Abstract
Objective
To determine if service members deployed to locations with open air burn pits have different serum microRNA (miRNA) profiles after deployment compared to length-of-service matched, non-deployed individuals. We also tested for correlations between miRNA and serum levels of Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans (PCDD/PCDFs).
Methods
MiRNAs were isolated and quantified by PCR array. Groups were analyzed for differences in miRNA expression. Correlation between serum miRNA and PCDD/PCDFs were assessed with a linear regression model.
Results
Several miRNAs were differentially expressed after deployment and a partially overlapping set of miRNAs were identified between deployed and non-deployed individuals. Significant correlations between miRNAs and PCDD/PCDFs were identified.
Conclusions
Serum miRNA levels show a link between deployment to locations with open burn pits and environmental exposures that can take place during deployment.
Introduction
Detection and quantification of serum microRNAs (miRNAs) for biomarker discovery or diagnostic purposes is a rapidly evolving field. MiRNAs are small, non-coding RNAs that can function to decrease gene expression by binding target mRNAs leading to translational repression or increased rates of mRNA degradation.(1) There are over 1500 reported human miRNAs with more being identified by multiple genomic approaches.(2) Many of these miRNAs are grouped in families that are similar in sequence and target the same or related genes and biological pathways.(3) On average, one miRNA can target approximately 400–500 different target mRNAs and thus miRNA expression is tightly controlled.(4, 5) Interestingly, miRNAs are abundant in the blood, as they are found in red blood cells, extracellular vesicles and/or in freely circulating protein complexes.(6, 7) MiRNAs have been shown to be quite stable in serum/plasma samples due to their small size, presence in protein complexes such as Argonaute-RISC complex and/or presence in small extracellular vesicles such as exosomes.(7–11)
Numerous reports have identified specific serum/plasma miRNAs to be correlated with diseases such as cancer, Alzheimer’s disease, Type 2 diabetes and lung diseases such as COPD.(12–15) We have previously shown that serum samples from the Department of Defense Serum Repository (DoDSR) are of sufficient quality to detect important biomarkers including over 150 different miRNA. Interestingly, specific miRNA show a strong correlation with other serum markers including cytokines, PAHs and immunoglobulin E (Woeller et al., this issue). Key remaining questions include whether the DoDSR samples can be used to detect specific changes or differences in biomarkers like miRNA expression that occur as a result of deployment, and whether these biomarkers are useful to identify past environmental exposures and/or potential resulting health issues.
One mission of the DoDSR is to collect and store serum samples from service members before and after deployment.(16) Service members have been deployed to various locations worldwide including recent war zone areas in Afghanistan and Iraq.(17) In these deployment zones, environmental exposures can be a major occupational hazard for service members.(18) Smoke from open burn pits, dust storms and diesel exhaust are potential harmful exposures for deployed service members and these insults may predispose individuals to respiratory diseases such as COPD, asthma and bronchiolitis.(19–22)
In this study, serum samples were obtained from service members deployed to sites with open burn pits, and service members who were not deployed (see Mallon et al, this issue). Importantly, serum samples were obtained from service members before and after deployment for the deployed group, and matched with control sera that were drawn from non-deployed service members who had similar times of service. The objectives of this study include: 1) identify a set of differentially expressed miRNAs in service members before and after deployment, 2) identify a set of miRNAs that are different between deployed (Case) or not deployed (Control) service members, and finally 3) determine whether specific miRNA levels significantly correlate with serum PAH, polychlorinated dibenzo-p-dioxin (PCDD) and polychlorinated dibenzofuran (PCDF) data reported in this issue (Xia and Hopke et al., this issue).
Currently, little is known about how serum miRNA status is affected by environmental PAH and dioxin/furan exposure. Recent studies have begun to address the relationship between miRNA expression and exposure to PAHs and dioxins. For example, Song et al treated HepG2 cells with the PAHs benzo[a]anthracene (BA) and benzo[k]fluoranthene (BF) in vitro and found an induction of the miR-181 family of miRNAs.(23) In another in vitro model Gordon et al demonstrated that two p53 targeting miRNAs (miR-25 and miR-92) were induced by benzo[a]pyrene and dioxin treatment of multiple myeloma cells.(24) Finally, Deng et al., reported a relationship between several plasma miRNAs and urinary PAH levels in coke oven workers.(25)
Herein, we sought to identify serum miRNAs that are differentially expressed after deployment to locations with open burn pits. We also aimed to identify correlations between serum miRNAs and serum PAH, PCDDs and/or PCDFs. Differentially expressed miRNAs may represent novel biomarkers of deployment status and/or deployment related environmental exposure.
Materials and Methods
Eight hundred de-identified serum samples were obtained from the DoDSR for targeted analysis of miRNA levels. Serum samples were collected from a cohort of service personnel who were deployed to Joint Base Balad in Iraq (2007–2008) or Bagram Airbase in Afghanistan (2011–2012). The 200 individuals selected for this study represent a subset of a larger study (19, 21) who had sufficient remaining sera for all the planned analyses at multiple sites (the Case group). Blood samples were drawn at medical facilities in the US pre- and post-deployment. The majority of deployments lasted between 6 and 12 months. The Control group consisted of 200 service members who were not deployed overseas, and who were selected based on the availability of serum samples collected from routine medical encounters that matched the Case group for time-in-service. The overall study design is described in more detail elsewhere (Mallon et al., this issue) and the serum samples were handled as previously described (Woeller et al., this issue).
