Abstract
The 2014 i2b2/UTHealth Natural Language Processing (NLP) shared task featured a new longitudinal corpus of 1,304 records representing 296 diabetic patients. The corpus contains three cohorts: patients who have a diagnosis of coronary artery disease (CAD) in their first record, and continue to have it in subsequent records; patients who do not have a diagnosis of CAD in the first record, but develop it by the last record; patients who do not have a diagnosis of CAD in any record. This paper details the process used to select records for this corpus and provides an overview of novel research uses for this corpus. This corpus is the only annotated corpus of longitudinal clinical narratives currently available for research to the general research community.
Graphical abstract
1. Introduction
The 2014 i2b21/UTHealth2 Natural Language Processing (NLP) shared task featured four tracks: 1) de-identification of medical records, 2) identifying risk factors for coronary artery disease (CAD) in diabetic patients over time, 3) assessment of software usability, and 4) novel uses of the i2b2/UTHealth data set. Of these, tracks 1, 2, and 4 relied on a new corpus and annotations created for that year's task.
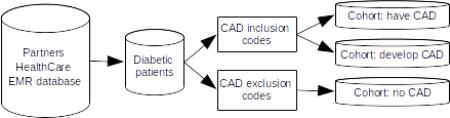
The goal of Track 2, the Risk Factor (“RF”) track was to look at CAD risk factors in patients over time. Accordingly, the corpus for this task included longitudinal data: multiple records for each patient, separated in time, that when put together would show changes in the patient's health status. In order to provide comparison points for patients with different medical histories, we chose for this corpus to represent three different diabetic patient cohorts. The first cohort contains patients who have a diagnosis of CAD in their first record, and continue to have it in subsequent records. The second cohort contains patients who do not have a diagnosis of CAD in the first record, but get a diagnosis of CAD by the last record. The third cohort contains patients who do not have a diagnosis of CAD in the first record and do not get it over the course of their records.
The 2014 i2b2/UTHealth NLP shared task tracks and annotations are all very different, but they all have the corpus in common. In this paper, we compare the 2014 corpus to other biomedical corpora and shared task data sets (Section 2), explain the source of the corpus and the process we used to select the records (Section 3), and examine the corpus in terms of data relating to represented groups (Section 4). This paper also discusses novel uses of this corpus outside of the tracks defined by the i2b2/UTHealth shared task organizers (Section 5), and closes with a discussion of our observations on the corpus-building process.
2. Related work
Shared tasks in the biomedical field have existed for over two decades: the OHSUMED corpus was used for an interactive task in 1994 (Hersh et al., 1994), the KDD Challenge Cup was held in 2002 (Yeh et al., 2002) and the TREC Genomics tracks started in 2003. However, the data for these shared tasks relied on abstracts from MEDLINE and similar sources, not clinical notes from medical facilities (Hersh and Vorhees, 2008). Laws pertaining to patient privacy make releasing clinical notes difficult, and as a result there are relatively few datasets of these notes available to researchers who are not affiliated with medical facilities (Chapman et al., 2011).
One of the most widely-used collections of clinical notes is the Mimic II Clinical Database (Clifford et al., 2012), a collection of medical records, including nursing notes and discharge summaries, gathered over 7 years from ICUs in the Beth Israel Deaconess Medical Center. These de-identified records have been used as a source for a variety of NLP shared tasks, such as the ShARe/CLEF eHealth Evaluation Labs 2013 (Suominen et al., 2013), and 2014 (Kelly et al., 2014).
Other notable collections of medical records include the THYME corpus, a collection of over 1,200 de-identified notes from the Mayo Clinic, representing patients from the oncology department, specifically those with brain or colon cancer (Styler et al., 2014); a recently created corpus of 3,503 de-identified medical records of 22 different types, including discharge summaries, progress notes, and referrals (Deleger et al., 2014); and TREC Medical Records corpora (Vorhees and Hersh, 2012).
