Abstract
Objective
Use high-resolution metabolomics (HRM) to identify metabolic pathways and networks associated with tobacco use in military personnel.
Methods
Four hundred de-identified samples obtained from the Department of Defense Serum Repository were classified as tobacco users or non-users according to cotinine content. HRM and bioinformatic methods were used to determine pathways and networks associated with classification.
Results
Eighty individuals were classified as tobacco users compared to 320 non-users based on cotinine levels ≥10 ng/mL. Alterations in lipid and xenobiotic metabolism, and diverse effects on amino acid, sialic acid and purine and pyrimidine metabolism were observed. Importantly, network analysis showed broad effects on metabolic associations not simply linked to well-defined pathways.
Conclusions
Tobacco use has complex metabolic effects which must be considered in evaluation of deployment-associated environmental exposures in military personnel.
Introduction
Despite nearly 50 years of antismoking campaigns, nicotine addiction remains a significant public health problem. Smokers lose at least one decade of life expectancy compared with those who have never smoked (1). According to the recent report by U.S. Department of Human and Health Service, smokers increase their risk of death from bronchitis and emphysema, cancers of the respiratory tract and coronary heart disease (2). These severe outcomes emphasize the important effects of smoking, which can complicate interpretations of studies to evaluate metabolic effects of occupational exposures (3, 4).
At least three components of cigarette smoke contribute to adverse health outcomes of smoking. First, nicotine itself is converted to carcinogens (5) and also causes abnormal fetal development due to aberrant activation of acetylcholine receptors (6). Secondly, cigarette smoke contains particulates that activate immune cells (7, 8) and also contain multiple polycyclic aromatic hydrocarbons (PAH) that are bioactivated to carcinogens in vivo (9). Thirdly, cigarettes contribute half of environmental exposure to cadmium, a carcinogenic metal ion (10). In addition, tobacco use is more prevalent among US service personnel than among the US general population; therefore, tobacco use must be included as a confounding factor in any analyses of health impacts of deployment-related exposures. The ability to identify active users following surveillance periods and the related biological response are important when attempting to reconstruct exposure to environmental hazards from archival samples, such as the Department of Defense Serum Repository (DoDSR).
Cotinine is the primary metabolic product of nicotine, a natural insecticide in plants of the Solanaceae family, including tobacco, potatoes, tomatoes and eggplant. Cotinine in the blood or urine is a useful biomarker for cigarette smoking use (or other tobacco use) because 87% of nicotine (t1/2 = 100-150 min) is converted to cotinine, which has a relatively long biological half-life in humans (t1/2 = 770-1130 min) (11). Nicotine from tobacco use is highly addictive, but low levels of nicotine derived from dietary sources and second hand cigarette smoke (12) are not. Most Americans have low levels of cotinine, probably derived from the dietary nicotine as well as second hand cigarette smoke exposure (12). Importantly, for health surveillance purposes, cotinine measures in the blood reflect nicotine exposure unaffected by reporting bias concerning nicotine product use (13, 14).
In the present study, we used high-resolution metabolomics (HRM) analysis of 400 serum samples from the DoDSR to determine metabolic pathway associations with tobacco use based upon cotinine levels. The samples were fully de-identified, and personal survey information was not available to the research team to evaluate the sources of nicotine. The analyses followed a workflow in which cotinine was quantified by HRM [liquid chromatography (LC) with ultra-high resolution mass spectrometry (MS)], samples for individuals that were classified as tobacco users according to cotinine levels were then compared to non-users using partial least squares-discriminant analysis (PLS-DA). Top metabolic features, defined in terms of the accurate mass m/z, LC retention time (RT) and intensity, were then annotated and mapped to metabolic pathways. Results show significant associations with pathways related to PAH exposure, cadmium exposure and inflammation, including pathways of amino acid and nucleic acid metabolism.
Methods
Source of Samples
Four-hundred de-identified serum samples from active duty Armed Forces personnel were obtained from the Department of Defense Serum Repository (DoDSR) as part of a larger study evaluating exposure and response biomarkers more fully described elsewhere in this issue (15). The repository consists of approximately 50 million serum samples originally collected for mandatory armed forces personnel HIV testing (16). Demographic information was available to one of the authors (P. Rorhbeck) but not to others. No information was linked to samples provided; therefore no demographic information was used in this study. Following collection, samples were stored at -30°C. Prior to analysis, specimens were thawed, 500 μL was aliquoted into separate microfuge tubes, refrozen and shipped on dry ice.