Isolation and analysis of miRNA from serum samples
Briefly, miRNA was extracted from 0.15 ml of serum and reverse-transcribed into corresponding miRNA cDNA as described previously (Woeller et al., this issue). The cDNA was analyzed on low density miRNA PCR arrays that contain serum/plasma focused miRNA panels (Exiqon, Boston, MA). The serum was processed in two batches of 400 samples each. PCR quantification cycles (Cq) values after quality control were compiled and data was normalized using the Normfinder.(26) Briefly, Normfinder ranks candidate normalization miRNAs (or combinations of miRNAs) based on their stability. Mestdagh et al. demonstrate that mean expression values of commonly expressed miRNAs are superior to other normalization methods in miRNA profiling studies.(27) In this study of 800 serum samples, the most stable normalizer was the mean expression of miRNAs common to all samples (n= 11 miRNAs: miR-451a, miR-16-5p, miR-144-3p, miR-24-3p, miR-23b-3p, miR-21-5p, miR-25-3p, miR-15a-5p, miR-23a-3p, miR-142-3p and let-7g, stability value = 5.92E-04). Thus, miRNA levels were normalized to the mean expression (Cq values) of these common miRNAs. After normalization, data was batch corrected using the Combat program and converted into relative miRNA levels for each specific miRNA.(28)
Cotinine analysis
Tobacco use was determined by cotinine analysis. Serum cotinine was measured by EIA as described previously (Woeller et al, this issue) and was used as a covariate in subsequent correlation tests. Values less than 1.0 ng/ml were defined as non-users and greater than 10ng/ml as tobacco users. Values between 1 and 10 ng/ml would meet the criteria for exposure to second hand tobacco smoke (9 of 800 samples, 1.1%). (29)
Statistical analysis
Statistically significant differences in the miRNA were tested using unpaired Student’s t-test. Benjamin-Hochberg (BH) method was used wherever indicated to adjust p-values for multiple hypothesis testing. To test the association of miRNA with PAH, PCDD and PCDF levels, a linear regression analysis was performed using cotinine and batch as a covariate. The levels of PAH, PCDD and PCDF were discretized into low and high or low, medium and high categories because of the zero-inflated distributions. The R package miRNApath(12) was used to perform the pathway analysis. Pathway information was obtained from KEGG(30, 31) and Biocarta(32) databases. All the analysis was performed in R and the heatmaps were created utilizing gplots.(33)
Results
Serum samples show distinct miRNA expression patterns based on deployment status
To determine if we could identify differentially expressed serum miRNAs in service members before and after deployment, serum samples were obtained from service members deployed to sites with open burn pits (Case) and service members who were never deployed (Control). Importantly, serum was collected before (Pre) and after deployment (Post) or at matched time-in-service from non-deployed individuals. In total, 800 serum samples were obtained with 200 samples in each of the four groups: 1) Control Pre, 2) Control Post, 3) Case Pre and 4) Case Post (See Figure 1). MiRNA isolation and detection was performed as previously described using the serum/plasma miRNA panel (version 3 or 4) from Exiqon (Woeller et al, this issue). All 800 samples were found to contain miRNA, however, five samples contained very low amounts of miRNA and were excluded from subsequent analysis. Another source of unwanted variation in serum sample miRNA expression is contamination from red blood cell lysis (hemolysis) during serum or plasma collection.(34) As in Blondal et al., we used the difference in expression between miR-451 (predominantly a red blood cell miRNA) and miR-23a (predominantly a non-cellular miRNA) to test for red blood cell miRNA contamination.(34) Here, four samples showed high levels of miR-451 which suggests some hemolysis and these samples were also omitted from subsequent analysis. Thus, 791 out of 800 (99%) of the samples were used for miRNA profiling.
Figure 1. Study design and sample nomenclature.
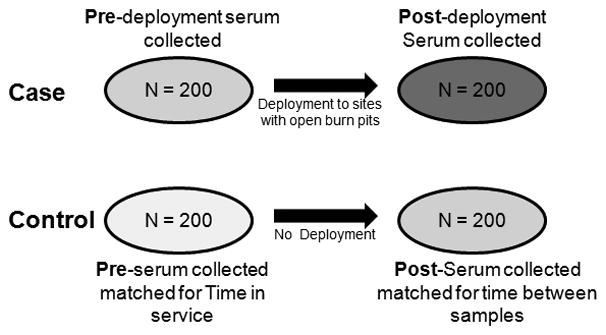
800 de-identified serum samples were obtained from the Department of Defense Serum Repository (DoDSR) for targeted analysis of microRNA (miRNA) levels. Serum samples were collected from a cohort of 200 service personnel who were deployed to either Joint Base Balad in Iraq (2006–2008) or Bagram Airbase in Afghanistan (2011–2012). These samples represent the Case group. Sera was collected Pre and Post deployment (N= 400 Case samples, 200 Pre and 200 Post). The Control group consisted of 200 service members who were not deployed overseas, and who were selected for matched time-in-service in both Pre and Post draws (N= 400 Control samples, 200 Pre and 200 Post). These groups were assigned and miRNA profiling was performed to identify changes in miRNA expression.
Expression profiling of serum miRNA revealed that on average, 120 miRNAs from the serum/plasma panel were detected per sample. 72 miRNAs were expressed in 95% of all samples indicating robust and consistent miRNA detection in analyzed sera. After quality control, normalization of miRNA expression and subsequent batch correction using Combat, the relative miRNA expression profiles were separated into their respective groups (Control Pre, Control Post, Case Pre and Case Post). Our first analysis was to determine if Case and Control samples expressed different serum miRNA levels before Case deployment. Thus a Case Pre vs. Control Pre comparison is provided in Table 1. 19 miRNAs were differentially expressed between the Case and Control group before Case deployment, with 8 miRNAs increased and 11 miRNAs decreased in expression in the Case Pre group, respectively (Table 1). These results reveal that, despite the controls being selected randomly, there were differences in serum miRNA between Control and Case groups before deployment, and thus, these miRNAs must be taken into account when comparing Control and Case groups after Case deployment.
Table 1. List of microRNAs significantly different in serum from service members in the Control Pre and Case Pre groups.
All microRNAs listed are significant with a p value cut off of < 0.05. Specific p values are shown for each miRNA. Changes denote increase (up) or decrease (down) in Case Pre deployment samples. MiRNAs shaded in gray are also significantly different in Control Post and Case Post groups and thus these differences highlight changes independent of both deployment and differences in Control and Case populations.
| Control Pre vs. Case Pre | Change | p-value |
|---|---|---|
| miR-101-3p | DOWN | 0.041 |
| miR-127-3p | UP | 0.0296 |
| miR-140-5p | UP | 0.0268 |
| miR-150-5p | DOWN | 0.0175 |
| miR-15a-5p | DOWN | 0.0171 |
| miR-18a-5p | UP | 0.0004 |
| miR-191-5p | UP | 0.0389 |
| miR-20a-5p | DOWN | 0.0142 |
| miR-21-5p | DOWN | 0.007 |
| miR-223-5p | UP | 0.0448 |
| miR-23a-3p | UP | 0.0349 |
| miR-301a-3p | UP | 0.0143 |
| miR-30b-5p | UP | 0.008 |
| miR-320b | DOWN | 0.006 |
| miR-34a-5p | DOWN | 0.0474 |
| miR-374a-5p | DOWN | 0.0161 |
| miR-451a | DOWN | 0.0078 |
| miR-486-5p | DOWN | 0.0047 |
| miR-92a-3p | DOWN | 0.0003 |
miRNAs significantly different in Control and Case groups independent of deployment (Significant in Control Pre vs. Case Pre and Control Post vs. Case Post)
The next analysis was to test for differences in miRNA expression in the deployed (Case) group (Table 2A). Excitingly, six miRNAs were differentially expressed in the Case group after deployment with 5 miRNAs increasing and 1 miRNA decreasing in expression, respectively. Next, we compared the Control Post and Case Post groups (Table 2B). Twenty-four miRNAs were differentially expressed in the Control Post vs. Case Post groups. Seven of the miRNAs that were significantly different between these two groups were also identified in the Control Pre vs. Case Pre comparison. These seven miRNAs were removed from Table 2B (7 miRNAs shaded in Table 1). Taken together, 6 miRNAs were significantly different in the Case group after deployment and 17 miRNAs were significantly different between the Case and Control groups after Case deployment to sites with open burn pits.
Table 2. List of microRNAs significantly changed in serum from service members post deployment and between deployed vs. non deployed groups.
