Abstract
The imaging of dysregulated cell-surface receptors (or biomarkers) is a potential means of identifying the presence of cancer with high sensitivity and specificity. However, due to heterogeneities in the expression of protein biomarkers in tumors, molecular imaging technologies should ideally be capable of visualizing a multiplexed panel of cancer biomarkers. Recently, surface-enhanced Raman-scattering (SERS) nanoparticles (NPs) have attracted wide interest due to their potential for sensitive and multiplexed biomarker detection. In this review, we focus on the most recent advances in tumor imaging using SERS-coded NPs. A brief introduction of the structure and optical properties of SERS NPs is provided, followed by a detailed discussion of key imaging issues such as the administration of NPs in tissue (topical versus systemic), the optical configuration and imaging approach of Raman imaging systems, spectral demultiplexing methods for quantifying NP concentrations, and the disambiguation of specific vs. nonspecific sources of contrast through ratiometric imaging of targeted and untargeted (control) NP pairs. Finally, future challenges and directions are briefly outlined.
Index Terms: biomarkers, biomedical optical imaging, cancer detection, fiberoptic probes, molecular imaging, nanomedicine, Raman spectroscopy, tumors
I. Introduction
A major focus of biomedical optics has been the development of technologies for the detection of cancers, as well as the imaging of surgical margins to guide the complete removal of tumors [1–6]. The general approach of optical diagnostics is to deduce tissue status through the measurement of optical signals generated either intrinsically by tissue [7–9] or extrinsically by probes that are targeted to disease biomarkers [10,11]. Since external contrast agents are not necessary for the imaging of intrinsic tissue signatures, such approaches have the advantage of increased simplicity in many cases, as well as potentially reduced regulatory complexity. However, exogenous contrast agents are often utilized to target molecular biomarkers of interest, which have the potential to provide a more direct and specific biological link to various diseases as well as enhanced contrast to detect such malignancies. A challenging issue for cancer detection is the large degree of heterogeneity in biomarker expression between tumors. This heterogeneity extends between cancer subtypes, between patients, within a single patient over time, and even between different locations within a single tumor (intratumoral heterogeneity) [13–15]. Therefore, to improve the sensitivity and specificity of detection, exogenous probes should ideally be capable of being multiplexed, allowing for the simultaneous detection of multiple biomarkers. In this article, we will discuss one class of contrast agent that can enable multiplexed molecular detection: surface-enhanced Raman-scattering (SERS) nanoparticles (NPs).
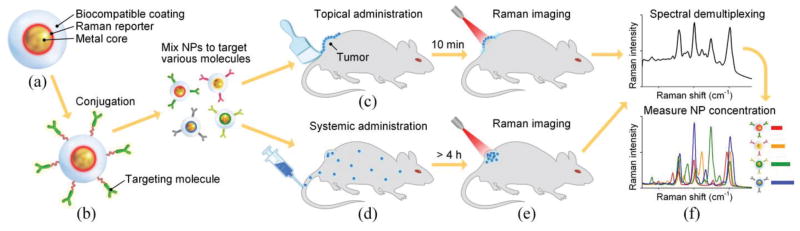
When molecules are adsorbed on metal nanostructures, their Raman scattering signal can be enhanced by more than 10 orders of magnitude [16–18]. Based on this effect, SERS NPs have been developed since the late 1990s [19–26], acting as extrinsic Raman labels for the optical detection of biomolecules [27–29]. Typical SERS NPs have a sandwich structure (Fig. 1(a)): a layer of Raman reporter molecules is permanently adsorbed onto the surface of a metal core to trigger the SERS effect, generating a characteristic Raman spectrum upon laser excitation at a specified wavelength. This characteristic SERS spectrum serves as a barcode or fingerprint for the identification of that particular “flavor” of NP, where different flavors of SERS NPs utilize different reporter molecules to generate specific spectral signatures. A biocompatible coating is generally used to surround the NP core and to stabilize the SERS NP signal, such that the SERS spectrum is constant for each NP flavor and is not influenced by the surrounding environment. For biomolecule detection, SERS NPs can be conjugated to a molecule such as an antibody or affibody to specifically target a biomolecule of interest (Fig. 1(b)) [30–34]. For example, popular cancer biomarker targets that have been imaged include the epidermal growth factor receptor (EGFR) and the human epidermal growth factor receptor 2 (HER2) [35–37]. Additional details about the design and synthesis of SERS NPs have been discussed in some recent reviews [38–40].
Fig. 1.
SERS NPs and their application for tumor detection. (a) Typical structure of SERS NPs. (b) Functionalization of SERS NPs with targeting molecules. Schematics showing (c) topical administration, (d) systemic administration, (e) imaging and (f) demultiplexing of multiplexed SERS NPs.
SERS NPs have several advantageous optical properties for biological and biomedical applications over other optical probes such as fluorescent dyes and quantum dots [41,42]. In comparison with broadband fluorescence spectra (~50 nm), the spectra of SERS NPs consist of a fixed combination of narrow emission peaks (1 2 nm) [34,43], which offer enhanced potential for spectral multiplexing. As mentioned previously, by utilizing different Raman reporter molecules, various flavors of SERS NPs can be engineered with unique spectral fingerprints [21,33,44]. Only a single laser excitation wavelength is needed to excite many different SERS NPs, such that those NPs can be multiplexed to target and detect a rich set of biomolecules. For the SERS NPs utilized in our studies, the excitation wavelength is in the near-infrared (NIR) region (785 nm), which minimizes background autofluorescence from cells and tissues. In addition, SERS NPs are photostable and do not bleach over time [33,45].
Over the past decade, researchers have been exploring the potential of SERS NPs for various applications ranging from pH and glucose sensing to circulating tumor cell (CTC) and tumor detection [32,39,40,46–50]. In this review, we focus on the most recent advances in tumor imaging using SERS NPs and discuss key imaging-related issues (Figs. 1(c)–1(f)) such as the administration of NPs in tissue (topical versus systemic), the optical configuration and scanned-imaging strategies employed in various Raman imaging systems, spectral demultiplexing methods for quantifying NP concentrations, and the ratiometric imaging of targeted and untargeted NP pairs to distinguish between specific vs. nonspecific sources of contrast.
II. Administration of SERS NPs in tissue
A. Systemic Administration
SERS NPs can be administered systemically or topically to label molecular biomarkers in tissues. Since SERS NPs are not currently approved for human use, systemic administration has only been employed for in vivo imaging studies in animals [33,34,44,51–53]. For the imaging of tumor xenografts, image contrast has been achieved 5 – 24 hours after injecting SERS NPs into the tail vein of mice (Figs. 1(d) and 1(e)). Tumor-to-normal contrast has been achieved either through the nonspecific accumulation of NPs in the tumor (i.e. the enhanced permeability and retention (EPR) effect [53,54]) orthrough specific binding of molecularly targeted NPs to their cancer biomarker targets [33,34,55]. SERS NPs have a relatively large size (20–120 nm [21,22,25,26,33,44,56]) compared with fluorescence dyes (~1 nm). Therefore, a large portion of them are taken up by the reticuloendothelial system and accumulate in the liver [33,44], where they are then cleared in one or two weeks [33,57]. In contrast, specifically bound NPs have been shown to remain at the tumor site with a constant concentration during the first week [33], suggesting that SERS NPs can be potentially used for short-term targeted imaging and therapy studies.
B. Topical Administration
The topical administration of SERS NPs has been extensively employed to image the expression of protein biomarkers at the surface of animal tissues (in vivo and ex vivo) and resected (ex vivo) human tissues [31,32,58–62]. A key advantage of topical application, for eventual clinical translation, is that toxicity issues are reduced, especially since the large size of the SERS NPs inhibits the penetration of topically applied NPs through deep tissue layers and into systemic circulatory pathways [57,63]. For tumor imaging, the topical application of biomarker-targeted NPs onto tissues, followed by the rinse removal of unbound NPs, has been shown to yield molecular image contrast between tumor and normal tissues in 5 – 60 min [30–32,62]. As mentioned, the large size of SERS NPs causes topically applied NPs to be confined to the tissue surface with a negligible penetration depth of ~20 μm (Fig. 2(a)), which is well-matched to the superficial imaging depth of optical imaging in general. In addition, it is hypothesized that the shallow penetration depth of topically applied targeted SERS NPs aids in the rinse removal of unbound NPs, rather than their becoming permanently trapped within the tissue. The efficient washout of unbound NPs, in turn, is key to maximizing the contrast between tumor and normal tissue regions. Note that since SERS NPs typically are not internalized by cells during the short incubation times utilized in our experiments (Fig. 2(b)), SERS NPs are generally limited to the targeting of exposed cell-surface receptors unless special procedures are performed, such as sectioning, to expose the intracellular receptors.