High-Resolution Mass Spectrometry and Data Pre-processing
For analysis, 65 μL serum aliquot was added to 130 μL of acetonitrile containing a mixture of stable isotope-labeled internal standards and prepared for metabolomics analysis using established methods (17-19). A quality control pooled reference sample (referred to as Q-Std3) was included at the beginning and end of each analytical batch of 20 samples for normalization and post hoc quantification (17). Samples were analyzed in triplicate by liquid chromatography with Fourier transform mass spectrometry (Dionex Ultimate 3000, Q-Exactive HF, Thermo Fisher) with C18 chromatography/positive electrospray ionization (ESI) mode and resolution of 70,000 (19). Spectral m/z features were acquired in scan range 85-1,250 m/z. Raw data files were extracted using apLCMS (20) with xMSanalyzer (21), followed by batch correction with ComBat (22). Triplicate injections were averaged, filtered for less than 70% non-missing values, log2 transformed and quantile normalized (17).
Annotation
Metabolites were annotated by matching the accurate mass m/z for adducts commonly formed under positive ESI conditions to the METLIN (https://metlin.scripps.edu/index.php) and KEGG (http://www.genome.jp/kegg/pathway.html) databases using a mass error threshold of 10 ppm (relative m/z error × 106). Identities of several metabolites of interest due to associations with cotinine (tryptophan, arginine, citrulline, 5-oxoproline, uracil, hypoxanthine, sphinganine and pyridoxine) were previously confirmed (17, 23). Glycerophosphocholine and acylcarnitines were verified by the characteristic ion dissociation (MS/MS) patterns. Other annotations are supported by pathway enrichment and metabolic correlations to improve annotation confidence (24) when authentic reference standards are not available and/or the abundance is too low for ion dissociation (MS/MS) analysis.
Classification of Tobacco Use Status
Cotinine levels for all samples were calculated from HRM data using reference standardization (17). In this framework, cotinine levels were first determined in Q-Std3 using methods of addition (25), providing a reference standard for cotinine that was subsequently analyzed twice in each batch. Average cotinine response factor was then determined for each batch based upon the H+ adduct (m/z= 177.1023, RT= 191 sec), with sample serum concentration calculated using single point calibration. An individual was considered an active tobacco user if serum cotinine concentration was greater than or equal to 10 ng/mL (11).
Selection of Tobacco Use Discriminatory Metabolites
Partial least squares discriminant analysis (PLSDA), which is a supervised multivariate dimensionality reduction method, was used to identify discriminatory features (26). Optimal number of latent components was determined using the plsgenomics package in R. MixOmics was used to build the PLSDA model and select discriminatory features based on variable importance for projection (VIP) measure (27). Metabolic features exhibiting VIP score ≥ 2.5 were selected for annotation and pathway enrichment.
Metabolic Pathway Enrichment of Tobacco Use Biomarkers
To evaluate systemic metabolic alterations due to tobacco use status, a metabolome-wide association analysis using PLS-DA was completed to capture any strongly correlated m/z features in the raw data. Spearman rank correlation coefficient and corresponding p-values were calculated pair-wise for PLS-DA selected features using the MetabNet() function in the R package MetabNet (28). Features exhibiting Spearman |r| ≥ 0.5 and Benjamini-Hochberg (29) FDR ≤ 20% were selected to test for metabolic pathway enrichment in Mummichog (24) and visualized using Cytoscape (30).