A.) Case Pre vs. Case Post analysis identified significant changes in miRNAs after the Case group returned from deployment. B.) Control Post vs. Case Post analysis lists significant miRNAs between Case and Control group after the Case group returned from deployment. All microRNAs listed are significant with a p value cut off of < 0.05. Specific p values are shown for each miRNA. Changes denote increase (up) or decrease (down) in Case Post groups. MicroRNAs in bold are also significant in both post deployment comparisons.
| A | ||
|---|---|---|
| Case Pre vs. Case Post | Change | p value |
| let-7c-5p | UP | 0.015 |
| miR-145-5p | UP | 0.006 |
| miR-17-5p | UP | 0.008 |
| miR-19a-3p | DOWN | 0.027 |
| miR-222-3p | UP | 0.031 |
| miR-29c-3p | UP | 0.048 |
| B | ||
|---|---|---|
| Control Post vs. Case Post | Change | p value |
| let-7a-5p | UP | 0.024 |
| let-7d-5p | UP | 0.001 |
| miR-126-3p | UP | 0.023 |
| miR-144-3p | DOWN | 0.015 |
| miR-144-5p | UP | 0.004 |
| miR-145-5p | UP | 1.28E-05 |
| miR-16-2-3p | DOWN | 0.042 |
| miR-16-5p | DOWN | 0.039 |
| miR-17-5p | UP | 0.037 |
| miR-23b-3p | UP | 0.043 |
| miR-24-3p | UP | 0.013 |
| miR-25-3p | DOWN | 0.028 |
| miR-30c-5p | UP | 0.047 |
| miR-32-5p | DOWN | 0.015 |
| miR-363-3p | DOWN | 0.007 |
| miR-374b-5p | UP | 0.046 |
| miR-424-5p | UP | 0.003 |
miRNAs significantly different in both comparisons
Figure 2A is a summary of the significantly different miRNA levels between the 4 sample sets. The most relevant differences to the study of service-related environmental exposures are Case Pre vs. Case Post and Case Post vs. Control Post (see Table 2). After filtering out differences between the Case and Control groups that existed pre-exposure, 21 miRNAs of interest remain that were changed by deployment to locations with open burn pits (Figure 2B). Interestingly, miR-145-5p and miR-17-5p were identified in both the Case Pre vs. Case Post analysis and the Control Post vs. Case Post analysis (shaded miRNAs in Tables 2A and 2B). Both of these miRNAs showed an increase in expression in the Case Post group when compared to either Control Post or Case Pre. MiR-145-5p and miR-17-5p have the lowest p values (0.006 and 0.008, respectively) in the Case Pre vs. Case Post analysis. Additionally, miR-145-5p has the lowest p-value in the Control Post vs. Case Post analysis (p = 1.28E-05). These data suggest that increased levels of miR-145-5p and miR-17-5p may be important markers of deployment to locations with open burn pits. Other miRNAs of interest as potential markers of deployment include let-7 miRNA family members. Let-7 miRNAs are a set of related miRNAs with similar biological functions. Let-7c-5p was significantly increased in the Case Pre vs. Case Post analysis while both let-7a-5p and let-7d-5p were significantly increased in the Control Post vs. Case Post analysis. (See Table 1 and 2B). These results suggest that an increase in let-7 miRNAs may also be an important marker of deployment status. Other miRNAs that are sequence related and found to be significantly associated with deployment were miR-25-3p and miR-32-5p. Both of these miRNAs were decreased in the Control Post vs. Case Post analysis.
Figure 2. Summary of serum microRNAs that were found to be significantly associated with deployment to sites with open burn pits.
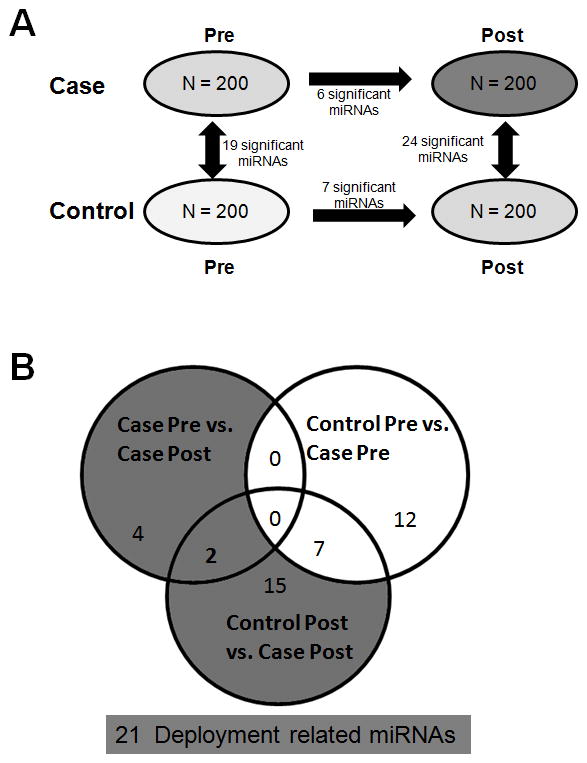
A.) The 4 groups were analyzed for significant changes in miRNA levels. Each group analyses revealed a number of significant changes as shown by the number next to each arrow for comparison. B.) Venn diagram revealing the subset of miRNAs that are potentially deployment related miRNAs. The significant miRNAs in Case Pre vs. Case Post and Control Post vs. Case Post comparisons but not in the Control Pre vs. Case Pre comparison totaled 21 miRNAs.
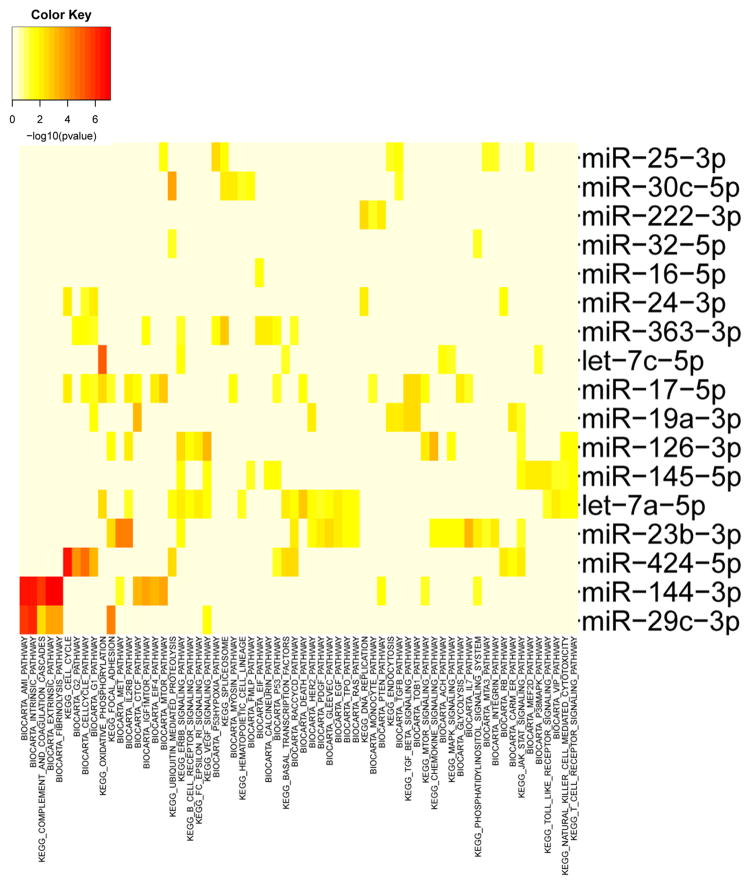
Many of the deployment related miRNAs are from different miRNA families and contain distinct seed sequences. Even with different seed sequences, some deployment related miRNAs may target similar signaling pathways that could be of significant biological relevance. To assess similarities, we grouped predicted miRNA target genes by cognate KEGG and Biocarta pathways using miRNApath (Figure 3).(12, 35) Interestingly, multiple cellular pathways are regulated by two or more miRNAs related to deployment (Figure 3). For example, multiple miRNAs target cell cycle, ErbB, focal adhesion and JAK-STAT signaling pathways. In particular, ErbB signaling (miR-363-3p, miR-145-5p, miR-23b-3p, miR-126-3p, let-7c and let-7a) and JAK-STAT signaling (miR-19a-3p, miR-126-3p, miR-145-5p, miR-23b-3p and miR-424-5p) have 5 or more common deployment related miRNAs targeting these pathways. These data suggest that strong relationships between deployment to areas with open burn pits and key cellular pathways are mediated by miRNAs.