Fig. 2.

Penetration of SERS NPs on tissue and cells. (a) Bright-field and fluorescence images showing that topically applied SERS NPs (conjugated to a fluorophore cyto-647 in this case) are localized to the tissue surface of a piece of mouse muscle that was topically stained with EGFR-targeted SERS NPs (300 pM, 10 min), rinsed in PBS (20 s) and embedded in O.C.T. (optimal cutting temperature) compound. The image shows a frozen section (10-μm thick) that was imaged with a fluorescence microscope. (b) Bright-field and fluorescence confocal microscopy images showing the distribution of EGFR-targeted NPs on A431 cells. In this thin confocal image section (~ 5 micron section thickness), the fluorescence signal is primarily localized at the periphery of the cells, indicating that the NPs are at the cell surface. The scale bars represent 50 μm..
The topical staining of real tissues using SERS NPs is more challenging than the topical staining of cultured cell lines due to considerable nonspecific accumulation in tissues as well as reduced access to cell-surface targets (compared to cultured cells in a dish). Therefore, it is important to optimize targeted SERS NPs to exhibit extremely high binding affinity/avidity to receptor-positive cell lines before tackling the greater challenge of detecting biomarkers in tissues. For example in Fig. 3, two flavors of SERS NPs were conjugated first to a fluorescent dye (to enable fluorescence microscopy of the NP concentrations) and then to either an anti-EGFR monoclonal antibody (mAb) or an isotype control (negative control) mAb (for details, refer to [31]). The two NPs were individually used to stain a control cell line (3T3) or an EGFR-positive cell line (A431), or mixed together in 1:1 ratio to stain normal mouse tissue or an A431 tumor xenograft (150 pM/each, 10 min staining followed by a 10-s rinse step in PBS). EGFR-positive cancer cells (A431) stained with EGFR-NPs exhibited 40× - 100× brighter fluorescence than the same cancer cells stained with isotype-NPs. EGFR-negative normal cells (3T3) stained with EGFR-NPs or isotype-NPs always showed low fluorescence. These experiments demonstrate a high binding affinity of the EGFR-NPs to the cell-surface receptor EGFR in cultured cells, which in turn allows for the successful staining of fresh tissues. In this case, tumor tissues (A431 xenograft) stained with EGFR-NPs were 2× brighter (Raman intensity) than the same tumor tissues stained with isotype-NPs.
Fig. 3.
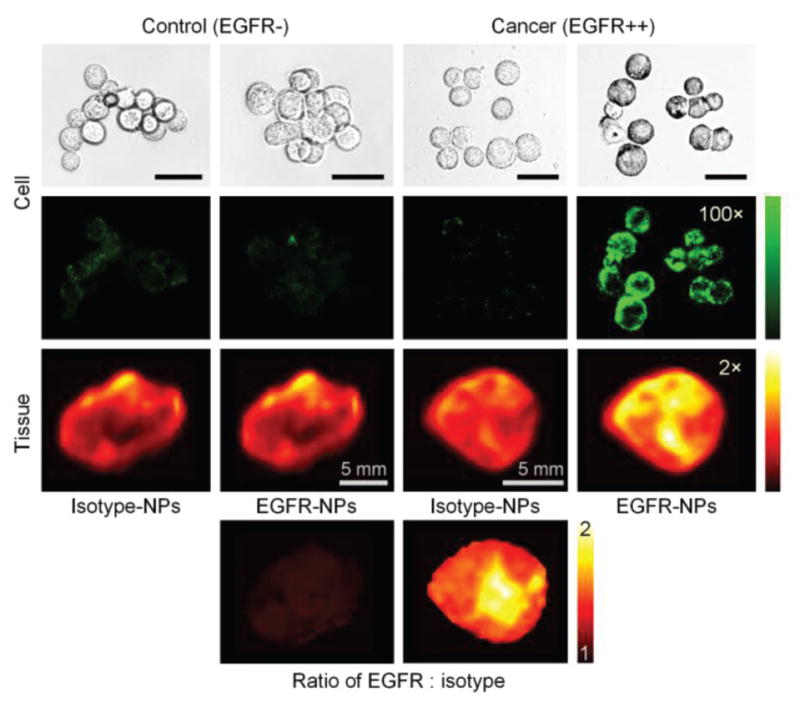
Nonspecific and specific binding of SERS NPs topically applied on cultured cells and tissues. The bright-field images (first row) and fluorescence images (second row) of NP-stained cells, and the Raman images of NP-stained tissues (third row) are shown. The green and red colormaps indicate the fluorescence intensity and Raman intensity from the SERS NPs, respectively. The bottom images show the measured concentration ratio of EGFR-NPs vs. isotype-NPs, in which the nonspecific accumulation of the NPs is normalized away and the EGFR expression is clearly revealed. The unlabeled scale bars represent 20 μm.
Adding 1% BSA to the NP staining solution can reduce the nonspecific accumulation of NPs within tissues and increase the contrast between targeted vs. untargeted NPs by 20%–40%. However, nonspecific accumulation is still high and can be extremely variable due to non-uniform tissue structures. In addition, uneven NP delivery/rinse removal at different tissue locations often results in misleading image contrast [31,61]. For example, we found that increased diffusion and passive retention can result in higher nonspecific accumulation of topically applied NPs in normal tissues in comparison to denser tumor tissues, i.e. inverse contrast [31]. One method that has been utilized to mitigate these nonspecific effects, and to accurately visualize the molecularly specific accumulation of NP concentrations, is to introduce a control NP flavor simultaneously with the targeted NP during imaging experiments in order to normalize for nonspecific effects. This method allows one to calculate the ratio of biomarker-specific NP accumulation exhibited by targeted NPs alone vs. the nonspecific accumulation exhibited by all NPs, as revealed by the control NP (e.g. see the bottom row of Fig. 3). This powerful strategy of ratiometric imaging will be discussed in Section V.
III. Raman imaging system
The development of Raman spectroscopy has provided a versatile tool for rapid cancer diagnoses, either through the analysis of intrinsic Raman signals from tissue that are associated with cancer progression [64–68] or through the detection of exogenously applied Raman contrast agents, such as SERS NPs, which are passively accumulated at a tumor site or targeted to cancer biomarkers [30–34,53,54]. Here we briefly discuss the optical set-ups and imaging approaches for the detection of exogenous SERS agents.
A. Optical configuration
Raman spectroscopy is performed by collecting a spectrum from an area that is illuminated at a precise wavelength in order to determine the relative concentration of various molecules or chemical constituents (e.g. hydrocarbons, nucleic acids, etc) of interest within the illuminated area. The size of the illumination spot therefore determines the spatial resolution of detection. Although commercial or custom Raman microscopes have been employed to image tissues at micron-level spatial resolution [29,33,34,47,69], a sub-millimeter- or millimeter-level resolution is preferred for imaging (scanning) a large tissue area [52,59,61] due to the relatively long acquisition times required for Raman imaging (typically 1 - 100 sec for unenhanced Raman spectroscopy and 1 – 100 ms for SERS). Furthermore, the detection of sub-millimeter-sized tumors is considered to be an acceptable size threshold for many clinical applications such as early cancer detection and surgical guidance.
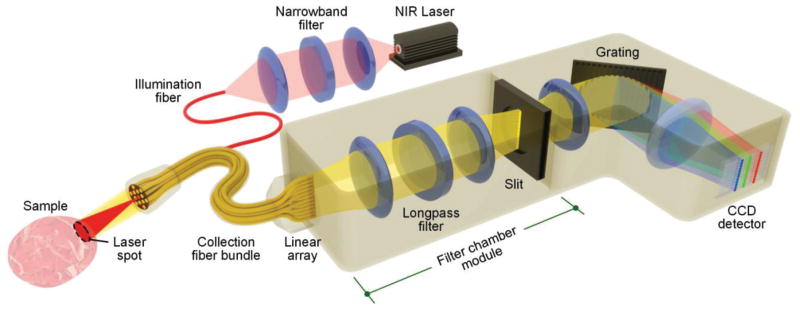
A standard optical system for macroscopic-resolution (>100-μm spatial resolution) imaging of SERS NPs includes an imaging probe and a spectrometer [31,58,59,70]. The light source is typically a NIR laser, which is delivered to the sample through a fiber-optic probe (Fig. 4). The optical signals (e.g. reflectance, scattering, and fluorescence) from the SERS NPs and tissue (background) are collected by multiple fibers in the imaging probe and are delivered through a fiber bundle to a spectrometer, where the collected light is dispersed by a diffraction grating onto a spectroscopic imaging array, such as a cooled deep-depletion CCD detector. Figure 4 provides some design details.
Fig. 4.
A common optical configuration for the imaging of SERS NPs in tissues.