Results
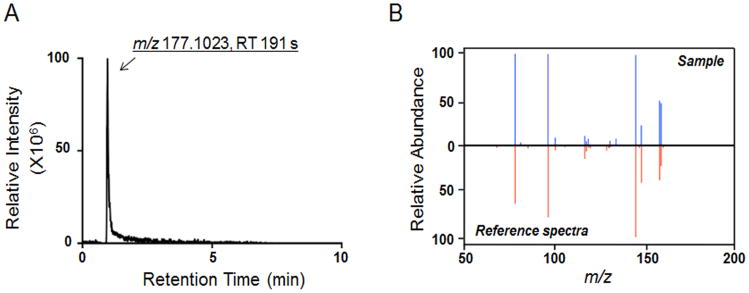
Cotinine was detected by HRM (m/z = 177.1023, RT= 191 sec, Fig 1A), and its identity was validated by ion dissociation (MS/MS) mass spectrometry and compared with authentic reference standard (Fig 1B). Concentration of cotinine in serum was quantified by HRM using reference standardization (Supplemental Table 1). Cotinine value above a threshold of 10 ng/mL was used to define regular tobacco use. Results showed 80 (20%) as tobacco users by this criterion (average cotinine values; 33 ng/mL, maximal cotinine value; 319 ng/mL). These values were within the range observed in active smokers in human metabolome database (HMDB01046, 19-799 ng/mL) and are consistent with tobacco use rates in the armed forces (31).
Figure 1.
Confirmation of cotinine in serum samples. A, Extracted ion chromatogram of m/z (M+H+) and retention time (sec) detected for cotinine. B, Comparison of MS2 of cotinine in serum sample and authentic reference sample.
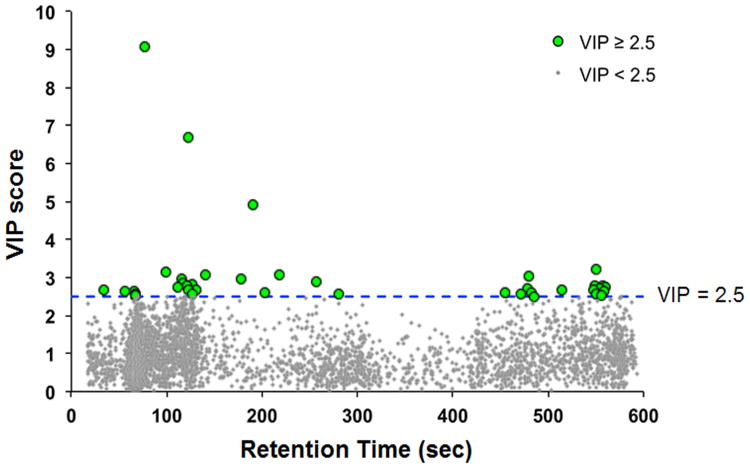
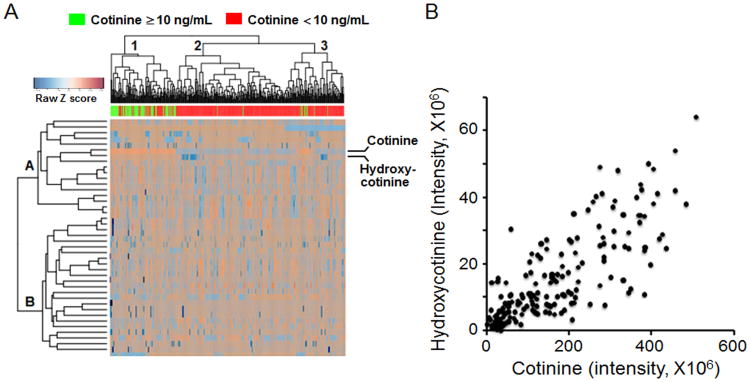
The 80 individuals classified as tobacco users were used for subsequent analysis of metabolic differences between individuals with high and low cotinine. PLS-DA analysis using a VIP threshold ≥ 2.5 identified 41 discriminatory metabolites (Figure 2, Supplemental Table 2), with hydroxycotinine (VIP= 9.07) and cotinine (VIP= 4.90) within the top three discriminatory metabolites. PLS-DA selected metabolites were then characterized using a 2-way hierarchical clustering analysis to 1) determine whether individuals classified as tobacco users clustered separately from non-users and 2) determine how metabolites clustered in distribution across individuals. The results showed there were 3 major clusters of individuals, with Cluster A containing mostly tobacco users, Cluster B containing almost exclusively non-users and Cluster C containing mostly non-users but also including several classified as tobacco users (Fig 3A). Metabolite grouping included two large clusters, with Cluster 1 containing cotinine, hydroxycotinine, and other metabolites with a tendency to be increased in tobacco users, and Cluster 2 containing metabolites that tended to be decreased in tobacco users. These metabolite distributions were very heterogeneous, however, emphasizing that differences in individual metabolism, extent of tobacco use, time course of cotinine clearance and other dietary and environmental exposures are likely to impact metabolic profiles.
