) |
| Pulmonary metastases | 9/18 (50%) | 24/82 (29%) | 5/29 (17%)c | 0/9 | 29/120 (24%)c |
| Mucinous histology | 7/21 (33%) | 3/82 (4%)a | 5/29 (17%) | 1/9 (11%) | 9/120 (8%)a |
| Poor differentiation | 3/21 (14%) | 10/82 (12%) | 10/29 (35%)b | 2/9 (22%) | 22/120 (18%) |
| NRAS or KRAS mutation | 11/21 (52%) | 55/82 (67%) | 13/29 (45%)d | 3/9 (33%) | 71/120 (59%) |
| BRAF V600E mutation | 7/21 (33%) | 5/82 (6%)a | 8/29 (28%)b | 0/9 | 13/120 (11%)a |
| MSI-H/MMR losse | 3/17 (18%) | 10/62 (16%) | 13/22 (59%)b | 3/8 (38%) | 26/92 (28%) |
| PTEN loss (IHC) | 6/16 (38%) | 6/17 (35%) | 5/15 (33%) | — | 11/32 (34%) |
| p-PRAS40 expression (IHC) | 10/16 (63%) | 8/17 (47%) | 8/15 (53%) | — | 16/32 (50%) |
P < 0.01, comparison with AKT1 E17K.
P < 0.01, comparison with PIK3CA E542K, E545K/G/D.
P < 0.05, comparison with AKT1 E17K.
P < 0.05, comparison with PIK3CA E542K, E545K/G/D.
The ratio of IHC to PCR testing (IHC:PCR) to assess MSI-H/MMR status for each PI3K mutation was as follows: AKT1 E17K (4:13), PIK3CA E542K, E545K/G/D (26:36); and PIK3CA H1047R/L/Y (13:9).
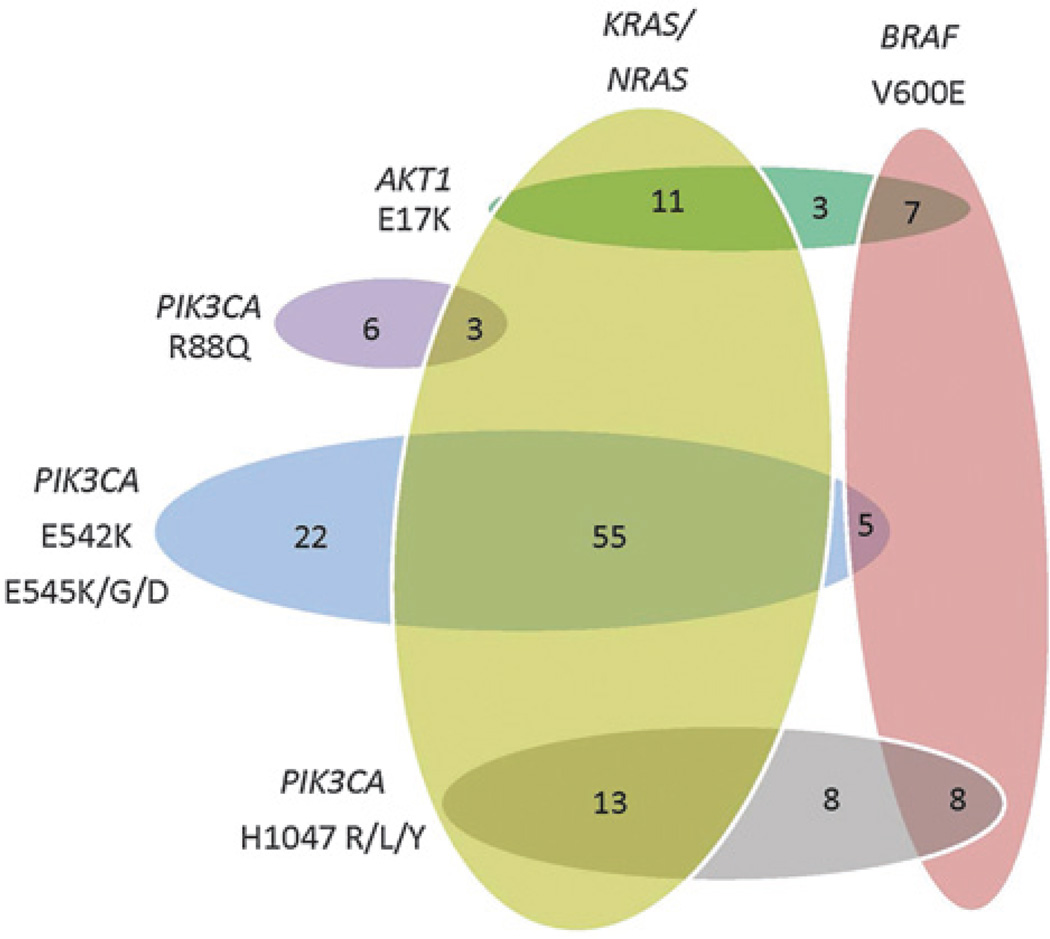
Figure 1.