Laser source
For biomedical applications, the American National Standards Institute (ANSI) maximum permissible exposure (MPE) limit is often referred to when selecting a laser power, in which one must take into account the laser wavelength, the exposure time, and the illumination area on the tissue. For example, for a 785-nm laser, the ANSI MPE limit for a 0.1-s and a 1-s exposure time are 0.915 W/cm2 and 1.63 W/cm2 respectively [58]. Assuming the illuminated spot has a diameter of 1 mm (or an area of 0.79 mm2), the laser power should be set below 71.5 mW (for a 0.1-s exposure) or 12.7 mW (for a 1-s exposure). A shutter is often used to control the laser exposure time to protect the tissue from overexposure damage [33,58].
Illumination path
A notch filter can be placed between the laser source and the tissue sample to eliminate broadband noise from the laser (e.g. amplified spontaneous emission) and to ensure a narrow illumination bandwidth, which is necessary to generate narrow high-resolution Raman spectra [31,52]. Either a singlemode fiber or multimode fiber can be used to deliver laser light to the tissue, depending upon the spot size that is required at the tissue. For example, a singlemode fiber should be used if a diffraction-limited focus is desired for high-resolution imaging applications, or if a smooth speckle-free Gaussian illumination profile is desired. Note that for intrinsic Raman spectroscopy, the illumination fiber itself can generate appreciable background autofluorescence and Raman scattering, which must often be removed by including a filter at the distal tip of the probe (after the illumination fiber) [71]. However, due to the high brightness of SERS NPs, it is not necessary to filter out these fiber-induced background components for SERS-based detection and imaging. Singlemode fibers exhibit lower bend losses than multimode fibers, which helps to minimize the leakage of illumination photons into surrounding fibers or jacketing materials. With proper coatings, multimode fiber bundles can also exhibit minimal crosstalk between illumination and collection fibers [72]. Unlike singlemode fibers, which emit light with a smooth Gaussian profile, multimode fibers can improve the overall uniformity of light distribution on the illuminated tissue area, but emit light with local nonuniformities due to speckle. A practical advantage for the use of multimode fibers is that they contain a much larger core diameter than singlemode fibers, which facilitates laser alignment and reduces variations in laser power when moving the imaging fibers in a detection probe. Multimode fibers also tend to randomize the polarization of the transmitted laser radiation, which can help with power stability through angled optical components and tissue interfaces, and are therefore preferred for designing portable devices unless high-resolution focusing and imaging is needed. In order to reduce specular reflections and the collection of tissue autofluorescence background, the illumination beam is often angled with respect to the direction normal to the tissue surface [12,31]. Note that while the penetration depth (diffuse) of NIR light in tissues can easily surpass several millimeters (depending upon the optical properties of the tissue) [58,60,70], topically applied SERS NPs are confined to the tissue surface and therefore provide only a superficial imaging technique.
Collection path
Collection fiber bundles normally consist of numerous (e.g. 10 – 60) multimode fibers (core diameter 100–200 μm) [58,61,64,72]. These fibers can be closely packed in a hexagonal arrangement at the probe tip to maximize signal collection and minimize space, and are often realigned into a linear array at the proximal side of the fiber bundle in order to interface with the entrance slit of a spectrometer (Fig. 4). Unless the spatial position of each fiber is important to track, full vertical binning of the detector array is often used to integrate the signals from all of the fibers positioned in a linear array at the entrance slit of the spectrometer. The height of the linear fiber array must be matched to the aperture heights of the spectrometer optics and the height of the detector array (through appropriate magnifying or demagnifying optics, in certain cases).
A longpass filter placed between the collection fibers and a secondary slit (at the proximal end of the filter chamber module) rejects reflected and Rayleigh-scattered illumination light as well as tissue autofluorescence at wavelengths that are shorter than the SERS peaks [70,73]. The secondary slit at the back end of the filter chamber module is used to spatially filter out the stray light in the filter chamber that originates from the large amount of light that is rejected (reflected) by the longpass interference filter. The goal of the slit (spatial filter) is to allow only the SERS photons to transmit towards the grating and ultimately onto the detector array.
When collimated light from an entrance slit (or linear array of fibers) is dispersed by a grating-based spectrometer and imaged onto a detector array, light from different positions along the length of the slit are incident on the grating at varying degrees of obliqueness, which results in a slightly curved image of the slit at the detector array and therefore a lower spectral resolution when full vertical binning of the detector array is utilized [74]. One solution to correct this image aberration is to utilize a collection fiber array that is slightly curved in the opposite direction to compensate for the image curvature, a strategy that has been shown to be effective for improving the signal-to-noise ratio (SNR) and spectral resolution when collecting intrinsic Raman signals from tissue [72]. SERS NPs generate Raman peaks that are bright and spectrally well separated. Therefore, many groups have found that the slight image curvature does not significantly reduce the measurement accuracy [59](Fig. 7).
Fig. 7.
Limit-of-detection (linearity) test by imaging four multiplexed SERS NPs. Four NP flavors were mixed in 1:1:1:1 ratio and diluted from 200 pM (per flavor) down to 1 pM. A 0.5-μL drop from each sample was imaged, and 3 spectra were acquired and demultiplexed to analyze the weight of each NP flavor. The concentrations were calculated based on the measured weights of stock NP mixture with known concentrations. (a) The reference spectra of the four NP flavors. (b) Raw spectra from NP mixtures at a concentration of 10 pM, 5 pM, 1 pM and 0 pM (water background). (c) Measured NP concentrations. (d) Concentration ratios. The measured concentrations show good linearity from 1 to 200 pM when 4 flavors of NPs are multiplexed, with larger errors in terms of concentration (~20%) and concentration ratio (~30%) appearing at 1 pM. (e–g) Imaging of 4-flavor NP mixtures. (e) A 2-μL drop from each dilution, and a drop from each undiluted NP flavor, were imaged to measure (f) the NP concentrations and (g) concentration ratios.
B. Imaging approaches
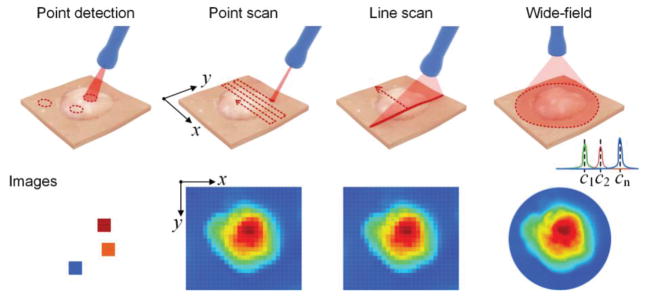
In certain medical applications, a diagnostic probe is used to analyze a suspicious lesion at a well-defined location on a patient [53,58,70] (Fig. 5). However, if the tumor’s location is unknown, a scanning approach is required to image and screen a larger area of tissue [44,52,59,61,75]. Raster scanning is often performed to image resected tissues (e.g. tissue data in Fig. 3, Fig. 6(f), 6(g), 7(f) and 7(g)) or surgically exposed tissues, while rotational scanning has been utilized in endoscopic approaches to image hollow organs such as the esophagus [12,61] (Fig. 8). To achieve rotational scanning, a glass prism may be employed at the probe tip to deflect the illumination beam to the tissue as well as to collect Raman signal from the illuminated tissue area. In animal models, rotational stepper motors have been utilized with linear stages to rotate an imaging probe and to translate it in the axial direction, respectively. While point-by-point scanning is typically utilized to create an image, line-wise scanning may also be exploited to simplify the scanning process, as has been utilized for food-inspection applications [76,77].
Fig. 5.
Schematic of imaging approaches. The labels c1 - cn represent the wavelength channels selected for wide-field imaging.
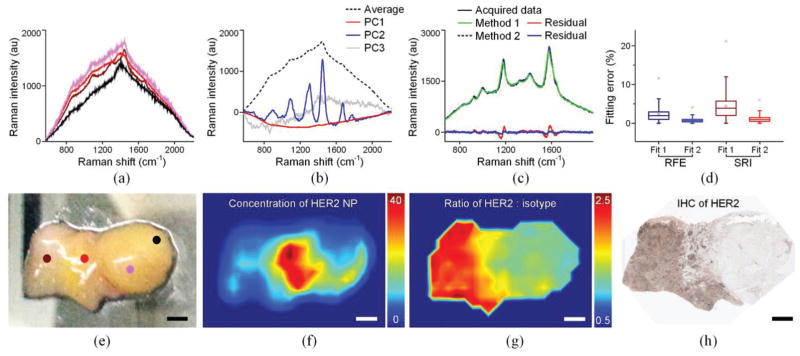
Fig. 6.