Figure 2.
PLS-DA-identified 41 metabolites at cotinine threshold of 10 ng/mL. Top metabolites included nicotine metabolites, cotinine and hydroxycotinine. Additional discriminatory metabolites included endogenous metabolic intermediates, xenobiotics and lipids.
Figure 3.
Hierarchical clustering and correlation of cotinine and hydroxycotinine. A, Unsupervised, hierarchical clustering showed clustering of individuals with high serum cotinine and hydroxycotinine levels. B, Correlation of cotinine and hydroxycotinine for 400 samples.
Direct comparison of cotinine with the m/z matching hydroxycotinine showed that these were highly correlated (Fig 3B). An m/z matching nicotine (m/z 163.1231, RT 472 s) had a weak correlation with cotinine (Pearson r = 0.3; data not shown); lack of strong correlation probably reflects known differences in pharmacokinetics (11). The average intensity for the m/z matching nicotine was significantly higher in those categorized as tobacco users (users; 7.07 × 106 ± 1.81 × 106, non-users; 5.74 × 106 ± 1.74 × 106), consistent with the known precursor-product relationship of nicotine and cotinine.
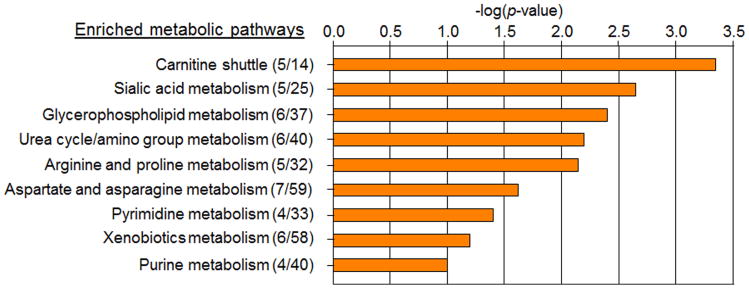
To improve pathway definition, MetabNet (28) was used to select other m/z features significantly correlated with the 41 PLS-DA discriminatory metabolites. Pathway and network analysis with Mummichog showed 61 m/z features matching metabolites with significant enrichment in nine pathways (Fig 4). The complete list of m/z features, annotations and pathway associations are given in Supplemental Table 3. The top pathway, carnitine shuttle, is associated with dyslipidemia and disruption of mitochondrial function, and included matches to multiple acylcarnitines, e.g., cervonylcarnitine (C22:6), docosapentaenoylcarnitine (C20:5), clupanodonylcarnitine (C22:5), and linoleylcarnitine (C18:2), all of which were increased in association with tobacco use. The third pathway from the Mummichog analysis was another lipid pathway, glycerophospholipid metabolism, and included matches to metabolites that were decreased with tobacco use, e.g., glycerophosphocholine, sphinganine, ethanolamine and linoleate.
Figure 4.
Pathway enrichment analysis. Metabolic pathways exhibiting a Mummichog significance score < 0.1 and with ≥ 4 metabolites present from either the PLS-DA selected or network correlation significant features. Significant pathways included fatty acid, amino acid, purine, pyrimidine and xenobiotic metabolism.
The second pathway from the Mummichog analysis (sialic acid metabolism) included five m/z features that matched to multiple sugars, N-acetyl sugars, and related chemicals. These include matches to multiple related isobaric species, with intensities of some being increased while others were decreased. The results suggest a disruption of related sialic acid metabolism.
The remaining significant pathways (Fig 4) included amino acid and urea metabolism, purine and pyrimidine metabolism, and xenobiotic metabolism. Key metabolites in the amino acid pathways, arginine and citrulline, were decreased in tobacco users. The purine pathway did not have a consistent pattern of change, with hypoxanthine increased and its oxidation product, xanthine, decreased. In the pyrimidine pathway, decreases were observed for uracil, dihydrouracil, and cytosine, and also for m/z matching deoxycytosine diphosphate (dCDP). The xenobiotic metabolism pathways included decreased levels of ions matching dihydroxynaphthalene, dihydroxy-epoxy-tetrahydronaphthalene, benzo[a]pyrene, hydroxybenzo[a]pyrene-4,5-oxide, trichloroethanol glucuronide, urochloralic acid, and chloral hydrate.