Figure 3. Pathway analysis of miRNAs significantly associated with deployment to sites with open burn pits.
Pathway analysis (see methods) of miRNAs associated with deployment to locations with burn pits based on ranking according to p-values was performed in R (R package miRNApath1 for pathway analysis). The color map p-values indicate the strength of the association between the miRNA and the relevant pathway. Pathways targeted by fewer than 2 miRNAs and miRNAs that targeted fewer than 2 pathways were omitted for clarity.
Serum PCDD and PCDF levels correlate with specific miRNA levels
Burning solid waste in open burn pits while adding JP-8 jet fuel as an accelerant causes numerous pollutants to be released into the surrounding air, including carbon monoxide, respirable particulate matter, polycyclic aromatic hydrocarbons (PAHs), polychlorinated dibenzo-p-dioxins (PCDDs) and polychlorinated dibenzofurans (PCDFs).(19) PCDDs and PCDFs are generated as the result of incomplete combustion of plastics and other materials present in open burn pits.(36–38) Using the serum PAH, PCDD and PCDF data from Xia et al., (This issue), we performed a linear regression analysis as described in the Methods section with serum miRNA data to identify significant associations. Importantly, since tobacco smoke can also lead to exposure, cotinine status was used as a covariate in the analysis.
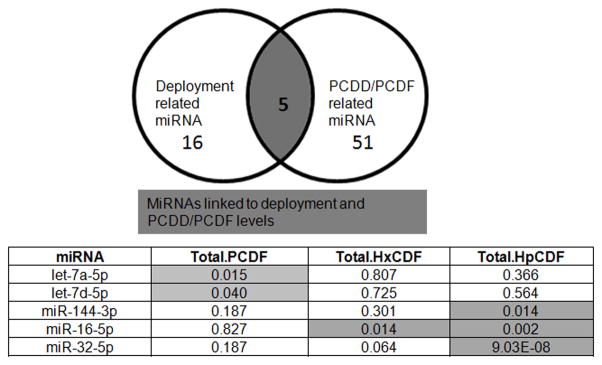
In this analysis, all 800 samples were treated as discreet data points, without regard to deployment or other sources of PAHs, PCDD and PCDF exposure, which are widespread in the environment.(39–41) Significant miRNAs were initially discovered to be associated with serum levels of PAHs, PCDDs and PCDFs. After false discovery rate correction, PAHs showed no significantly associated miRNAs in this set. However, 56 miRNAs were found to have significant correlations with PCDDs and/or PCDFs (Table 3). Since PCDDs and PCDFs could accumulate in serum as the result of many environmental exposures, not just deployment and open burn pit smoke, we determined if there were any overlapping miRNAs that showed significant relationships with both PCDD/PCDFs and deployment to locations with open burn pits (Figure 4A). We identified 5 miRNAs that were altered with deployment and with PCDD/PCDFs (Figure 4B). Thus, while 56 miRNAs are impacted by PCDDs and PCDFs in general, 5 were specifically affected by PCDDs and PCDFs in the context of deployment to sites with open burn pits. Interestingly, in this analysis two let-7 miRNAs (let-7a-5p and let-7d-5p) were significantly correlated with total PCDFs, while miR-144-3p, miR-16-5p and miR-32-5p are associated with heptachlorodibenzofuran (HpCDF).
Table 3.
List of serum microRNAs that were found to be significantly correlated with specific or total PCDD/PCDFs. All sample groups were merged and used in analysis and significance was analyzed after controlling for cotinine status as a covariate (q<0.25). Highlighted columns show significant p-values (p < 0.05) after Benjamin-Hochberg false discovery rate correction.