Imaging of human breast tissues stained with a 2-flavor NP mixture (HER2-NPs and isotype-NPs, 150 pM/flavor). (a) Background spectra collected at different locations from the tissue specimen marked in (e). (b) The first three principal components and average background spectrum of the 162 background spectra collected from the specimen. (c) Example of fitted spectra and corresponding residuals when using Method 1 and Method 2. (d) The fitting error for all 2000 spectra when using the two fitting algorithms and two assessment metrics. (e) Photograph of a freshly resected breast tissue specimen. (f) Image of the concentration of HER2-NPs. (g) Image of the concentration ratio of HER2-NPs vs. isotype-NPs. (h) IHC staining with an anti-HER2 mAb. The scale bars represent 2 mm.
Fig. 8.
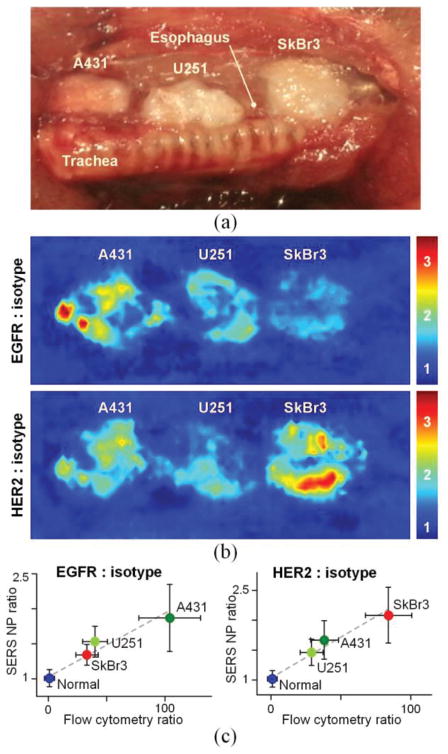
In vivo endoscopic molecular imaging performed with multiplexed SERS NPs delivered via oral gavage. (a) Photograph of a surgically exposed rat esophagus implanted with three tumor xenografts. (d) Images showing the concentration ratio of EGFR-NPs vs. isotype-NPs and HER2-NPs vs. isotype-NPs. (c) Plots showing the correlation between the image-derived ratios from various tissue types (normal esophagus and three tumors) and the corresponding fluorescence ratio (targeted-NP vs. isotype-NP) from flow-cytometry experiments with the cell lines used to generate the various tumor xenografts. All values in the figures are presented as mean ± standard deviation. R > 0.95. Reproduced with permission from [12] © Biomedical Optics Express (2015).
The approaches mentioned above are all based on acquiring full spectra. A direct wide-field imaging method can also be performed by introducing a tunable filter, or multiple dichroic beam splitters, to enable the acquisition of two-dimensional images at multiple wavelengths over time (in the case of a tunable filter) or simultaneously (in the case of dichroic beam splitters) [32,60]. The advantage of this approach is that it eliminates the need for mechanical scanning and can enable rapid spectral imaging if only a few wavelength channels are required. However, due to the speed limitations of tunable filters, the number of spectral “bins” (i.e. the spectral resolution) is limited if rapid imaging is desired, which limits the degree of multiplexing that can be accurately handled. Theoretically, a spectrometer-based approach has the advantage of fully utilizing all of the Raman photons that are collected into a probe, and dispersing them onto a detector array. A tunable filter only analyzes a small spectral window at each moment in time and discards the rest of the photons, which is not efficient. Nevertheless, recent studies have demonstrated that it is possible to utilize this approach for the multiplexed imaging of multiple NP flavors applied in mouse tissues over a field of view of 2.2 cm2 and a pixel resolution of 50 μm [32,60]. Ideally, this approach requires the optimal selection of suitable NP flavors with minimal spectral overlap in certain wavelength regions. In addition, the determination and removal of background spectral components is challenging, and requires a number of assumptions and interpolation methods [60,78].
IV. Spectral demultiplexing
A. Least squares algorithm
A critical aspect of SERS imaging is the use of an appropriate algorithm to analyze the acquired spectra and to quantify the abundance of the detected SERS NPs. The most popular algorithm is the classical least squares algorithm due to its speed and ease of implementation [44]. Generally, other than sources of random noise, it is assumed that each measured spectrum (S) is a linear combination of known reference spectra (Rn) (1), which include the reference spectra of all of the SERS NP flavors, and the tissue-background spectra when imaging/detecting SERS NPs in tissues. This algorithm can accurately predict the weight of each spectral component (wn) when the spectra of the SERS NP flavors and background components are stable.
In practice, variations in tissue background spectra are often observed due to heterogeneities in the tissue components. For example, muscle, fat and tumor exhibit different autofluorescence spectra. In addition, variations in hemoglobin concentration in muscle, or variations in collagen, NADH, and/or melanin in skin, can alter tissue background spectra [79]. These variations in the tissue background spectra can compromise the accuracy of the classical least squares algorithm. Many methods have been proposed to increase the robustness of the demultiplexing algorithms to such variations, such as adding a low-order polynomial term to compensate for slight background changes [80], allowing the baseline to vary [81–83], allowing the peaks of the reference spectra to vary [84], and incorporating the principal components of variable background spectra as reference spectra [75,85]. Among these various methods, two methods are commonly used for the imaging of SERS NPs, as discussed in the following subsection.
| (1) |
| (2) |
| (3) |
S = measured spectral data
wn = weight of spectral component n (here the spectral components include the different SERS NP flavors and tissue backgrounds)
Rn = reference spectrum of component n
am = weight of mth-order polynomial term
Pm = mth-order polynomial term (to compensate for slight variations in the background)
bln = weight of lth principal component for spectral component n
Cln = lth principal component for spectral component n
B. Least squares algorithm with a method to compensate for background variations
One method to deal with background variations (referred to as Method 1) is to allow the background spectra to vary according to a low-order polynomial model, equation (2) [80]. This method can effectively account for minor broadband background variations without acquiring numerous background spectra [31,58]. The data in Figs. 7 were demultiplexed using this method, based on data from SERS NPs placed on a glass slide (with a relatively uniform background spectrum). The other method (referred to as Method 2) assumes that the background is a linear combination of principal components derived from experimental measurements (training sets) from many potential sources of tissue background (tumor, normal, fat, skin, etc.), equation (3). Although it requires the acquisition of additional background measurements, the combination of least squares and principal component analysis (PCA) has been shown to be a robust method to accommodate for moderate variations in the background spectra [12,61,75]. The spectra in Figs. 6 and 8 were demultiplexed using this method based on data from breast or esophagus tissues (with variable background spectra).
Fig. 6 is an example showing how PCA improves demultiplexing accuracy. A freshly resected breast specimen (containing both tumor and normal tissue, size = 2 cm × 1.5 cm) shows various tissue backgrounds due to tissue heterogeneity and incomplete rinse removal of hemoglobin and surgical inks (Figs. 6(a) and 6(e)). The specimen was raster scanned using a 15-mW 785-nm laser to acquire 162 background spectra for the calculation of their principal components and an average background spectrum (Fig. 2(b)). After background acquisition, the specimen was stained with an equimolar mixture of HER2-NPs and isotype-NPs (150 pM/each with 1% BSA) for 10 min, followed by a 10-s rinse step in PBS. The NP-stained specimen was raster-scanned with a 0.5-mm laser spot in 3 min (integration time 0.1 s). The entire procedure, including background acquisition, staining, and imaging, was performed in ~15 min. The acquired spectra were demultiplexed using the two methods as discussed in the previous paragraph. Fig. 6(c) shows the post-demultiplexing fit to one of the 2000 acquired spectra. Method 2 is clearly superior to Method 1 because Method 2 includes three principal components that are responsible for 99.4% of the variation seen in all of the tissue-background measurements.
C. Metrics to assess demultiplexing accuracy
The accuracy of spectral fitting can be quantified by two metrics: the relative fitting error (RFE), which evaluates the fit of the full spectrum [44]; and the spectral reliability index (SRI), which ignores the background components and evaluates only the SERS NP components [73]. SRI is better at evaluating the demultiplexing accuracy, especially when the NP signals are weak in comparison to the broadband background signals. This is because the RFE may be biased towards 1 (a perfect fit) when the background signals are fit with high fidelity even if the fit for the NP components is poor. Fig. 6(d) shows the fitting errors of all 2000 spectra in the form of a box plot. The fitting metrics (RFE and SRI) show that Method 2 outperforms Method 1. With accurate demultiplexing using Method 2, the ratiometric imaging of HER2-NP concentrations vs. control-NP concentrations exhibits a HER2 expression pattern that is in agreement with IHC validation data (Figs. 6(g) and 6(h)).