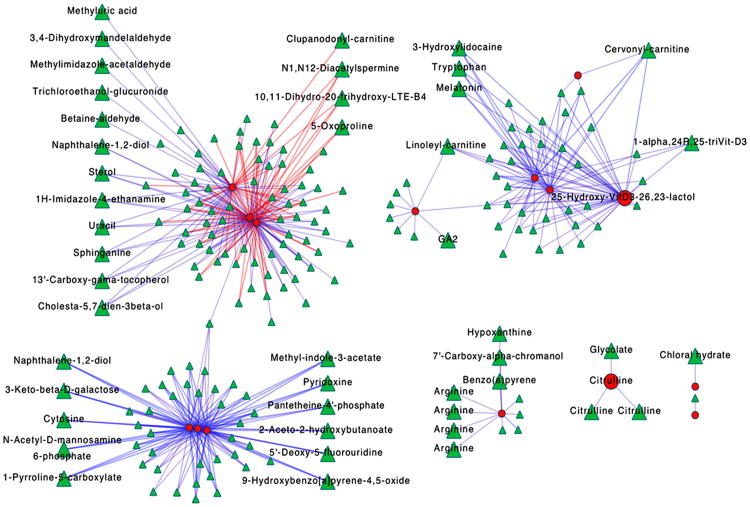
As an alternative way to look at metabolic effects associated with tobacco use, network associations for PLS-DA discriminatory metabolites in Figure 4 were plotted (Fig 5). In total, 225 m/z features from the raw metabolomics data correlated with 15 PLS-DA identified metabolites at the Spearman rank correlation threshold |r| ≥ 0.5 and FDR ≤ 20%. Unlike the pathway enrichment analysis, these clusters include all 225 features regardless of annotation. A large number of the m/z features provided no matches to the KEGG database using common adducts for positive ESI and a 10 ppm error threshold, and annotations of network correlation features included additional amino acids, xenobiotics, vitamins and cofactors that were not captured in the pathway enrichment analysis. Surprisingly, these network structures did not recapitulate the pathway associations, but rather showed three larger hubs connecting metabolites from different pathways, and separate smaller networks associated with urea formation (Fig 5). The top left cluster included lipid metabolites, secondary amino acid metabolites, xenobiotics, urate and a polyamine. The lower left cluster included xenobiotics, enzyme cofactors, sugar derivatives and secondary amino acid metabolites. These two major hubs were linked by one non-annotated metabolite. The major cluster on the upper right is centered on a vitamin D metabolite and included acylcarnitines and tryptophan. Together, the network patterns suggest that dominant associations are influenced by factors other than metabolic pathway-specific effects, such as could occur from miRNA, cytokine or other central transcriptional regulators, epigenetic mechanisms or cellular or organ system responses.
Figure 5.
Network correlation analysis. 225 m/z features (green triangles) were correlated with 15 metabolites (red circles) selected using PLS-DA (blue edges; r ≥ 0.5, red edges; r ≤ 0.5). Annotation of network correlation features included amino acids, xenobiotics, vitamins and cofactors.
Discussion
Prior metabolomics research in smokers used whole blood and identified 21 smoking-related metabolites (32). Of these, 19 were found to be reversible in their concentrations in former smokers suggesting a reduced risk of smoking-related diseases after stopping smoking (32). More recently, Wen et al. showed that low bilirubin in serum was associated with higher risk of lung cancer and death in male smokers and suggested that serum bilirubin level can be used as biomarker to improve risk stratification in smokers (33). Direct comparison of intensities for the m/z matching bilirubin did not differ (p=0.58) between those classified as tobacco users and non-users and was not pursued (data not shown).