Distribution of PI3K pathway mutations in colorectal carcinoma. AKT1 E17K– and PIK3CA H1047R/L/Y–mutant colorectal carcinoma had similar mutational profiles, harboring BRAF V600E co-mutations (33% and 28%, respectively) more frequently than PIK3CA E542K or E545K/G/D mutants (6%).
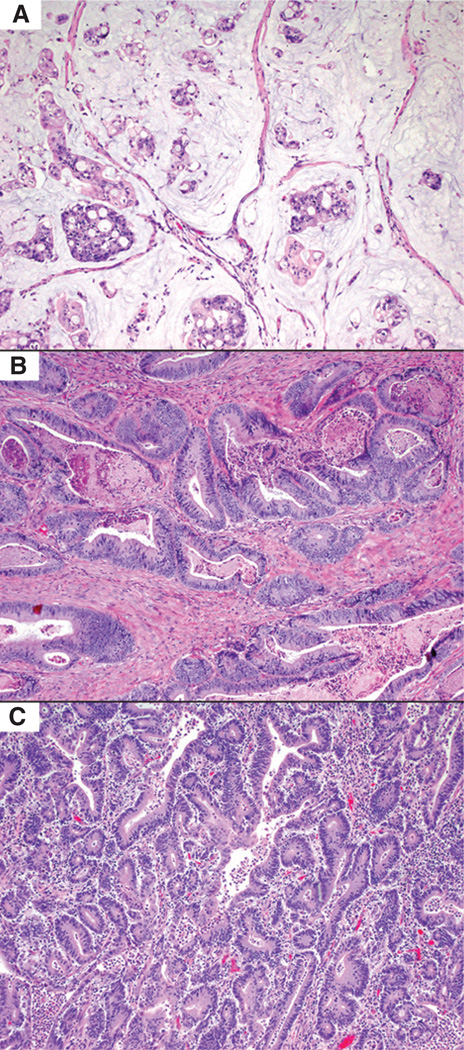
Figure 2.
Histologic features of AKT1 E17K- and PIK3CA-mutant colorectal carcinoma. A, AKT1 E17K–mutant colorectal carcinomas more frequently had mucinous histology (33% of cases), while PIK3CA E542K and E545K/G/D mutants (B) had conventional histology in 84% of cases. C, PIK3CA H1047R/L/Y–mutant colorectal carcinoma more frequently were MSI-H/mismatch repair protein deficient (59% of cases), with histologic features including increased intratumoral lymphocytes (A–C: H&E, ×100).
Table 2.
Response to anti-EGFR therapy in patients with stage IV KRAS/BRAF wild-type, PI3K/Akt pathway–mutant colorectal carcinomas
| PI3K pathway mutation | Synchronous interventions/chemotherapies | Outcome |
|---|---|---|
| PIK3CA R88Q | Fluorouracil, irinotecan, floxuridine, hepatic resection | No evidence of disease for at least 24 months |
| PIK3CA E542K | Irinotecan | Radiologic response for at least 5 months, including disappearance of multiple 1- to 2-cm hepatic metastases with stable size of peritoneal metastases |
| PIK3CA E542V | None | Stable disease for 6 months |
| PIK3CA E542K | Irinotecan | Stable disease at 1 month, progression of disease at 3 months |
| PIK3CA E545K | Irinotecan, fluorouracil, leucovorin, radiofrequency ablation, hepatic resection and infusion pump |
Stable disease for at least 9 months |
| PIK3CA E545K | Irinotecan | Stable disease for 10 months before progression |
| PIK3CA E545K | Irinotecan | No evidence of disease for at least 14 months (macroscopic disease resected before cetuximab) |
| PIK3CA H1047R | Yttrium-90 | Stable disease for 6 months |
| AKT1 E17K | Irinotecan | Progression of disease without any period of stabilization or response |
| AKT1 E17K | Irinotecan | Progression of disease without any period of stabilization or response |
AKT1 E17K
All 18 institutional AKT1 E17K cases had distant metastases. The site tested was primary tumor for 12 (67%) and distant metastasis for 6 (33%) cases. AKT1-mutated colorectal carcinoma was significantly associated with mucinous morphology (P = 0.0006) compared with PIK3CA-mutated colorectal carcinoma. Comparing exon-specific subsets, AKT1 mutants, like PIK3CA exon 21 mutants, were more likely to have mucinous differentiation and BRAF V600E co-mutation than PIK3CA exon 10 mutants (P = 0.0001).