V. Ratiometric quantification of specific vs. nonspecific contrast
Recently, a ratiometric imaging approach has been demonstrated to mitigate the ambiguity caused by nonspecific NP accumulation and uneven NP delivery [31,32,59,61], a universal problem in the molecular imaging field that often leads to erroneous image interpretation. In this method, an untargeted agent is mixed with a targeted agent during tissue staining, in which the untargeted agent controls for the nonspecific behavior of the targeted agent. Instead of assessing the pure targeted-NP signal, the ratio of the targeted-NP signal vs. untargeted-NP signal is used to quantify specific vs. nonspecific binding. Figs. 6(f) and 6(g) show how ratiometric quantification helps to reveal the specific binding of HER2-targeted NPs to a HER2-positive human breast tumor. The absolute NP concentrations are misleading (Fig. 6(f)): the HER2-NPs accumulate more (nonspecifically) on normal tissue regions than on the tumor regions because the tumors are denser and less porous than the surrounding normal tissues (similar results have been observed with tumor xenografts [31,86]). However, the ratiometric calculation of the HER2-NP vs. control-NP concentrations allow one to normalize away the nonspecific NP accumulation and reveals a HER2 expression pattern that is in agreement with IHC validation data (Figs. 6(g) and 6(h)).
A. Limit of detection
Figure 7 shows the linearity and detection limit of our Raman imaging system when imaging four multiplexed SERS NPs. Although the SERS peaks are barely resolvable at 1 pM by visual inspection of the raw spectra (Fig. 7(b)), the demultiplexing algorithm is still capable of calculating the concentration of each NP flavor with a detection error <20% (Fig. 7(c)). Since the ratio combines two noisy measurements, a slightly higher detection limit of 5 pM is necessary to ensure that the error in the ratio is < 20% (Fig. 7(d), 7(g)). The detection limit depends on the specific NP flavors employed, how many NP flavors are being multiplexed (e.g. a detection limit of 1–2 pM can be achieved when analyzing two or three multiplexed NP flavors [31,61]), and the concentration ratio of the NPs (e.g. the error/crosstalk in NP measurements increases when the NP concentration ratio reaches 10:1). However, based on our data with tumor xenografts and human breast tumor specimens (Figs. 3,6 and 8), the ratio of targeted vs. untargeted NPs rarely exceeds 2–3 for tumors, and the measured NP concentrations are typically in the range of 10–100 pM, where we have shown that the NP measurements are accurate and unaffected by crosstalk between NP flavors [12,31]. To ensure reliable measurments, a linearity test should be performed prior to all tissue-staining and imaging experiments in which new conditions are encountered.
B. Advantages of SERS NPs for ratiometric quantification
A number of previous studies have demonstrated the efficacy of a ratiometric (or “dual-tracer”) approach to mitigate the effects of nonspecific accumulation of systemically administered fluorescent agents [87–91]. Compared to fluorescent agents, SERS NPs offer several important advantages for ratiometric imaging (Table 1). First, all SERS NP flavors possess an identical geometry and outer coating material, ensuring the identical nonspecific behavior of different flavors of NPs in tissue. This is especially true if the NPs are conjugated to isotype-matched monoclonal antibodies, and multiplexed together with an isotype-control antibody to normalize for nonspecific effects. Second, multiple flavors of SERS NPs can be excited at a single illumination wavelength, ensuring that all NP flavors are interrogated identically in terms of optical irradiance and penetration depth. In contrast, different fluorescent agents need to be imaged at different excitation and/or emission wavelengths, where complications may arise due to variations in tissue optical properties as a function of wavelength [5,92–94]. Finally, due to the excellent multiplexing capabilities of SERS NPs, one untargeted NP flavor can serve as a control for a large panel of targeted NP flavors for multiplexed biomarker detection.
Table 1.
Comparison of fluorescence dyes and SERS NPs for ratiometric imaging.
| Properties | Fluorescence dyes | SERS NPs |
|---|---|---|
| Chemical composition | different | same |
| Excitation and/or emission wavelength range | different | same |
| Photon distribution in tissue | wavelength-dependent | same |
| Tissue autofluorescence | wavelength-dependent | same |
| Photostability | bleaches over time | stable |
| Multiplexing capability | typically 2–4 | >10 |
We have demonstrated the ratiometric quantification of two cell-surface receptors, EGFR and HER2, by topically applying an equimolar mixture of 3 NP flavors - EGFR-targeted NPs, HER2-targeted NPs and isotype control NPs [31] (Fig. 8), in which the ratiometric results agree with flow-cytometry results [12].
C. Kinetic modeling to derive receptor densities
Although ratiometric imaging of cell-surface receptors has shown good agreement with IHC and flow cytometry (Figs. 6 and 8), the measured ratio is not necessarily proportional to the receptor density. Kinetic modeling has been demonstrated as an effective approach to estimate the receptor density by analyzing the retention and washout of exogenous contrast agents over time [95]. Most recently, a kinetic model has been developed to quantify the binding potential (BP: proportional to receptor densities) of cell-surface receptors in fresh tissues based on SERS NP retention kinetics [62]. More specifically, this model accounts for the different binding and dissociation rates of SERS NPs (both specific binding and nonspecific binding), and takes into account the movement of NPs between “bound” compartments and “free” compartments. By modeling the decrease of measured concentrations of targeted and untargeted NPs as a function of serial rinsing steps, the kinetic model aimed to calculate the BP of targeted receptors. Experiments with tumor xenografts have shown that this kinetic model is capable of discriminating between different tissue types with known differences in EGFR expression [62].
VI. Challenges and future perspectives
Increasing numbers of studies have demonstrated the feasibility of using SERS NPs to detect tumors and to guide tumor-resection surgeries. For example, the in vivo imaging of untargeted SERS NPs injected in mice has been shown to allow for the visualization of brain tumor xenografts through passive (non-chemical) EPR effects [53]. The detection of subcutaneous or orthotopic tumor xenografts has been demonstrated through the in vivo imaging of targeted SERS NPs, either administered systemically or topically [12,31–34]. Unfortunately, one obstacle is that SERS NPs are not yet approved for human use. However, one potential approach to use SERS NPs to guide surgery is to intraoperatively image freshly resected tissues, which circumvents toxicity issues and has the potential to receive expedited regulatory approval.
One example of how SERS NPs may be utilized on ex vivo tissues to guide surgery is for breast cancer lumpectomy surgeries. We have demonstrated that the topical application and ratiometric quantification of SERS NPs allows for the rapid (<15 min) molecular phenotyping of freshly excised tissues (Fig. 6) to potentially enable the intraoperative detection of residual tumor at lumpectomy margins. This surface-imaging approach enables the rapid intraoperative assessment of large tissue surfaces without the sampling errors that are inevitable for conventional post-operative pathology, in a time frame that is consistent with current intraoperative guidance techniques such as 2D specimen X-ray and frozen-section pathology (not typically done for breast tissues due to their high lipid content). In terms of clinical translation, it is important to note that the application of targeted SERS NPs does not interfere with downstream IHC (Fig. 6). While the aim of SERS imaging would be to greatly reduce the number of re-excision surgeries, post-operative histopathology can still be performed on tissue afterwards as a gold standard to identify microscopic tumor burden or close margins.
Additional work is necessary to further promote the SERS imaging technique for surgical guidance. Recent advances in demultiplexing algorithms, especially the combination of least squares fitting and PCA, have improved the accuracy of SERS NP measurements by accounting for background variations caused by tissue heterogeneities, residual hemoglobin, surgical inks, or unknown chemical components (Fig. 6)[75]. However, brighter NPs would also help to improve signal to background ratios, detection sensitivities, and imaging speeds [69]. Another potential challenge is that cauterization of tissues may denature proteins, including the biomarkers targeted by SERS NPs. Note that cautery, used by many but not all surgeons, is also a challenge for conventional pathology, where it has been observed that certain protein targets and epitopes are more adversely affected than others. A major advantage of SERS NPs is their potential to simultaneously image a large panel of biomarkers (potentially 5 to 10 [58]), which may mitigate the effects of cautery since certain biomarkers have been shown to be less affected by cauterization damage [96]. Future work is needed to demonstrate the ability to quantify a larger panel of biomarkers (perhaps up to 10 or more) with targeted SERS NPs, which will require additional optimization of the targeted SERS NPs themselves as well as the detection hardware and tissue-staining, rinsing, and imaging protocols. Finally, the SERS NP imaging technique provides a wide-area macroscopic imaging strategy to identify millimeter/submillimeter-sized tumors and may potentially be combined with high-resolution imaging techniques, such as confocal mosaicing microscopy [97] and light reflectance spectroscopy [98], to provide surgeons with information about both biomarker distribution and cell/tissue morphology.
Acknowledgments
This work was primarily supported by the NIH / NIBIB – R21 EB015016 (Liu), the Department of Mechanical Engineering at the University of Washington, and the department of education GAANN fellowship program (S.K.). Additional support for J.T.C.L. and A.K.G. was provided by the NIH / NCI – R01 CA175391 (Liu and Sanai) and the NIH / NIDCR – R01 DE023496 (Liu).