The results of the current study present a global view of the metabolic effects of tobacco use and lead to the conclusion that effects are broad and complex. The results for nicotine, cotinine and hydroxycotinine are consistent with previous toxicokinetics studies showing that nicotine from smoking is mostly converted to cotinine and hydroxycotinine. The results are also consistent with low levels of nicotine being present within the population due to consumption of common foods containing nicotine. At low levels, tissue distribution determines rates of conversion of nicotine to cotinine, consistent with the relatively poor correlation between the two.
Significant pathway enrichment for acylcarnitine and glycerophospholipid metabolism is consistent with known perturbations associated with smoking. Cigarette smoke contains particulates with PAH, which are associated with dyslipidemia (34) and fatty liver (35). These associations, along with the enrichment for xenobiotic pathways, are in agreement with the literature on the toxicity characteristics of cigarette smoking (36). Similarly, arginine, purine and pyrimidine pathway effects are consistent with known inflammation and tissue repair responses to smoking (37, 38).
In contrast to enriched metabolic pathways, however, network associations provide a very different and somewhat unexpected result. Instead of correlations according to biochemically defined pathways, the network associations appear to reflect a very different organizational structure than defined by traditional metabolic models. While some uncertainty exists for pathway associations because of incomplete metabolite identification, many of the metabolite identities have been confirmed in previous studies (17). For instance, arginine, tryptophan and other amino acids have been confirmed; acylcarnitines have been confirmed; sphinganine, urate and enzyme cofactors have been confirmed (17). PAH metabolites have been linked to authentic PAH measures by significance in metabolic correlation analyses (39). Thus, the overall evidence supports the conclusion that the central metabolic network structures that discriminate tobacco users from non-users is complex and not simply linked to biochemically defined metabolic pathways.
Limitations of the study include lack of tobacco use history to evaluate source of cotinine. For instance, elevated blood levels of cotinine can be derived from nicotine patches or other smoking cessation products. Additionally, pharmacokinetics for nicotine and cotinine differ among individuals, which could affect the comparability of data among individuals. This effect could account for the lack of complete separation of tobacco users and non-users in the hierarchical cluster analysis (Fig 3). Finally, the cotinine threshold (10 ng/mL) is a cutoff for a heavy smoker, so some individuals considered “light” smokers are likely to have been classified as non-tobacco users by this criterion. Thus, the analysis as performed does not allow conclusions concerning effects of “light” tobacco use on metabolism.
In summary, the present study shows that classification of personnel as tobacco users according to plasma cotinine levels >10 ng/mL allowed study of metabolic pathway and network analysis of 80 users compared to 320 non-users. Results showed metabolic differences previously associated with PAH and/or Cd exposure, as well as differences in lipid and xenobiotic metabolism and diverse effects on amino acid, sialic acid and purine and pyrimidine metabolism. Importantly, network analysis showed associations not simply linked to these well-defined pathways, supporting the interpretation that metabolic effects of tobacco use are also determined by integrative regulatory mechanisms, such as involving miRNA, cytokines, transcriptional regulation, epigenetic mechanisms and changes in cell populations.
Conclusions
The results show complex metabolic effects associated with tobacco use and indicate that metabolic effects of tobacco use will need to be considered in evaluation of deployment-associated environmental exposures in military personnel. For instance, the carnitine pathway changes associated with tobacco use are also found in association with cadmium exposure (40) and in association with PAH levels in the blood [Walker et al. this volume, (39)]. Several metabolic pathways changed with tobacco use, including arginine and proline metabolism, aspartate and asparagine metabolism, and uria cycle/amino group metabolism are also altered association with strenuous physical exercise condition (41). These complex patterns require that appropriate design and biostatistical analyses be performed to distinguish exposure-related effects from those caused by tobacco use and other behavioral factors.
Supplementary Material
Supplemental Table 1: The data includes cotinine levels in serum samples measured by HRM for the 400 pre-deployment individuals.
Supplemental Table 2: The top 41 discriminatory HRM-identified m/z features/metabolites were determined from PLS-DA.
Supplemental Table 3: The list of m/z features, annotations and pathway associations.
Acknowledgments
This public health surveillance project was supported by funding from the Department of Defense award (306889-1.00-64239), and National Institute of Health (award R01 ES023485 and P30 ES019776).