Of the 18 AKT1 E17K cases, 15 (83.3%) cases had concurrent RAS/RAF–activating mutations in the same sample, including 6 (33.3%) KRAS exon 2 mutations, 5 (27.8%) BRAF V600E mutations, 2 (11.1%) KRAS exon 4 mutation, 1 (5.9%) NRAS exon 3 mutation, and 1 (5.9%) case with both a KRAS exon 2 and an NRAS exon 3 mutation. The mutation status of the last case was confirmed by repeat extraction and Sanger sequencing. In 4 patients, material was available to correlate the AKT1 status of both primary and metastasis. All 4 patients had the same RAS/RAF–activating mutation in the primary and metastasis. Three of these 4 patients had AKT1 E17K mutations in both primary and metastasis. Interestingly, the fourth patient harbored an AKT1 E17K mutation (concurrent with KRAS) in a liver metastasis and a PIK3CA E542K mutation in the primary tumor (with the same KRAS mutation). These findings are suggestive of convergent, or parallel, evolution in the PI3K pathway (14). Two patients with AKT1 E17K mutations who had wild-type KRAS and BRAF received cetuximab, and both demonstrated primary resistance.
Three additional AKT1 E17K–mutated cases were identified via the cBioPortal of The Cancer Genome Atlas, among 491 total colorectal carcinoma. One case had mucinous features and harbored a BRAF V600E mutation. The other two cases were moderately differentiated, one with a BRAF V600E mutation and the other with a KRAS A146T mutation.
PIK3CA exon 10
Seventy of 82 (85.4%) PIK3CA exon 10 mutations, including E542K (32), E545K (44), E545G (5), and E545D (1), were identified in stage IV patients. The site tested was the primary tumor for 51 (62.2%) and distant metastasis for 31 (37.8%) cases. Primary tumors were located in the proximal colon (n = 32, 39.0%), transverse (n = 8, 9.8%), and distal colon (n = 42, 51.2%). In comparison with PIK3CA exon 21 mutants, these cases less frequently had poor differentiation (P = 0.007) and more frequently had hepatic metastases (P = 0.0001).
Sixty (72.3%) cases had activating RAS/RAF mutations detected in the same sample. Of these, 49 harbored KRAS exon 2 mutations, 5 harbored BRAF V600E mutations, 3 had KRAS exon 4 mutations, 1 had a KRAS exon 3 mutation, and 1 had mutation in both KRAS and NRAS at exon 2. Compared with exon 21 kinase domain mutations, PIK3CA exon 10–mutated colorectal carcinomas showed a higher prevalence of NRAS/KRAS mutations and a lower prevalence of BRAF mutations (P = 0.035 and 0.002, respectively). Twenty-three (23.7%) cases were wild-type for KRAS, BRAF, and NRAS.
PIK3CA exon 21
A total of 29 cases had PIK3CA exon 21 mutations: H1047R (25), H1047L (3), and H1047Y (1). Two cases with H1047R mutations had a second PIK3CA mutation: C420R in one and R88Q in the other. Twenty-two cases (75.9%) were stage IV. The site tested was the primary tumor for 23 (79.3%) and distant metastasis for 6 (20.7%) cases.
Twenty-one (72.4%) patients had RAS/RAF co-mutations. These included 12 (41.4%) KRAS exon 2 mutations, 8 (27.6%) with BRAF V600E mutations, and 1 (3.5%) KRAS exon 3 mutation. Eight patients were wild-type for KRAS, NRAS, and BRAF. Thirteen of 21 (59%) cases with available material for microsatellite instability polymerase chain reaction or mismatch repair protein immunohistochemistry were microsatellite instability-high/mismatch repair deficient (P = 0.0002), compared with all other PIK3CA mutants.
PIK3CA R88Q
The clinicopathologic features of the 9 PIK3CA mutation R88Q are summarized in Table 1. Five patients were stage IV. Four patients had liver metastases and 2 had intra-abdominal metastases. The site tested was primary tumor for 4 patients and metastasis for 5 patients. Only 3 patients (33%) in this group also harbored a RAS/RAF mutation. These included 2 KRAS exon 2 mutations and 1 KRAS exon 4 mutation. This contrasts with the helical and kinase domain PIK3CA mutants as well as the AKT1 E17K mutants, which had RAS/RAF driver mutations in over 70% of cases.