Biographies

Yu Wang received his Ph.D. degree in mechanical engineering from Beihang University, Beijing, China, in 2013. After graduation, he joined Dr. Jonathan Liu’s research group as a postdoctoral associate to develop SERS nanoparticles and a Raman-imaging system for visualizing cancer biomarkers in tissues. His research interests include biomedical optics and molecular imaging.

Soyoung Kang received the B.S. degree in Biomedical Engineering from University of Southern California in 2010 and the M.S. degree in Medical Systems Engineering from University of Magdeburg, Germany in 2014. She is currently a GAANN fellow and pursuing a PhD degree in the Mechanical Engineering Department at the University of Washington. Her work aims to develop a Raman-imaging system for visualizing cancer biomarkers intraoperatively during tumor resection procedures.

Joshua D. Doerksen was born in Lyon, France in 1993. He will be completing his B.S. in Mechanical Engineering at the University of Washington in 2016. In 2013 and 2014, he worked on the 3D modeling of Submarines and cargo ships for the UW Human Powered Submarine Team and the Westwood Shipping Lines respectively. In 2015, he developed an automated tissue-staining device for SERS nanoparticle staining and imaging. His areas of interest include biomedical research and sustainable and low-energy housing.

Adam Glaser received his Ph.D. degree in Engineering Science from the Thayer School of Engineering at Dartmouth College in 2015 under the guidance of Dr. Brian Pogue exploring the biomedical applications of Cherenkov radiation. After graduation, he joined Dr. Jonathan Liu’s research group at the University of Washington as a post-doctoral research associate to work on the development of a miniature dual-axis confocal microscope and investigate the feasibility of performing light-sheet microscopy in biological tissue. His research interests include Monte Carlo methods for modeling radiation and light transport, biomedical optics, molecular imaging, and medical physics.

Jonathan Liu was born in Albany, NY and raised in Honolulu, HI. Jonathan received degrees in mechanical engineering at Princeton (B.S.E., 1999) and Stanford (M.S., 2000 & Ph.D., 2005). He was a postdoctoral fellow in the department of electrical engineering (Ginzton Labs) and the Molecular Imaging Program at Stanford (2005–2009), and was later appointed as an instructor within the Stanford University School of Medicine (2009–2010).
From 2010 to 2014, he was an assistant professor in the biomedical engineering department at SUNY Stony Brook. Since 2014, he has been an assistant professor in the mechanical engineering department at the University of Washington in Seattle.
Liu received an award as the top graduate in mechanical engineering at Princeton, an NSF graduate research fellowship, a Canary Foundation / American Cancer Society postdoctoral fellowship, and a K99/R00 career-development award from the NIH. His laboratory for molecular biophotonics develops optical strategies for improving the diagnosis and treatment of diseases. https://washington-seattle.digication.com/jonliu
Contributor Information
Yu Wang, Email: yuwang2@uw.edu.
Soyoung Kang, Email: soyoungk@uw.edu.
Josh D. Doerksen, Email: doerksenjosh@yahoo.fr.
Adam K. Glaser, Email: akglaser@uw.edu.
Jonathan T.C. Liu, Email: jonliu@uw.edu.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. International journal of cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–60. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 3.Bergholt MS, Zheng W, Lin K, Ho KY, Teh M, Yeoh KG, et al. Characterizing variability in in vivo Raman spectra of different anatomical locations in the upper gastrointestinal tract toward cancer detection. Journal of Biomedical Optics. 2011;16:037003–10. doi: 10.1117/1.3556723. [DOI] [PubMed] [Google Scholar]
- 4.Jo JA, Applegate BE, Park J, Shrestha S, Pande P, Gimenez-Conti IB, et al. In vivo simultaneous morphological and biochemical optical imaging of oral epithelial cancer. Biomedical Engineering, IEEE Transactions on. 2010;57:2596–2599. doi: 10.1109/TBME.2010.2060485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dam GM, Themelis G, Crane LM, Harlaar NJ, Pleijhuis RG, Kelder W, et al. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-[alpha] targeting: first in-human results. Nature medicine. 2011;17:1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 6.Bird-Lieberman EL, Neves AA, Lao-Sirieix P, O'Donovan M, Novelli M, Lovat LB, et al. Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett's esophagus. Nature Medicine. 2012;18:315–21. doi: 10.1038/nm.2616. [DOI] [PubMed] [Google Scholar]
- 7.Le Q, Turzhitsky V, Chuttani R, Pleskow DK, Goldsmith JD, Lianyu G, et al. Spectral Imaging With Scattered Light: From Early Cancer Detection to Cell Biology. Selected Topics in Quantum Electronics, IEEE Journal of. 2012;18:1073–1083. doi: 10.1109/JSTQE.2011.2161575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergholt MS, Zheng W, Lin K, Ho KY, Teh M, Yeoh KG, et al. In vivo diagnosis of gastric cancer using Raman endoscopy and ant colony optimization techniques. International journal of cancer. 2011;128:2673–2680. doi: 10.1002/ijc.25618. [DOI] [PubMed] [Google Scholar]
- 9.Kong K, Kendall C, Stone N, Notingher I. Raman spectroscopy for medical diagnostics — From in-vitro biofluid assays to in-vivo cancer detection. Advanced Drug Delivery Reviews. 2015;89:121–134. doi: 10.1016/j.addr.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Frangioni JV. New Technologies for Human Cancer Imaging. Journal of Clinical Oncology. 2008;26:4012–4021. doi: 10.1200/JCO.2007.14.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fass L. Imaging and cancer: A review. Molecular Oncology. 2008;2:115–152. doi: 10.1016/j.molonc.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YW, Kang S, Khan A, Bao PQ, Liu JTC. In vivo multiplexed molecular imaging of esophageal cancer via spectral endoscopy of topically applied SERS nanoparticles. Biomedical Optics Express. 2015;6:3714–3723. doi: 10.1364/BOE.6.003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 15.Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol. 2015;12:381–394. doi: 10.1038/nrclinonc.2015.73. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chemical Physics Letters. 1974;26:163–166. [Google Scholar]
- 17.Le Ru EC, Blackie E, Meyer M, Etchegoin PG. Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study. The Journal of Physical Chemistry C. 2007;111:13794–13803. [Google Scholar]
- 18.Sharma B, Frontiera RR, Henry AI, Ringe E, Van Duyne RP. SERS: Materials, applications, and the future. Materials Today. 2012;15:16–25. [Google Scholar]
- 19.Ni J, Lipert RJ, Dawson GB, Porter MD. Immunoassay Readout Method Using Extrinsic Raman Labels Adsorbed on Immunogold Colloids. Analytical Chemistry. 1999;71:4903–4908. doi: 10.1021/ac990616a. [DOI] [PubMed] [Google Scholar]
- 20.Graham D, Mallinder BJ, Smith WE. Detection and identification of labeled DNA by surface enhanced resonance Raman scattering. Biopolymers. 2000;57:85–91. doi: 10.1002/(SICI)1097-0282(2000)57:2<85::AID-BIP5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–40. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 22.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Femtomolar Detection of Prostate-Specific Antigen: An Immunoassay Based on Surface-Enhanced Raman Scattering and Immunogold Labels. Analytical Chemistry. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 23.Cao YC, Jin R, Nam JM, Thaxton CS, Mirkin CA. Raman Dye-Labeled Nanoparticle Probes for Proteins. Journal of the American Chemical Society. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 24.Su X, Zhang J, Sun L, Koo TW, Chan S, Sundararajan N, et al. Composite Organic Inorganic Nanoparticles (COINs) with Chemically Encoded Optical Signatures. Nano Letters. 2005;5:49–54. doi: 10.1021/nl0484088. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Chen Y, Wang T, Ma Z, Su Z. Monodispersed Gold Nanorod-Embedded Silica Particles as Novel Raman Labels for Biosensing. Advanced Functional Materials. 2008;18:355–361. [Google Scholar]
- 26.Kustner B, Gellner M, Schutz M, Schoppler F, Marx A, Strobel P, et al. SERS labels for red laser excitation: silica-encapsulated SAMs on tunable gold/silver nanoshells. Angew Chem Int Ed Engl. 2009;48:1950–3. doi: 10.1002/anie.200804518. [DOI] [PubMed] [Google Scholar]