Footnotes
Conflicts of interest: None to declare
References
- 1.Jha P, Ramasundarahettige C, Landsman V, et al. 21st-century hazards of smoking and benefits of cessation in the United States. N Engl J Med. 2013;368:341–350. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- 2.Defense Do, State U. 2011 Executive Summary of Health Related Behaviors Survey of Active Duty Military Personnel. 2013 [Google Scholar]
- 3.Gomez Raposo C, De Castro Carpeno J, Gonzalez Baron M. Causes of lung cancer: smoking, environmental tobacco smoke exposure, occupational and environmental exposures and genetic predisposition. Med Clin (Barc) 2007;128:390–396. doi: 10.1157/13099973. [DOI] [PubMed] [Google Scholar]
- 4.Pallasaho P, Kainu A, Sovijarvi A, Lindqvist A, Piirila PL. Combined effect of smoking and occupational exposure to dusts, gases or fumes on the incidence of COPD. Copd. 2014;11:88–95. doi: 10.3109/15412555.2013.830095. [DOI] [PubMed] [Google Scholar]
- 5.Hecht SS, Hochalter JB, Villalta PW, Murphy SE. 2′-Hydroxylation of nicotine by cytochrome P450 2A6 and human liver microsomes: formation of a lung carcinogen precursor. Proc Natl Acad Sci U S A. 2000;97:12493–12497. doi: 10.1073/pnas.220207697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang LZ, Winzer-Serhan UH. Chronic neonatal nicotine upregulates heteromeric nicotinic acetylcholine receptor binding without change in subunit mRNA expression. Brain Res. 2006;1113:94–109. doi: 10.1016/j.brainres.2006.06.084. [DOI] [PubMed] [Google Scholar]
- 7.Hockertz S, Emmendorffer A, Scherer G, et al. Acute effects of smoking and high experimental exposure to environmental tobacco smoke (ETS) on the immune system. Cell Biol Toxicol. 1994;10:177–190. doi: 10.1007/BF00757561. [DOI] [PubMed] [Google Scholar]
- 8.Lee WK, Ramanathan M, Jr, Spannhake EW, Lane AP. The cigarette smoke component acrolein inhibits expression of the innate immune components IL-8 and human beta-defensin 2 by sinonasal epithelial cells. Am J Rhinol. 2007;21:658–663. doi: 10.2500/ajr.2007.21.3094. [DOI] [PubMed] [Google Scholar]
- 9.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 10.Lewis GP, Coughlin LL, Jusko WJ, Hartz S. Contribution of cigarette smoking to cadmium accumulation in man. Lancet. 1972;1:291–292. doi: 10.1016/s0140-6736(72)90294-2. [DOI] [PubMed] [Google Scholar]
- 11.Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 12.Domino EF, Hornbach E, Demana T. The nicotine content of common vegetables. N Engl J Med. 1993;329:437. doi: 10.1056/NEJM199308053290619. [DOI] [PubMed] [Google Scholar]
- 13.Connor Gorber S, Schofield-Hurwitz S, Hardt J, Levasseur G, Tremblay M. The accuracy of self-reported smoking: a systematic review of the relationship between self-reported and cotinine-assessed smoking status. Nicotine Tob Res. 2009;11:12–24. doi: 10.1093/ntr/ntn010. [DOI] [PubMed] [Google Scholar]
- 14.Shakleya DM, Huestis MA. Simultaneous and sensitive measurement of nicotine, cotinine, trans-3′-hydroxycotinine and norcotinine in human plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3537–3542. doi: 10.1016/j.jchromb.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallon TM. Introduction to Department of Defense research on burn pits, biomarkers and health outcomes related to deployment in Iraq and Afghanistan. Journal of Occupational and Environmental Medicine. 2016;(1) doi: 10.1097/JOM.0000000000000775. [DOI] [PubMed] [Google Scholar]
- 16.Perdue CL, Cost AA, Rubertone MV, Lindler LE, Ludwig SL. Description and utilization of the United States department of defense serum repository: a review of published studies, 1985-2012. PLoS One. 2015;10:e0114857. doi: 10.1371/journal.pone.0114857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Go YM, Walker DI, Liang Y, et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol Sci. 2015;148:531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park YH, Lee K, Soltow QA, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295:47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9:S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu T, Park Y, Li S, Jones DP. Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J Proteome Res. 2013;12:1419–1427. doi: 10.1021/pr301053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uppal K, Soltow QA, Strobel FH, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 23.Go YM, Liang Y, Uppal K, et al. Metabolic Characterization of the Common Marmoset (Callithrix jacchus) PLoS One. 2015;10:e0142916. doi: 10.1371/journal.pone.0142916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd B, Basic C, Bethem R. In: Trace quantitative analysis by mass spectrometry. Hoboken NJ, editor. Chichester, England: John Wiley & Sons; 2008. p. 724. [Google Scholar]