Discussion
This study is the first to characterize the clinical, pathologic, molecular, and pathway activation properties of colorectal carcinoma harboring AKT1 E17K mutations, with comparison to PIK3CA mutations. We find that colorectal carcinoma with AKT1 E17K mutation has a higher incidence of concurrent BRAF V600E mutations and mucinous histology in comparison with PIK3CA exon 10–mutated colorectal carcinoma. Our finding that PIK3CA exon 21 mutations have a higher prevalence of BRAF mutations and MSI-H status than PIK3CA exon 10 mutations is in agreement with recent reports from other groups. The prevalence of mucinous histology by PIK3CA mutation in our cohort was also in line with previous work (4).
The AKT1 mutations were concurrent with activating mutations in the RAS/RAF pathway in the majority (85%) of cases, but were consistently mutually exclusive with PIK3CA mutations. The 3 cases of AKT1 E17K mutation that occurred in the absence of RAS/RAF mutations shared clinical features with other AKT1-mutant cases, including mucinous histology and microsatellite stable status.
The impact of AKT1 mutation on response to anti-EGFR therapy has not been previously studied. Two of these 3 patients received anti-EGFR therapy (cetuximab), and both patients were resistant to this treatment. In comparison, of 8 patients with PIK3CA-mutant and KRAS/BRAF wild-type colorectal carcinoma, 7 (88%) exhibited either tumor regression or stability when treated with anti-EGFR therapy, either alone or in combination with other therapies (Table 2). A possible explanation for this different response to anti-EGFR therapy is that mutant PIK3CA leads to increased phosphorylation and dependence on ERK, which can be inhibited by cetuximab, while AKT1 E17K mutation does not increase phosphorylation of ERK1/2 (15). While anti-EGFR therapy may not be an effective therapeutic option for these patients, targeted therapy against the AKT/mTOR pathway may be an emerging option in patients with metastatic disease.
Similar to PIK3CA H1047R/L/Y–mutated colorectal carcinoma, AKT1 E17K colorectal carcinoma showed a high rate of concurrent BRAF V600E mutation (28% for both PIK3CA exon 21 and AKT1 institutional cases), yet not as high of a rate of MSI (59% for PIK3CA exon 21 vs. 18% for AKT1). This relative enrichment of microsatellite-stable BRAF V600E mutants might account for the higher number of colorectal carcinoma with distant metastases (75.9% vs. 100%) seen in our AKT1 E17K colorectal carcinoma cohort as BRAF-mutant microsatellite-stable colorectal carcinoma has been associated with more aggressive disease and poor survival (16).
Unlike AKT1 E17K– and PIK3CA H1047R/L/Y–mutated colorectal carcinoma, PIK3CA R88Q–, E542K–, and E545K/G/D–mutated colorectal carcinoma had a very low incidence of BRAF V600E co-mutation and relatively higher incidences of KRAS co-mutations. Recent work has shown that PIK3CA helical domain mutations in codons 542 and 545 require activation of the RAS–GTP pathway, while kinase domain mutations in codon 1047 do not (17). This may explain why we found that the PIK3CA exon 10–mutant cases had more RAS and fewer BRAF co-mutations in comparison with the PIK3CA exon 21–mutant cases.
These associations are in line with previous work by Day and colleagues (4) for PIK3CA E542, E545, and H1047 mutations. To our knowledge, this is also the first clinicopathologic report of PIK3CA R88Q in colorectal carcinoma. PIK3CA R88Q mutations have previously been reported in endometrial carcinoma. This mutation is thought to activate PI3K through the formation of a hydrogen bond with D746 that results in conformational change of the encoded protein and increased kinase activity (18). While our numbers were too low for statistical analysis, the histologic and molecular profile of this subset seems more similar to helical domain (E542 and E545) mutant cases than kinase domain (H1047) mutant cases.
Our data also show that PTEN loss of expression and activating mutations within other areas of the PI3K mutation are not mutually exclusive as approximately one third of all AKT1 or PIK3CA mutants lost PTEN expression. We found no difference in downstream activation of p-PRAS40 by IHC between AKT1 E17K, PIK3CA helical domain, PIK3CA kinase domain, RAS, and BRAF mutants in this study.