The i2b2 NLP shared tasks have existed since 2006, with the first challenges in de-identification (Uzuner et al., 2007) and smoking status classification (Uzuner et al., 2008). Other i2b2 challenges included identifying obesity and its comorbidities (Uzuner 2009), extracting medications and associated information (Uzuner et al., 2010), identifying concepts, assertions, and relations (Uzuner et al., 2011), coreference (Uzuner et al., 2012), and temporal relations (Sun et al., 2013). Each of these shared tasks included a corpus of clinical narratives, each annotated in accordance with the task description and split into training and test sets; some of these corpora reuse documents from previous years. These corpora are available from http://i2b2.org/NLP/DataSets with a data use agreement.
The 2014 i2b2/UTHealth corpus is unique among the existing corpora in that it consists entirely of longitudinal clinical records (as opposed to scientific papers and abstracts) that represent a particular medical population. The longitudinal nature of the data provides an interesting challenge for de-identification, and allows us to glean information about changes to patients over time. As part of the de-identification process, we generated realistic surrogates that maintained the narrative nature of the clinical records, the temporal relationships between dates in the patients' timelines, and co-references between locations (Stubbs et al., 2015). Other corpora use generic placeholders for identifiable information, which can change the narrative structure, or swap the information between patients, breaking the continuity of the records. The i2b2/UTHealth corpus avoids these problems and is therefore suited for both de-identification work and medical research.
Institutional review boards of MIT, Partners Healthcare, and SUNY Albany approved the collection, annotation, and distribution of this corpus. The 2014 i2b2/UTHealth corpus will be available in November 2015 at https://i2b2.org/NLP/DataSets with a data use agreement.
3. i2b2/UTHealth 2014 corpus selection
The medical records for this corpus came from the Partners HealthCare Electronic Medical Records (EMR). EMR at Partners HealthCare comprises a platform shared by two large academic tertiary hospitals – Massachusetts General Hospital (MGH) and Brigham and Women's Hospital (BWH). First, we created a datamart of 314,000 possible Type II Diabetes (T2D) patients using highly sensitive and lenient criteria based on codified data such as the T2D ICD-9-CM billing codes (250.xx), T2D medications, abnormal glycated hemoglobin or plasma glucose. This datamart is thus enriched for patients in whom diabetes is suspected, but the majority of patients in fact do not have T2D.
To identify the actual cases of T2D in this population, an expert provided terms related to T2D. The cTAKES NLP platform (Savova et al., 2010) analyzed the narrative notes for the presence of these terms, including diabetes medications, complications (e.g. retinopathy, dialysis) and mentions of the term “diabetes mellitus”. This narrative data was used, along with the codified data, by a logistic regression with LASSO penalty to develop a T2D classification algorithm. Algorithm development and validation used a training set of 400 random patients (180 T2D cases by physician gold standard, manual chart review), and a test set of 200 patients (170 T2D cases). This method successfully identified a cohort of 65,099 T2D patients at 0.97 specificity and positive predictive value (PPV) 0.96.
The smaller and validated set of patients (n=65,099) greatly narrowed the search space, but identifying patients who fit our cohorts and finding records that met our criteria was still a challenge. In order to further reduce the number of eligible records, we searched for all of the following criteria to identify patients who have or develop CAD, applied to the patient's entire medical history:
at least 3 CAD codes or 1 procedure code for a coronary revascularization
at least 4 codified mentions of beta-adrenergic inhibitor medications
at least 4 codified mentions of anti-platelet agents (such as aspirin)
at least 4 codified mentions of statins (cholesterol lowering drugs)
These criteria returned 6,382 patients, and helped us to ensure that the selected patients would have information in their records relevant to the RF task. We used similar restrictions for the patients without CAD:
no CAD procedure
no CAD codes at all
at least 4 beta blockers
at least 4 codes of anti-platelet (aspirin)
at least 4 statins
Final section of patients and notes for the NLP challenge required manual review of the records, and selection of 2-5 records per patient based on the following:
Each record had to be longer than 300 words,
The notes needed to contain information about different aspects of the patient's health related to CAD risk factors and diabetes.