- 26.Wold S, Sjostrom M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometrics and intelligent laboratory systems. 2001;58:109–130. [Google Scholar]
- 27.Le Cao KA, Gonzalez I, Dejean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uppal K, Soltow QA, Promislow DE, Wachtman LM, Quyyumi AA, Jones DP. MetabNet: An R Package for Metabolic Association Analysis of High-Resolution Metabolomics Data. Front Bioeng Biotechnol. 2015;3:87. doi: 10.3389/fbioe.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 30.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with cytoscape 3. Curr Protoc Bioinformatics. 2014;47:8 13 11–18 13 24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Services USDoHaH, Prevention CfDCa, Promotion NCfCDPaH, Health OoSa. The Health Consequences of Smoking-50 years of Progress A Report of the Surgeon General. Atlanta: 2014. [Google Scholar]
- 32.Xu T, Holzapfel C, Dong X, et al. Effects of smoking and smoking cessation on human serum metabolite profile: results from the KORA cohort study. BMC Med. 2013;11:60. doi: 10.1186/1741-7015-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen CP, Zhang F, Liang D, et al. The ability of bilirubin in identifying smokers with higher risk of lung cancer: a large cohort study in conjunction with global metabolomic profiling. Clin Cancer Res. 2015;21:193–200. doi: 10.1158/1078-0432.CCR-14-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranjbar M, Rotondi MA, Ardern CI, Kuk JL. Urinary Biomarkers of Polycyclic Aromatic Hydrocarbons Are Associated with Cardiometabolic Health Risk. PLoS One. 2015;10:e0137536. doi: 10.1371/journal.pone.0137536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Go YM, Sutliff RL, Chandler JD, et al. Low-Dose Cadmium Causes Metabolic and Genetic Dysregulation Associated With Fatty Liver Disease in Mice. Toxicol Sci. 2015;147:524–534. doi: 10.1093/toxsci/kfv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright WR, Parzych K, Crawford D, Mein C, Mitchell JA, Paul-Clark MJ. Inflammatory transcriptome profiling of human monocytes exposed acutely to cigarette smoke. PLoS One. 2012;7:e30120. doi: 10.1371/journal.pone.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amato M, Frigerio B, Castelnuovo S, et al. Effects of smoking regular or light cigarettes on brachial artery flow-mediated dilation. Atherosclerosis. 2013;228:153–160. doi: 10.1016/j.atherosclerosis.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 38.Speina E, Zielinska M, Barbin A, et al. Decreased repair activities of 1,N(6)-ethenoadenine and 3,N(4)-ethenocytosine in lung adenocarcinoma patients. Cancer Res. 2003;63:4351–4357. [PubMed] [Google Scholar]
- 39.Walker DI. Pilot metabolome-wide association study of benzo(a)pyrene in serum from military personnel. Journal of Occupational and Environmental Medicine. 2016;(1) doi: 10.1097/JOM.0000000000000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Go YM, Roede JR, Orr M, Liang Y, Jones DP. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of cd toxicity. Toxicol Sci. 2014;139:59–73. doi: 10.1093/toxsci/kfu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JH, Banton SA, Awad M, et al. Training-related metabolic adaptations in American-style football participants. Annals of sports medicine and research. 2015;2:1048–1054. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: The data includes cotinine levels in serum samples measured by HRM for the 400 pre-deployment individuals.
Supplemental Table 2: The top 41 discriminatory HRM-identified m/z features/metabolites were determined from PLS-DA.
Supplemental Table 3: The list of m/z features, annotations and pathway associations.