A potential bias of our study is the preponderance of metastatic cases. In the vast majority of colorectal carcinoma submitted for clinical molecular analysis at our institution, testing is performed to determine candidacy for anti-EGFR therapy, and, therefore, almost all of our cases (including all 17 AKT1 E17K) were late stage. Whether these results are applicable across all stages remains to be seen. Another shortfall pertains to the material tested: The technical sensitivity of our assay is 10%, thus false negatives due to low percentages of tumor cells or subclonal mutations may not be excluded. Finally, although the correlation between MSI testing by PCR and mismatch repair protein expression is very good, rare cases with mismatch repair abnormalities may be missed by either method.
In summary, the mutation AKT1 E17K in colorectal carcinoma is associated with mucinous morphology, pulmonary metastases, and co-mutation of BRAF V600E. They occur in mutual exclusivity of PIK3CA mutations. Although AKT1 E17K mutations, even without concurrent BRAF or RAS mutations, seem to confer primary resistance to anti-EGFR therapy in our limited series, this observation warrants further studies. In addition, it may serve as a targetable alteration in both RAS/RAF–mutated and RAS–RAF wild-type colorectal carcinoma.
Implications.
This first systematic study of AKT1 and PIK3CA hotspot mutations and their association with cetuximab resistance and BRAF V600E mutation has important ramifications for the development of personalized medicine, particularly in identifying patient candidates for PI3K or AKT inhibitors.
Acknowledgments
The MSKCC Sequenom facility was supported by the Anbinder Fund. The authors thank Angela Yannes for assistance with Sequenom assays and Dr. Khedoudja Nafa, Ph.D., for assistance with MSI PCR.
Footnotes
Disclosure of Potential Conflicts of Interest
J.F. Hechtman is a consultant/advisory board member for Navigant. R. Yaeger is a consultant/advisory board member for Amgen. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: J.F. Hechtman, M. Ladanyi, M.E. Arcila
Development of methodology: J.F. Hechtman, J.T. Huse, J. Shia, M.E. Arcila
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): J.F. Hechtman, J.T. Huse, L. Borsu, R.D. Yaeger, J. Shia, E. Vakiani, M.E. Arcila
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.F. Hechtman, J. Sadowska, J.T. Huse, R.D. Yaeger, J. Shia, E. Vakiani, M.E. Arcila
Writing, review, and/or revision of the manuscript: J.F. Hechtman, J. Sadowska, J.T. Huse, R.D. Yaeger, J. Shia, E. Vakiani, M. Ladanyi, M.E. Arcila
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J. Sadowska, M.E. Arcila
Study supervision: M. Ladanyi, M.E. Arcila
References
- 1.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 2.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 3.Rosty C, Young JP, Walsh MD, Clenndenning M, Sanderson K, Walters RJ, et al. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8:e65479. doi: 10.1371/journal.pone.0065479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day FL, Jorissen RN, Lipton L, Mouradov D, Sakthianandeswaren A, Christie M, et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clin Cancer Res. 2013;19:3285–3296. doi: 10.1158/1078-0432.CCR-12-3614. [DOI] [PubMed] [Google Scholar]
- 5.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 6.Fan CD, Lum MA, Xu C, Black JD, Wang X. Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J Biol Chem. 2013;288:1674–1684. doi: 10.1074/jbc.M112.416339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Purohit R. Cancer associated E17K mutation causes rapid conformational drift in AKT1 pleckstrin homology (PH) domain. PLoS One. 2013;8:e64364. doi: 10.1371/journal.pone.0064364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landgraf KE, Pilling C, Falke JJ. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry. 2008;47:12260–12269. doi: 10.1021/bi801683k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosman F, Carneiro F. Lyon, France: IARC Press; 2010. World Health Organization classification of tumours, pathology and genetics of tumours of the digestive system. [Google Scholar]
- 11.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shia J, Klimstra DS, Nafa K, Offit K, Guillem JG, Markowitz AJ, et al. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colorectal neoplasms. Am J Surg Pathol. 2005;29:96–104. doi: 10.1097/01.pas.0000146009.85309.3b. [DOI] [PubMed] [Google Scholar]
- 13.Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18:2490–2501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 14.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multi-region sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebucci M, Peixoto P, Dewitte A, Wattez N, DeNuncques MA, Rezvoy N, et al. Mechanisms underlying resistance to cetuximab in the HNSCC cell line: role of AKT inhibition in bypassing this resistance. Int J Oncol. 2011;38:189–200. [PubMed] [Google Scholar]
- 16.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, et al. Poor survival associated with the BRAF V600E mutation in micro-satellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino MJ, et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17:1331–1340. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]