| Tetrachloro | Pentachlorodi | Hexachlorodi | Heptachlorodi | Octachlorodi | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| dibenzo.p.di | benzo.p.dioxi | benzo.p.dioxi | benzo.p.dioxi | benzo.p.dioxi | Total | Total | Total | Total | Total | |
| miRNA | oxins | ns | ns | ns | ns | Dioxins | TCDF | PCDF | HxCDF | HpCDF |
| let-7a-5p | 0.954 | 0.981 | 0.868 | 0.966 | 0.47 | 0.985 | 0.929 | 0.01 | 0.807 | 0.366 |
| let-7b-5p | 2.43E-04 | 0.04 | 0.626 | 0.378 | 0.03 | 0.116 | 0.122 | 0.397 | 0.888 | 0.02 |
| let-7d-5p | 0.954 | 0.604 | 0.56 | 0.44 | 0.562 | 0.985 | 0.772 | 0.04 | 0.725 | 0.564 |
| let-7g-5p | 0.954 | 0.499 | 0.04 | 0.297 | 0.097 | 0.985 | 0.04 | 0.46 | 0.672 | 5.02E-06 |
| let-7i-5p | 0.954 | 0.138 | 0.083 | 0.01 | 0.065 | 0.985 | 0.02 | 0.651 | 0.01 | 1.45E-06 |
| miR-1 | 0.954 | 0.04 | 0.208 | 0.544 | 0.595 | 0.985 | 0.03 | 0.827 | 0.923 | 0.21 |
| miR-103a-3p | 0.954 | 0.901 | 0.457 | 0.03 | 0.02 | 0.996 | 0.743 | 0.04 | 0.168 | 0.351 |
| miR-106a-5p | 0.937 | 0.499 | 0.068 | 0.01 | 0.01 | 0.912 | 0.05 | 0.01 | 0.01 | 0.02 |
| miR-107 | 0.954 | 0.419 | 0.258 | 0.01 | 0.01 | 0.902 | 0.276 | 0.134 | 0.147 | 0.292 |
| miR-127-3p | 0.05 | 0.499 | 0.609 | 0.707 | 0.996 | 0.985 | 0.595 | 0.75 | 0.888 | 0.886 |
| miR-130a-3p | 0.761 | 0.266 | 0.609 | 0.05 | 0.01 | 0.996 | 0.088 | 0.827 | 0.168 | 0.603 |
| miR-130b-3p | 0.768 | 0.445 | 0.388 | 0.467 | 0.114 | 0.902 | 0.04 | 0.863 | 0.8 | 1.62E-03 |
| miR-136-5p | 0.441 | 0.579 | 3.92E-03 | 0.05 | 0.065 | 0.902 | 4.75E-03 | 0.332 | 0.15 | 2.75E-08 |
| miR-140-3p | 0.954 | 0.969 | 0.03 | 8.36E-04 | 1.78E-05 | 0.985 | 0.504 | 0.827 | 2.93E-05 | 1.35E-06 |
| miR-140-5p | 0.993 | 0.01 | 0.379 | 0.414 | 0.1 | 0.942 | 0.171 | 0.837 | 0.696 | 0.263 |
| miR-142-3p | 0.667 | 0.03 | 1.63E-07 | 0.01 | 1.78E-05 | 0.116 | 1.81E-06 | 0.994 | 0.01 | 1.23E-17 |
| miR-142-5p | 0.761 | 0.91 | 0.56 | 0.01 | 0.01 | 0.902 | 0.57 | 0.827 | 0.01 | 0.213 |
| miR-144-3p | 0.01 | 0.475 | 0.03 | 0.054 | 0.173 | 0.74 | 0.01 | 0.187 | 0.301 | 0.01 |
| miR-146a-5p | 0.993 | 0.077 | 0.156 | 0.99 | 0.996 | 0.764 | 0.01 | 0.827 | 0.47 | 0.061 |
| miR-148b-3p | 0.993 | 0.493 | 0.171 | 0.544 | 0.814 | 0.902 | 0.101 | 0.994 | 0.913 | 0.04 |
| miR-150-5p | 0.737 | 0.327 | 0.986 | 0.966 | 0.753 | 0.985 | 0.894 | 0.75 | 0.515 | 0.05 |
| miR-151a-5p | 0.973 | 0.995 | 0.705 | 0.04 | 0.02 | 0.996 | 0.877 | 0.04 | 0.111 | 0.745 |
| miR-15a-5p | 0.803 | 0.772 | 0.083 | 0.445 | 0.04 | 0.785 | 0.401 | 0.46 | 0.8 | 0.01 |
| miR-15b-3p | 0.954 | 0.182 | 0.04 | 0.736 | 0.141 | 0.985 | 0.101 | 0.897 | 0.587 | 0.15 |
| miR-16-5p | 0.993 | 0.888 | 0.12 | 0.05 | 0.02 | 0.985 | 0.249 | 0.827 | 0.01 | 1.62E-03 |
| miR-191-5p | 0.993 | 0.729 | 0.129 | 5.22E-04 | 1.78E-05 | 0.902 | 0.158 | 0.376 | 0.01 | 0.02 |
| miR-195-5p | 0.704 | 0.888 | 0.083 | 0.03 | 0.74 | 0.902 | 0.805 | 0.663 | 0.115 | 0.167 |
| miR-199a-5p | 0.993 | 0.73 | 0.02 | 0.237 | 0.223 | 0.985 | 0.088 | 0.801 | 0.137 | 0.01 |
| miR-19b-3p | 0.519 | 0.812 | 0.833 | 0.115 | 0.118 | 0.996 | 0.929 | 0.827 | 0.01 | 0.102 |
| miR-200a-3p | 0.931 | 0.735 | 0.511 | 0.196 | 0.359 | 0.985 | 0.929 | 0.332 | 0.8 | 0.02 |
| miR-2110 | 7.48E-07 | 0.01 | 0.129 | 0.284 | 0.703 | 0.74 | 0.01 | 0.969 | 0.507 | 0.05 |
| miR-215-5p | 0.618 | 0.499 | 0.652 | 0.225 | 0.151 | 0.985 | 0.399 | 0.75 | 0.03 | 0.04 |
| miR-21-5p | 0.799 | 0.604 | 0.199 | 0.246 | 0.052 | 0.902 | 0.57 | 0.603 | 0.338 | 1.62E-03 |
| miR-221-3p | 0.993 | 0.604 | 0.03 | 0.03 | 0.1 | 0.902 | 2.56E-04 | 0.827 | 0.01 | 9.05E-06 |
| miR-22-3p | 0.993 | 0.598 | 0.509 | 0.063 | 0.01 | 0.985 | 0.929 | 0.376 | 0.054 | 0.02 |
| miR-28-5p | 0.448 | 0.604 | 0.509 | 0.063 | 0.28 | 0.764 | 0.651 | 0.463 | 0.07 | 0.04 |
| miR-29b-3p | 0.848 | 0.347 | 0.03 | 0.782 | 0.989 | 0.985 | 0.784 | 0.827 | 0.917 | 0.02 |
| miR-301a-3p | 0.954 | 0.04 | 0.482 | 0.32 | 0.561 | 0.985 | 0.158 | 0.827 | 0.766 | 0.746 |
| miR-30e-5p | 0.954 | 0.59 | 0.379 | 0.03 | 0.01 | 0.985 | 0.236 | 0.46 | 0.064 | 0.02 |
| miR-324-3p | 0.05 | 0.04 | 0.771 | 0.342 | 0.794 | 0.764 | 0.05 | 0.75 | 0.807 | 0.208 |
| miR-32-5p | 0.993 | 0.138 | 2.05E-04 | 4.54E-03 | 0.01 | 0.985 | 3.79E-03 | 0.187 | 0.064 | 9.03E-08 |
| miR-326 | 0.649 | 9.11E-06 | 4.57E-06 | 0.059 | 0.057 | 0.04 | 2.29E-12 | 0.332 | 0.111 | 3.28E-09 |
| miR-328-3p | 0.931 | 0.05 | 1.08E-06 | 0.225 | 0.116 | 0.785 | 4.64E-15 | 0.656 | 0.148 | 1.58E-08 |
| miR-339-3p | 0.803 | 0.566 | 0.414 | 0.115 | 0.03 | 0.985 | 0.14 | 0.292 | 0.426 | 0.547 |
| miR-33a-5p | 0.799 | 0.179 | 2.93E-03 | 0.02 | 0.03 | 0.985 | 1.29E-05 | 0.987 | 0.185 | 1.40E-05 |
| miR-342-3p | 0.448 | 0.731 | 0.136 | 0.115 | 0.359 | 0.985 | 0.714 | 0.292 | 0.05 | 0.619 |
| miR-382-5p | 0.179 | 0.874 | 0.04 | 0.634 | 0.6 | 0.996 | 0.623 | 0.964 | 0.725 | 0.732 |
| miR-409-3p | 0.448 | 0.812 | 0.136 | 0.417 | 0.773 | 0.942 | 0.311 | 0.757 | 0.111 | 4.10E-03 |
| miR-423-3p | 0.954 | 0.04 | 5.79E-06 | 0.428 | 0.385 | 0.785 | 1.05E-06 | 0.994 | 0.538 | 9.03E-08 |
| miR-423-5p | 0.993 | 0.731 | 0.04 | 0.04 | 1.05E-03 | 0.785 | 0.09 | 0.827 | 0.087 | 1.66E-03 |
| miR-484 | 0.803 | 0.106 | 7.26E-04 | 0.03 | 0.01 | 0.58 | 1.31E-07 | 0.292 | 0.04 | 9.03E-08 |
| miR-501-3p | 0.847 | 0.731 | 0.951 | 0.981 | 0.794 | 0.985 | 0.77 | 0.827 | 0.8 | 0.03 |
| miR-584-5p | 0.993 | 0.138 | 0.414 | 0.417 | 0.376 | 0.902 | 0.01 | 0.987 | 0.173 | 0.208 |
| miR-766-3p | 0.847 | 0.604 | 0.89 | 0.04 | 0.223 | 0.902 | 0.684 | 0.105 | 0.504 | 0.54 |
| miR-92b-3p | 0.993 | 0.931 | 0.083 | 0.44 | 0.694 | 0.985 | 0.939 | 0.603 | 0.634 | 0.05 |
| miR-93-5p | 0.332 | 0.731 | 0.457 | 0.01 | 4.28E-03 | 0.985 | 0.734 | 0.063 | 0.148 | 0.02 |
Figure 4. Summary of serum microRNAs that were found to be significantly correlated with specific or total PCDD or PCDFs and associated with deployment.