By the last record, the patient's medical status should be different than in the first (i.e., different medications, more risk factors present), and these changes should occur over the course of the intervening records.
One of the authors examined the records (VK) for suitability for the NLP challenge, and estimates that for every five records he selected for the corpus, he had to review and reject 80-100 records. In total, it took approximately 120 hours to select the records of 301 patients for the 2014 corpus. Of those, we later removed five patients from the corpus as their file formats were incompatible with our de-identification systems.
4. 2014 i2b2/UTHealth shared task corpus
In total, the 2014 i2b2/UTHealth shared task corpus contains 1,304 medical records describing 296 patients. The records are a mixture of discharge summaries and correspondences between medical professionals. For training and testing purposes, we used a 60/40 split: 790 records in the training set and 514 records in the test set. The entire corpus contains 805,124 whitespace-separated tokens, an average of 617.4 tokens per record. Each cohort (those with CAD, those who develop CAD, or those without CAD) is equally represented in the training and test sets.
The records in this corpus reflect a variety of different tones and styles. One of the represented styles in this corpus are letters between medical professionals, which take a familiar tone to report on updates to the patient. For example, a common record of this type might be “Dear [Dr]: I had the pleasure of seeing [patient] today for a follow up visit. I last saw him in November of last year. As you know, [patient] is a 72 year old man...” This record tone provides background information on the patient, as well as details on the patient's current health and latest check-up. There are roughly 150 records of this type in the corpus.
The majority of remaining records in the corpus are discharge summaries and notes written by the doctor after a patient visit. Most of these records discuss the patient in terms similar to those found in the correspondences. For example: “The patient is a 40-year-old woman with history of coronary disease and has had intermittent chest pain over the past 4 days...” These records express information about the patient in complete sentences, usually with minimal jargon and abbreviations. However, some of these summary records employ a much more terse tone, with short, incomplete sentences, and heavy use of abbreviations. For example, “72M with DM on oral agents who noticed DOE x2 weeks [...] at NH, EKG with q-waves V1-4. The tone of the record often correlates with certain characteristics of the content: records written with complete sentences generally have additional information about the patient's medical and life history, while terse records lack that extra context and focus on the medically pertinent facts at hand, probably because the basic necessary context for those records comes from the structured portions (excluded from this corpus) of those records.
For example, of the 1304 records in the corpus, 330 contain no information on the age of the patient at the time of the visit. For 153 of the patients with records missing ages, the age at those visits can be inferred (with a small margin of error) by examining the dates and ages in other records for that patient. However, 12 of the patients in the corpus have no age stated in any of their records: for 17 of the patients, ages listed in different records are not internally consistent. These gaps and inconsistencies in the records reflect a duality present in the clinical narratives: while on the one hand, the narratives contain information about the patient that would not be captured by ICD codes or databases (e.g., smoking status and family history of disease), on the other hand the narratives are not necessarily accurate in all particulars, and can contain errors that may be difficult to identify and correct.
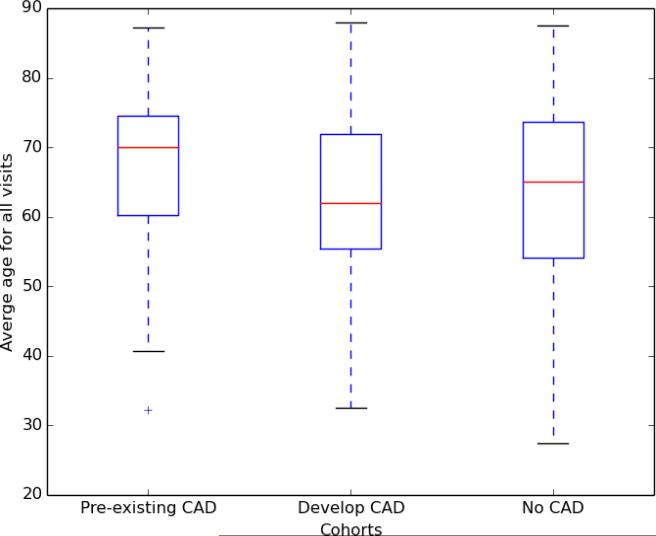
Figure 1 shows the average age of each of the cohorts, over all visits per patient (excluding the 12 patients whose ages could not be inferred). Overall, the median ages for all cohorts are above 60: pre-existing CAD = 70 (standard deviation = 10.67; Q1 = 60.29; Q3 = 74.53); develop CAD = 62.75 (standard deviation = 11.05; Q1 = 55.50; Q3 = 71.88); no CAD = 65 (standard deviation = 12.47; Q1 = 54.00; Q3 = 73.63). All of the cohorts have a maximum possible age of 89, as HIPAA prohibits releasing records with ages over 90 (45 CFR 164.514).