- 27.Doering WE, Piotti ME, Natan MJ, Freeman RG. SERS as a Foundation for Nanoscale, Optically Detected Biological Labels. Advanced Materials. 2007;19:3100–3108. [Google Scholar]
- 28.Porter MD, Lipert RJ, Siperko LM, Wang G, Narayanan R. SERS as a bioassay platform: fundamentals, design, and applications. Chemical Society Reviews. 2008;37:1001–1011. doi: 10.1039/b708461g. [DOI] [PubMed] [Google Scholar]
- 29.Schlucker S. SERS microscopy: nanoparticle probes and biomedical applications. Chemphyschem. 2009;10:1344–54. doi: 10.1002/cphc.200900119. [DOI] [PubMed] [Google Scholar]
- 30.Jokerst JV, Miao Z, Zavaleta C, Cheng Z, Gambhir SS. Affibody-Functionalized Gold–Silica Nanoparticles for Raman Molecular Imaging of the Epidermal Growth Factor Receptor. Small. 2011;7:625–633. doi: 10.1002/smll.201002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YW, Khan A, Som M, Wang D, Chen Y, Leigh SY, et al. Rapid ratiometric biomarker detection with topically applied SERS nanoparticles. Technology. 2014;02:118–132. doi: 10.1142/S2339547814500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallia RJ, McVeigh PZ, Fisher CJ, Veilleux I, Wilson BC. Wide-field multiplexed imaging of EGFR-targeted cancers using topical application of NIR SERS nanoprobes. Nanomedicine. 2014:1–13. doi: 10.2217/nnm.14.80. [DOI] [PubMed] [Google Scholar]
- 33.Maiti KK, Dinish US, Samanta A, Vendrell M, Soh KS, Park SJ, et al. Multiplex targeted in vivo cancer detection using sensitive near-infrared SERS nanotags. Nano Today. 2012;7:85–93. [Google Scholar]
- 34.Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nature biotechnology. 2007;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 35.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Yarden Y. The EGFR family and its ligands in human cancer: signalling mechanisms and therapeutic opportunities. European Journal of Cancer. 2001;37(Supplement 4):3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 37.Krishnamurti U, Silverman JF. HER2 in Breast Cancer: A Review and Update. Advances in Anatomic Pathology. 2014;21:100–107. doi: 10.1097/PAP.0000000000000015. [DOI] [PubMed] [Google Scholar]
- 38.Wang YL, Schlucker S. Rational design and synthesis of SERS labels. Analyst. 2013;138:2224–2238. doi: 10.1039/c3an36866a. [DOI] [PubMed] [Google Scholar]
- 39.Wang YQ, Yan B, Chen LX. SERS Tags: Novel Optical Nanoprobes for Bioanalysis. Chemical Reviews. 2013;113:1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 40.Samanta A, Jana S, Das RK, Chang YT. Biocompatible surface-enhanced Raman scattering nanotags for in vivo cancer detection. Nanomedicine. 2014;9:523–535. doi: 10.2217/nnm.13.222. [DOI] [PubMed] [Google Scholar]
- 41.Lakowicz JR. Principles of fluorescence spectroscopy. Springer Science & Business Media; 2013. [Google Scholar]
- 42.Wang Y, Chen L. Quantum dots, lighting up the research and development of nanomedicine. Nanomedicine. 2011;7:385–402. doi: 10.1016/j.nano.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 43.McCreery RL. Raman spectroscopy for chemical analysis. Vol. 225. John Wiley & Sons; 2005. [Google Scholar]
- 44.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, et al. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13511–13516. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doering WE, Nie S. Spectroscopic Tags Using Dye-Embedded Nanoparticles and Surface-Enhanced Raman Scattering. Analytical Chemistry. 2003;75:6171–6176. doi: 10.1021/ac034672u. [DOI] [PubMed] [Google Scholar]
- 46.Bantz KC, Meyer AF, Wittenberg NJ, Im H, Kurtulus O, Lee SH, et al. Recent progress in SERS biosensing. Phys Chem Chem Phys. 2011;13:11551–67. doi: 10.1039/c0cp01841d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vendrell M, Maiti KK, Dhaliwal K, Chang YT. Surface-enhanced Raman scattering in cancer detection and imaging. Trends in Biotechnology. 2013;31:249–257. doi: 10.1016/j.tibtech.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Shi W, Paproski RJ, Moore R, Zemp R. Detection of circulating tumor cells using targeted surface-enhanced Raman scattering nanoparticles and magnetic enrichment. J Biomed Opt. 2014;19:056014. doi: 10.1117/1.JBO.19.5.056014. [DOI] [PubMed] [Google Scholar]
- 49.Nima ZA, Mahmood M, Xu Y, Mustafa T, Watanabe F, Nedosekin DA, et al. Circulating tumor cell identification by functionalized silver-gold nanorods with multicolor, super-enhanced SERS and photothermal resonances. Sci Rep. 2014;4:4752. doi: 10.1038/srep04752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farhadi A, Roxin Á, Wilson BC, Zheng G. Nano-Enabled SERS Reporting Photosensitizers. Theranostics. 2015;5:469–476. doi: 10.7150/thno.10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keren S, Zavaleta C, Cheng Z, de la Zerda A, Gheysens O, Gambhir SS. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5844–9. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bohndiek SE, Wagadarikar A, Zavaleta CL, Van de Sompel D, Garai E, Jokerst JV, et al. A small animal Raman instrument for rapid, wide-area, spectroscopic imaging. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12408–13. doi: 10.1073/pnas.1301379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karabeber H, Huang RM, Iacono P, Samii JM, Pitter K, Holland EC, et al. Guiding Brain Tumor Resection Using Surface-Enhanced Raman Scattering Nanoparticles and a Hand-Held Raman Scanner. Acs Nano. 2014;8:9755–9766. doi: 10.1021/nn503948b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E, et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nature medicine. 2012;18:829–834. doi: 10.1038/nm.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samanta A, Maiti KK, Soh KS, Liao X, Vendrell M, Dinish US, et al. Ultrasensitive Near-Infrared Raman Reporters for SERS-Based In Vivo Cancer Detection. Angewandte Chemie International Edition. 2011;50:6089–6092. doi: 10.1002/anie.201007841. [DOI] [PubMed] [Google Scholar]
- 56.Su X, Zhang J, Sun L, Koo TW, Chan S, Sundararajan N, et al. Composite organic-inorganic nanoparticles (COINs) with chemically encoded optical signatures. Nano Lett. 2005;5:49–54. doi: 10.1021/nl0484088. [DOI] [PubMed] [Google Scholar]
- 57.Thakor AS, Luong R, Paulmurugan R, Lin FI, Kempen P, Zavaleta C, et al. The fate and toxicity of Raman-active silica-gold nanoparticles in mice. Science Translational Medicine. 2011;3:79ra33. doi: 10.1126/scitranslmed.3001963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zavaleta CL, Garai E, Liu JTC, Sensarn S, Mandella MJ, Van de Sompel D, et al. A Raman-based endoscopic strategy for multiplexed molecular imaging. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2288–E2297. doi: 10.1073/pnas.1211309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garai E, Sensarn S, Zavaleta CL, Sompel DVd, Loewke NO, Mandella MJ, et al. High-sensitivity, real-time, ratiometric imaging of surface-enhanced Raman scattering nanoparticles with a clinically translatable Raman endoscope device. Journal of Biomedical Optics. 2013;18:096008–1–106008–13. doi: 10.1117/1.JBO.18.9.096008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McVeigh PZ, Mallia RJ, Veilleux I, Wilson BC. Widefield quantitative multiplex surface enhanced Raman scattering imaging in vivo. Journal of Biomedical Optics. 2013;18:046011. doi: 10.1117/1.JBO.18.4.046011. [DOI] [PubMed] [Google Scholar]
- 61.Wang YW, Khan A, Leigh SY, Wang D, Chen Y, Meza D, et al. Comprehensive spectral endoscopy of topically applied SERS nanoparticles in the rat esophagus. Biomedical Optics Express. 2014;5:2883–2895. doi: 10.1364/BOE.5.002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinha L, Wang Y, Yang C, Khan A, Brankov JG, Liu JTC, et al. Quantification of the binding potential of cell-surface receptors in fresh excised specimens via dual-probe modeling of SERS nanoparticles. Scientific Reports. 2015;5:8582. doi: 10.1038/srep08582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zavaleta CL, Hartman KB, Miao Z, James ML, Kempen P, Thakor AS, et al. Preclinical evaluation of Raman nanoparticle biodistribution for their potential use in clinical endoscopy imaging. Small. 2011;7:2232–2240. doi: 10.1002/smll.201002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Shenk R, Wang N, et al. Diagnosing breast cancer using Raman spectroscopy: prospective analysis. Journal of Biomedical Optics. 2009;14:054023. doi: 10.1117/1.3247154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao J, Lui H, Kalia S, Zeng H. Real-time Raman spectroscopy for automatic in vivo skin cancer detection: an independent validation. Analytical and Bioanalytical Chemistry. 2015:1–7. doi: 10.1007/s00216-015-8914-9. [DOI] [PubMed] [Google Scholar]