A.) Diagram illustrating the five miRNAs that were correlated with both the deployment related miRNA group and the PCDD/PCDF miRNA group. B.) List of the five miRNAs from the above Venn diagram. Highlighted columns show significant p-values (p < 0.05) after Benjamin-Hochberg false discovery rate correction.
Discussion
Here, using sera from time-in-service matched service members that were either deployed to locations with open burn pits or not deployed, we identified several miRNAs that were associated with environmental exposures and deployment. Even in this large cohort of serum samples from the DoDSR, the overwhelming majority of samples expressed robust levels of many different miRNA species. These results suggest that serum miRNA levels are indeed powerful biomarkers to gauge deployment and deployment related environmental exposures.
Although the Control subjects were chosen at random (from a database of non-deployed personnel with available serum samples drawn at matching times-in-service), their miRNA profile differed significantly from the pre-deployment miRNA profile of the Case subjects. While the full explanation for this is currently unclear, one potential factor is that the never deployed group was never deployed, while half of the Case group had one or more prior deployments to Iraq or Afghanistan area of operations (See Rohrbeck et al., this issue). Prior deployments may have changed their miRNA profile through deployment-related factors such as stress, diet, travel and environmental exposures. Furthermore, some of the differences may be due to differences in age, gender and general health status in the Control and Case cohorts. For example, a recent study by Noren Hooten et al., revealed that certain serum miRNA levels change depending on subject age.(42) We could not further break down the Case and Control groups based on age and gender as the detailed demographic data was under privacy restrictions, however, such subgroup analysis may be fruitful at a later date.
Despite the possible confounding effect of prior deployment in the Case group, we were still able to identify 21 miRNAs that were associated with deployment to sites with open burn pits. Some of these miRNAs found to be associated with deployment are members of the same family. MiRNA families target the same or similar mRNAs and often regulate related biological pathways.(43) For example, the related miRNAs, miR-25-3p and miR-32-5p, are predicted to target pathways including MAPK and TGFβ signaling.(44) Both miR-25-3p and miR-32-5p are decreased in serum from deployed personnel and thus these biological pathways may be affected by these two miRNAs.
While we cannot yet explain differences between the Control and Case cohorts due to restrictions of necessary demographic data, we did find significant changes in miRNA expression in the Case group before and after deployment to bases with open burn pits. In this analysis, the Case group served as its own reference and thus, even without demographic data, changes are likely due to deployment. Six miRNAs were found to be significantly different in this analysis and two of these miRNAs, miR-145-5p and miR-17-5p, have a p-value of less than 0.01. Interestingly, miR-145-5p is predicted to target numerous biological pathways that are important in exposure and inflammatory related insults including: TLR signaling, JAK-STAT signaling, p38 signaling and T cell receptor signaling (Figure 3). Predicted target mRNAs of miR-145-5p that are associated with inflammation include: IFNB1, MAPK14, and STAT3.(45) A recent report found that miR-145-5p is upregulated in plasma from asthma patients.(46) Thus, since recent reports suggest an increase in respiratory problems with personnel deployed to areas with open air burn pits (20), one mechanism may include induction of miR-145-5p levels which can alter inflammatory gene responses and promote onset of respiratory diseases such as asthma. Likewise, miR-17-5p has been shown to be upregulated in various disease states including: pulmonary artery hypertension, hepatocellular carcinoma and lung cancer.(47–49) Elevated serum miR-17-5p levels have been proposed as a diagnostic biomarker for lung cancer.(47) Biological pathways predicted to be regulated by miR-17-5p include: p53 signaling, mTOR signaling and oxidative phosphorylation (Figure 3). Target mRNAs of miR-17-5p include: PCAF, SMAD7 and PTEN, which can limit cell proliferation.(48, 50, 51) Additionally, miR-17-5p levels increased arginase II expression in pulmonary artery smooth muscle cells, which led to an increase in cell proliferation, a hallmark of pulmonary artery hypertension.(49) Thus, induction of miR-17-5p expression may serve to alter biological pathways leading to aberrant cellular proliferation. While miR-145-5p and miR-17-5p may be playing a mechanistic role in increasing respiratory health problems in personnel deployed to open air burn pits, further validation studies are needed.
Finally, we identified 56 miRNAs that were strongly associated with PCDD and PCDF exposure regardless of source. To our knowledge, this novel finding represents the first identification of miRNAs associated with PCDD and PCDF levels from human sera. Even after false discovery rate analysis, we were surprised to identify that PCDDs and PCDFs affected approximately 1/3 of the total number of measured serum miRNAs. PCDDs and PCDFs are long lasting environmental contaminants, with half-lives of months to years.(52) It is not surprising that some non-deployed Controls and pre-deployment Case subjects had high levels of PCDD and PCDF as exposure sources include combustion, industrial processes, air pollution and soil contamination.(53–56) PCDDs and PCDFs can profoundly activate cellular detoxification pathways and have effects on important regulatory pathways including the cell cycle, cell differentiation, and carcinogenesis.(57–60) It may be that one mechanism whereby these compounds have such broad effects is by altering a large subset of miRNAs to influence these crucial cellular pathways.