Figure 1.
Average ages of patients at visit times by cohort
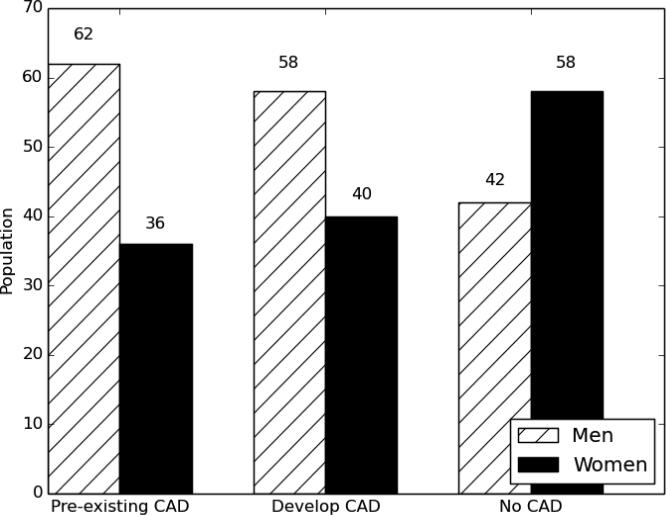
Similar to information on ages, information on patient genders is not always present in the clinical narratives. A manual review shows that 24 of the records in our corpus do not contain any information about the patient's gender. However, given the longitudinal nature of the corpus, it is sufficient for each patient to have just one record that indicates his or her gender. Again, the records employing a terse style are less likely to use pronouns or other indications of gender. Figure 2 shows the gender distributions between cohorts. These numbers show that women are a majority in the cohort that does not develop CAD but a minority in the pre-existing CAD and developing CAD cohorts. This is in line with recent statistics on heart disease, which indicate that men are at greater risk for heart disease than women (Mozaffarian et al., 2015).
Figure 2.
Population by group and gender
The gaps in knowledge in the 2014 i2b2/UTHealth corpus reflect the reality that clinical narratives, while excellent sources of information that would not be found in a database, are nevertheless imperfect. This observation should be taken into account when using any corpus of clinical narratives.
5. Track 4: Novel data use
Each record in this corpus was annotated both for Protected Health Information (PHI), for use in the de-identification track (Stubbs and Uzuner, this issue), and for CAD-related risk factors (Stubbs and Uzuner, this issue), for use in the Risk Factor track. However, the data sets and annotations created for our shared tasks are often later put to other uses for NLP research. For example, past i2b2 shared task corpora have been used for modeling relationships between medications and symptoms (Ling et al., 2014); syntactic parsing (Fan et al., 2013); and assertion classification in phenotype extraction (Bejan et al., 2013). These unsolicited uses of the past shared task data inspired Track 4, the novel data use track, of the i2b2/UTHealth 2014 shared task. Track 4, accordingly, allowed the shared task participants to use the data for any of their research projects and to share with us their task information and results. We received 5 submissions for this track, spanning a wide variety of topics. The systems submitted to this track are summarized below.
Grouin (2014) focused on the identification of medication side effects, including both positive and negative reactions. They annotated 1,014 of the records from the corpus for two types of information: section headers (i.e., “Past Medical History”, “Assessment”) and side effects (and their corresponding medications). They then built a series of systems to test against their human-annotated records: for a baseline they used a rule-based system; the test systems were all based on conditional random fields (CRFs) with different configurations. Overall, their best f-measure for identifying side effects was .480, with a system that used a combination of CRFs and the rule-based system.