- 66.Vargis E, Tang YW, Khabele D, Mahadevan-Jansen A. Near-infrared Raman Microspectroscopy Detects High-risk Human Papillomaviruses. Transl Oncol. 2012;5:172–9. doi: 10.1593/tlo.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fenn MB, Xanthopoulos P, Pyrgiotakis G, Grobmyer SR, Pardalos PM, Hench LL. Raman spectroscopy for clinical oncology. Advances in Optical Technologies. 2011;2011 [Google Scholar]
- 68.Teh SK, Zheng W, Ho KY, Teh M, Yeoh KG, Huang Z. Near-infrared Raman spectroscopy for early diagnosis and typing of adenocarcinoma in the stomach. Br J Surg. 2010;97:550–7. doi: 10.1002/bjs.6913. [DOI] [PubMed] [Google Scholar]
- 69.Harmsen S, Huang R, Wall MA, Karabeber H, Samii JM, Spaliviero M, et al. Surface-enhanced resonance Raman scattering nanostars for high-precision cancer imaging. Science Translational Medicine. 2015;7:271ra7. doi: 10.1126/scitranslmed.3010633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohs AM, Mancini MC, Singhal S, Provenzale JM, Leyland-Jones B, Wang MD, et al. Hand-held spectroscopic device for in vivo and intraoperative tumor detection: contrast enhancement, detection sensitivity, and tissue penetration. Analytical chemistry. 2010;82:9058–9065. doi: 10.1021/ac102058k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahadevan-Jansen A, Richards-Kortum R. Raman spectroscopy for cancer detection: a review. Engineering in Medicine and Biology Society, 1997. Proceedings of the 19th Annual International Conference of the IEEE; 1997. pp. 2722–2728. [Google Scholar]
- 72.Short MA, Lam S, McWilliams A, Zhao J, Lui H, Zeng H. Development and preliminary results of an endoscopic Raman probe for potential in vivo diagnosis of lung cancers. Opt Lett. 2008;33:711–3. doi: 10.1364/ol.33.000711. [DOI] [PubMed] [Google Scholar]
- 73.Leigh SY, Som M, Liu JTC. Method for Assessing the Reliability of Molecular Diagnostics Based on Multiplexed SERS-Coded Nanoparticles. PLoS ONE. 2013;8:e62084. doi: 10.1371/journal.pone.0062084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang Z, Zeng H, Hamzavi I, McLean DI, Lui H. Rapid near-infrared Raman spectroscopy system for real-time in vivo skin measurements. Optics Letters. 2001;26:1782–1784. doi: 10.1364/ol.26.001782. [DOI] [PubMed] [Google Scholar]
- 75.Van de Sompel D, Garai E, Zavaleta C, Gambhir SS. A Hybrid Least Squares and Principal Component Analysis Algorithm for Raman Spectroscopy. PLoS ONE. 2012;7:e38850. doi: 10.1371/journal.pone.0038850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.ElMasry G, Sun D-W, Allen P. Near-infrared hyperspectral imaging for predicting colour, pH and tenderness of fresh beef. Journal of Food Engineering. 2012;110:127–140. [Google Scholar]
- 77.Gowen AA, O'Donnell CP, Cullen PJ, Downey G, Frias JM. Hyperspectral imaging – an emerging process analytical tool for food quality and safety control. Trends in Food Science & Technology. 2007;18:590–598. [Google Scholar]
- 78.Mallia RJ, McVeigh PZ, Veilleux I, Wilson BC. Filter-based method for background removal in high-sensitivity wide-field-surface-enhanced Raman scattering imaging in vivo. Journal of Biomedical Optics. 2012;17:076017. doi: 10.1117/1.JBO.17.7.076017. [DOI] [PubMed] [Google Scholar]
- 79.Han X, Lui H, McLean DI, Zeng H. Near-infrared autofluorescence imaging of cutaneous melanins and human skin in vivo. J Biomed Opt. 2009;14:024017. doi: 10.1117/1.3103310. [DOI] [PubMed] [Google Scholar]
- 80.Lutz BR, Dentinger CE, Nguyen LN, Sun L, Zhang J, Allen AN, et al. Spectral analysis of multiplex Raman probe signatures. ACS Nano. 2008;2:2306–14. doi: 10.1021/nn800243g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao J, Lui H, McLean DI, Zeng H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl Spectrosc. 2007;61:1225–32. doi: 10.1366/000370207782597003. [DOI] [PubMed] [Google Scholar]
- 82.Lieber CA, Mahadevan-Jansen A. Automated method for subtraction of fluorescence from biological Raman spectra. Appl Spectrosc. 2003;57:1363–7. doi: 10.1366/000370203322554518. [DOI] [PubMed] [Google Scholar]
- 83.Zhang ZM, Chen S, Liang YZ. Baseline correction using adaptive iteratively reweighted penalized least squares. Analyst. 2010;135:1138–46. doi: 10.1039/b922045c. [DOI] [PubMed] [Google Scholar]
- 84.Kode K, Shachaf C, Elchuri S, Nolan G, Paik DS. Raman Labeled Nanoparticles: Characterization of Variability and Improved Method for Unmixing. J Raman Spectrosc. 2012;43:895–905. doi: 10.1002/jrs.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berger AJ, Koo TW, Itzkan I, Feld MS. An Enhanced Algorithm for Linear Multivariate Calibration. Analytical Chemistry. 1998;70:623–627. doi: 10.1021/ac970721p. [DOI] [PubMed] [Google Scholar]
- 86.Tichauer KM, Samkoe KS, Sexton KJ, Hextrum SK, Yang HH, Klubben WS, et al. In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging. Molecular Imaging and Biology. 2012;14:584–92. doi: 10.1007/s11307-011-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baeten J, Haller J, Shih H, Ntziachristos V. In vivo investigation of breast cancer progression by use of an internal control. Neoplasia. 2009;11:220–7. doi: 10.1593/neo.08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pogue BW, Samkoe KS, Hextrum S, O'Hara JA, Jermyn M, Srinivasan S, et al. Imaging targeted-agent binding in vivo with two probes. J Biomed Opt. 2010;15:030513. doi: 10.1117/1.3449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tichauer KM, Samkoe KS, Sexton KJ, Gunn JR, Hasan T, Pogue BW. Improved tumor contrast achieved by single time point dual-reporter fluorescence imaging. Journal of Biomedical Optics. 2012;17:066001. doi: 10.1117/1.JBO.17.6.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tichauer KM, Samkoe KS, Gunn JR, Kanick SC, Hoopes PJ, Barth RJ, et al. Microscopic lymph node tumor burden quantified by macroscopic dual-tracer molecular imaging. Nat Med. 2014;20:1348–1353. doi: 10.1038/nm.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu JTC, Helms MW, Mandella MJ, Crawford JM, Kino GS, Contag CH. Quantifying Cell-Surface Biomarker Expression in Thick Tissues with Ratiometric Three-Dimensional Microscopy. Biophysical Journal. 2009;96:2405–2414. doi: 10.1016/j.bpj.2008.12.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacques SL. Optical properties of biological tissues: a review. Physics in Medicine and Biology. 2013;58:R37. doi: 10.1088/0031-9155/58/11/R37. [DOI] [PubMed] [Google Scholar]
- 93.Hilderbrand SA, Weissleder R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol. 2010;14:71–9. doi: 10.1016/j.cbpa.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 94.Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotech. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 95.Tichauer KM, Wang Y, Pogue BW, Liu JTC. Quantitative in vivo cell-surface receptor imaging in oncology: kinetic modeling and paired-agent principles from nuclear medicine and optical imaging. Physics in Medicine and Biology. 2015;60:R239. doi: 10.1088/0031-9155/60/14/R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nayak A, Bhuiya TA. Utility of cytokeratin 5/6 and high-molecular-weight keratin in evaluation of cauterized surgical margins in excised specimens of breast ductal carcinoma in situ. Ann Diagn Pathol. 2011;15:243–9. doi: 10.1016/j.anndiagpath.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 97.Abeytunge S, Li Y, Larson B, Peterson G, Seltzer E, Toledo-Crow R, et al. Confocal microscopy with strip mosaicing for rapid imaging over large areas of excised tissue. J Biomed Opt. 2013;18:61227. doi: 10.1117/1.JBO.18.6.061227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharma V, Shivalingaiah S, Peng Y, Euhus D, Gryczynski Z, Liu H. Auto-fluorescence lifetime and light reflectance spectroscopy for breast cancer diagnosis: potential tools for intraoperative margin detection. Biomed Opt Express. 2012;3:1825–40. doi: 10.1364/BOE.3.001825. [DOI] [PMC free article] [PubMed] [Google Scholar]