Five miRNAs were co-associated with deployment and with all-source PCDD/PCDFs. These data suggest that these miRNAs may be activated by combinations of pathways impacted by deployment such as PCDD/PCDF in combination with non-anthropogenic air pollution (dust, etc.) or other deployment related factors (pesticides, allergens, stress). Two of these miRNAs are let-7a and let-7d, both members of the let-7 family. (61) Let-7 miRNAs are predicted to target many pathways including cell stress, cell proliferation and differentiation and carcinogenesis.(62–64) These biological pathways are also influenced by PCDD/PCDFs, suggesting that these are important pathways to monitor in environmentally exposed, deployed service members.
In conclusion, we identified several miRNAs that were differentially expressed after deployment to sites with open burn pits. Additionally, when comparing service members deployed to those not deployed, we found a partially overlapping set of miRNAs. Furthermore, we discovered significant, novel correlations between serum miRNAs and serum levels of PCDDs and PCDFs. Interestingly, five miRNAs that were differentially expressed in the deployed group also correlated with serum PCDD and/or PCDF status. We conclude that certain serum miRNA levels show a link between deployment and strong correlation with environmental exposures that can occur during deployment such as exposure to open air burn pits.
Acknowledgments
This work was supported by The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. grant number HT9404-13-1-0030, and the National Institute of Environmental Health Sciences Grant # P30-ES01247.
Footnotes
There are no conflicts of interest.
References
- 1.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153(3):516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci U S A. 2015;112(10):E1106–1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs. target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, I, Shih H, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Koberle V, Kakoschky B, Ibrahim AA, Schmithals C, Peveling-Oberhag J, Zeuzem S, et al. Vesicle-associated microRNAs are released from blood cells on incubation of blood samples. Transl Res. 2015 doi: 10.1016/j.trsl.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Kowdley KV. Method for microRNA isolation from clinical serum samples. Anal Biochem. 2012;431(1):69–75. doi: 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacLellan SA, MacAulay C, Lam S, Garnis C. Pre-profiling factors influencing serum microRNA levels. BMC Clin Pathol. 2014;14:27. doi: 10.1186/1472-6890-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grasedieck S, Scholer N, Bommer M, Niess JH, Tumani H, Rouhi A, et al. Impact of serum storage conditions on microRNA stability. Leukemia. 2012;26(11):2414–2416. doi: 10.1038/leu.2012.106. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, et al. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012;7(10):e46957. doi: 10.1371/journal.pone.0046957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnecke-Eberz U, Chon SH, Holscher AH, Drebber U, Bollschweiler E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015;36(6):4643–4653. doi: 10.1007/s13277-015-3112-0. [DOI] [PubMed] [Google Scholar]
- 12.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Dai X, Zhan J, Zhang Y, Zhang H, Zhang H, et al. Profiling peripheral microRNAs in obesity and type 2 diabetes mellitus. APMIS. 2015;123(7):580–585. doi: 10.1111/apm.12389. [DOI] [PubMed] [Google Scholar]
- 14.Ovchinnikova ES, Schmitter D, Vegter EL, Ter Maaten JM, Valente MA, Liu LC, et al. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail. 2015 doi: 10.1002/ejhf.332. [DOI] [PubMed] [Google Scholar]
- 15.Ebrahimi A, Sadroddiny E. MicroRNAs in lung diseases: Recent findings and their pathophysiological implications. Pulm Pharmacol Ther. 2015;34:55–63. doi: 10.1016/j.pupt.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Mancuso JD, Mallon TM, Gaydos JC. Maximizing the Capabilities of the DoD Serum Repository to Meet Current and Future Needs: Report of the Needs Panel. Mil Med. 2015;180(10 Suppl):13–24. doi: 10.7205/MILMED-D-14-00732. [DOI] [PubMed] [Google Scholar]
- 17.Lindler LE. Building a DoD Biorepository for the Future: Potential Benefits and Way Forward. Mil Med. 2015;180(10 Suppl):90–94. doi: 10.7205/MILMED-D-14-00724. [DOI] [PubMed] [Google Scholar]
- 18.Lindler LE. Enhancing the Department of Defense’s Capability to Identify Environmental Exposures Into the 21st Century. Mil Med. 2015;180(10 Suppl):5–9. doi: 10.7205/MILMED-D-14-00723. [DOI] [PubMed] [Google Scholar]
- 19.Smith B, Wong CA, Boyko EJ, Phillips CJ, Gackstetter GD, Ryan MA, et al. The effects of exposure to documented open-air burn pits on respiratory health among deployers of the Millennium Cohort Study. J Occup Environ Med. 2012;54(6):708–716. doi: 10.1097/JOM.0b013e31825107f9. [DOI] [PubMed] [Google Scholar]
- 20.Falvo MJ, Osinubi OY, Sotolongo AM, Helmer DA. Airborne hazards exposure and respiratory health of Iraq and Afghanistan veterans. Epidemiol Rev. 2015;37:116–130. doi: 10.1093/epirev/mxu009. [DOI] [PubMed] [Google Scholar]
- 21.Abraham JH, DeBakey SF, Reid L, Zhou J, Baird CP. Does deployment to Iraq and Afghanistan affect respiratory health of US military personnel? J Occup Environ Med. 2012;54(6):740–745. doi: 10.1097/JOM.0b013e318252969a. [DOI] [PubMed] [Google Scholar]
- 22.Smith B, Wong CA, Smith TC, Boyko EJ, Gackstetter GD, Margaret AKRftMCST. Newly reported respiratory symptoms and conditions among military personnel deployed to Iraq and Afghanistan: a prospective population-based study. Am J Epidemiol. 2009;170(11):1433–1442. doi: 10.1093/aje/kwp287. [DOI] [PubMed] [Google Scholar]
- 23.Song MK, Park YK, Ryu JC. Polycyclic aromatic hydrocarbon (PAH)-mediated upregulation of hepatic microRNA-181 family promotes cancer cell migration by targeting MAPK phosphatase-5, regulating the activation of p38 MAPK. Toxicol Appl Pharmacol. 2013;273(1):130–139. doi: 10.1016/j.taap.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Gordon MW, Yan F, Zhong X, Mazumder PB, Xu-Monette ZY, Zou D, et al. Regulation of p53-targeting microRNAs by polycyclic aromatic hydrocarbons: Implications in the etiology of multiple myeloma. Mol Carcinog. 2015;54(10):1060–1069. doi: 10.1002/mc.22175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Q, Huang S, Zhang X, Zhang W, Feng J, Wang T, et al. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect. 2014;122(7):719–725. doi: 10.1289/ehp.1307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 27.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 29.Max W, Sung HY, Shi Y. Who is exposed to secondhand smoke? Self-reported and serum cotinine measured exposure in the U.S., 1999–2006. Int J Environ Res Public Health. 2009;6(5):1633–1648. doi: 10.3390/ijerph6051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199, 205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentleman RC, V, Carey J, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1–6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Thakar J, Hartmann BM, Marjanovic N, Sealfon SC, Kleinstein SH. Comparative analysis of anti-viral transcriptomics reveals novel effects of influenza immune antagonism. BMC Immunol. 2015;16:46. doi: 10.1186/s12865-015-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamal A, Cincinelli A, Martellini T, Malik RN. A review of PAH exposure from the combustion of biomass fuel and their less surveyed effect on the blood parameters. Environ Sci Pollut Res Int. 2015;22(6):4076–4098. doi: 10.1007/s11356-014-3748-0. [DOI] [PubMed] [Google Scholar]
- 37.Kim KH, Jahan SA, Kabir E, Brown RJ. A review of airborne polycyclic aromatic hydrocarbons (PAHs) and their human health effects. Environ Int. 2013;60:71–80. doi: 10.1016/j.envint.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Farmer PB, Singh R, Kaur B, Sram RJ, Binkova B, Kalina I, et al. Molecular epidemiology studies of carcinogenic environmental pollutants. Effects of polycyclic aromatic hydrocarbons (PAHs) in environmental pollution on exogenous and oxidative DNA damage. Mutat Res. 2003;544(2–3):397–402. doi: 10.1016/j.mrrev.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q, Jiang X, Hedgeman E, Knutson K, Gillespie B, Hong B, et al. Estimation of age- and sex-specific background human serum concentrations of PCDDs, PCDFs, and PCBs in the UMDES and NHANES populations. Chemosphere. 2013;91(6):817–823. doi: 10.1016/j.chemosphere.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 40.Demond A, Franzblau A, Garabrant D, Jiang X, Adriaens P, Chen Q, et al. Human exposure from dioxins in soil. Environ Sci Technol. 2012;46(3):1296–1302. doi: 10.1021/es2022363. [DOI] [PubMed] [Google Scholar]
- 41.Franzblau A, Hedgeman E, Jolliet O, Knutson K, Towey T, Chen Q, et al. Case report: the University of Michigan dioxin exposure study: a follow-up investigation of a case with high serum concentration of 2,3,4,7,8-pentachlorodibenzofuran. Environ Health Perspect. 2010;118(9):1313–1317. doi: 10.1289/ehp.0901723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noren Hooten N, Fitzpatrick M, Wood WH, 3rd, De S, Ejiogu N, Zhang Y, et al. Age-related changes in microRNA levels in serum. Aging (Albany NY) 2013;5(10):725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016;7(2):100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Huang Y, Liang Z, Liu D, Lu Y, Dai Y, et al. Plasma miRNAs might be promising biomarkers of chronic obstructive pulmonary disease. Clin Respir J. 2016;10(1):104–111. doi: 10.1111/crj.12194. [DOI] [PubMed] [Google Scholar]
- 47.Chen Q, Si Q, Xiao S, Xie Q, Lin J, Wang C, et al. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30(1):353. doi: 10.1007/s12032-012-0353-2. [DOI] [PubMed] [Google Scholar]
- 48.Shan SW, Fang L, Shatseva T, Rutnam ZJ, Yang X, Du W, et al. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J Cell Sci. 2013;126(Pt 6):1517–1530. doi: 10.1242/jcs.122895. [DOI] [PubMed] [Google Scholar]
- 49.Jin Y, Jin Y, Chen B, Tipple TE, Nelin LD. Arginase II is a target of miR-17-5p and regulates miR-17-5p expression in human pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2014;307(2):L197–204. doi: 10.1152/ajplung.00266.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu X, et al. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp Mol Med. 2014;46 doi: 10.1038/emm.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong AY, Eischeid AN, Xiao J, Zhao J, Chen D, Wang ZY, et al. miR-17-5p targets the p300/CBP-associated factor and modulates androgen receptor transcriptional activity in cultured prostate cancer cells. BMC Cancer. 2012;12:492. doi: 10.1186/1471-2407-12-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milbrath MO, Wenger Y, Chang CW, Emond C, Garabrant D, Gillespie BW, et al. Apparent half-lives of dioxins, furans, and polychlorinated biphenyls as a function of age, body fat, smoking status, and breast-feeding. Environ Health Perspect. 2009;117(3):417–425. doi: 10.1289/ehp.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Caneghem J, Block C, Vandecasteele C. Destruction and formation of dioxin-like PCBs in dedicated full scale waste incinerators. Chemosphere. 2014;94:42–47. doi: 10.1016/j.chemosphere.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Pieters R, Focant JF. Dioxin, furan and PCB serum levels in a South African Tswana population: comparing the polluting effects of using different cooking and heating fuels. Environ Int. 2014;66:71–78. doi: 10.1016/j.envint.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 55.Lundin L, Jansson S. The effects of fuel composition and ammonium sulfate addition on PCDD, PCDF, PCN and PCB concentrations during the combustion of biomass and paper production residuals. Chemosphere. 2014;94:20–26. doi: 10.1016/j.chemosphere.2013.08.059. [DOI] [PubMed] [Google Scholar]
- 56.Amand LE, Kassman H. Decreased PCDD/F formation when co-firing a waste fuel and biomass in a CFB boiler by addition of sulphates or municipal sewage sludge. Waste Manag. 2013;33(8):1729–1739. doi: 10.1016/j.wasman.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 57.Gasiewicz TA, Singh KP, Bennett JA. The Ah receptor in stem cell cycling, regulation, and quiescence. Ann N Y Acad Sci. 2014;1310:44–50. doi: 10.1111/nyas.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh KP, Wyman A, Casado FL, Garrett RW, Gasiewicz TA. Treatment of mice with the Ah receptor agonist and human carcinogen dioxin results in altered numbers and function of hematopoietic stem cells. Carcinogenesis. 2009;30(1):11–19. doi: 10.1093/carcin/bgn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vezina CM, Allgeier SH, Moore RW, Lin TM, Bemis JC, Hardin HA, et al. Dioxin causes ventral prostate agenesis by disrupting dorsoventral patterning in developing mouse prostate. Toxicol Sci. 2008;106(2):488–496. doi: 10.1093/toxsci/kfn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bemis JC, Alejandro NF, Nazarenko DA, Brooks AI, Baggs RB, Gasiewicz TA. TCDD-induced alterations in gene expression profiles of the developing mouse paw do not influence morphological differentiation of this potential target tissue. Toxicol Sci. 2007;95(1):240–248. doi: 10.1093/toxsci/kfl132. [DOI] [PubMed] [Google Scholar]
- 61.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22(9):474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J, Li S, Jia W, Deng H, Chen K, Zhu L, et al. Reduced Let-7a Is Associated with Chemoresistance in Primary Breast Cancer. PLoS One. 2015;10(7):e0133643. doi: 10.1371/journal.pone.0133643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crippa S, Cassano M, Sampaolesi M. Role of miRNAs in muscle stem cell biology: proliferation, differentiation and death. Curr Pharm Des. 2012;18(13):1718–1729. doi: 10.2174/138161212799859620. [DOI] [PubMed] [Google Scholar]
- 64.Saleh AD, Savage JE, Cao L, Soule BP, Ly D, DeGraff W, et al. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS One. 2011;6(10):e24429. doi: 10.1371/journal.pone.0024429. [DOI] [PMC free article] [PubMed] [Google Scholar]