Ling et al. (2014) used the annotations provided for the risk factors track of the i2b2/UTHealth shared task to create visualizations. They represented each patient as a vector of the 39 attributes identified in the risk factors track annotations, and created a matrix of all the patients and the number of times each attribute was mentioned in each patient's set of records. The authors then applied non-negative matrix factorization (NMF) to create clusters of patients and their risk factors. The resulting clusters are visualized in a unique and interesting way.
Jonnagaddala et al. (this issue) also used the risk factors annotations, but this time to calculate the Framingham Risk Score (Wilson et al., 1998), which is used to assess the risk of coronary heart disease (CHD) in patients. The authors modified their risk factors system developed for Track 2 (Chang et al., 2014) to extract the factors used to calculate the risk scores (i.e., age, sex, total cholesterol, HDL, blood pressure, diabetes history and smoking history). The authors used spot-checks to determine that their system's output was generally reliable, but they found that many sets of patient records did not include all the information necessary to calculate the risk scores. In the end, 98 patients had sufficient information for calculating risk scores, out of the 297 included in the data set.
Shivade et al. (this issue) examined the problem of identifying patients who are qualified to participate in a clinical trial based on the eligibility criteria for that trial. The authors created an annotated dataset that labeled sentences in medical records which identified whether a particular criterion was met or not. Once they had a gold standard data set, they used four automated methods to identify the relevant sentences and documents. Of the two lexical and two semantic methods implemented, the system that used Unified Medical Language System (UMLS) similarity matching to identify relevant sentences performed the best.
Solomon and Nielsen (this issue) used the risk factors track annotations to train a linear regression that predicts changes in systolic blood pressure over time. The authors identified patients whose longitudinal records contained at least two blood pressure measurements, and used the data from the most recent note as the measurement that the system attempted to predict. Using a collection of features, including features that took into account the time-sensitive nature of the measurements, the authors trained a logistic regression to predict whether the final measurement would be unchanged, go down by 10, or go up by 10. The final system selected the correct change 49.4% of the time, an improvement over the baseline accuracy of 36.1%. Two other novel data uses were published after the end of the shared task: Chen et al. (2014) used the corpus for developing a system to recognize section headers in clinical texts, and Zweigenbaum and Grouin (2014) reformatted these medical records to match a global standard.
Overall, the novel uses of the data represent a variety of tasks and are a testimony to the applicability of the 2014 i2b2/UTHealth data to tasks outside of those originally intended by the creators of the data set.
6. Discussion
The 2014 i2b2/UTHealth NLP shared task corpus provides snapshots in time of diabetic patients and their progress, with particular attention to CAD-related health concerns. The records included in the corpus were selected to provide maximal amounts of information about the patients, their family histories, and other relevant medical facts. The corpus provides ample information about these concerns; in addition, it reflects various styles of clinical narratives and the different tones used by medical practitioners. Some narratives styles of records eschew what might appear to be basic information about the patient, such as age and gender. This represents the true nature of clinical narratives in general.
7. Conclusions
The 2014 corpus is a new and novel resource that, given the information it contains, can support longitudinal studies of diabetes and CAD. To the best of our knowledge, this is the first annotated longitudinal corpus that represents medically-defined patient cohorts and is available for research. This corpus provides sufficient information to support not only an NLP task for identifying CAD-related risk factors, but also a de-identification shared task.
Evidence from the 2014 and earlier shared tasks indicate that the i2b2 corpora can also support some research unrelated to their original intent. As a result, we expect that the medical records gathered, de-identified, and distributed by i2b2 will remain useful to the community even in the absence of any new annotations generated by i2b2.
Highlights.
New corpus of 1,304 de-identified medical records
Longitudinal records represent 296 patients
Three patient cohorts: with Coronary Artery Disease (CAD), no CAD, develop CAD
Overview of novel uses for this corpus
Acknowledgements
This project was funded by NIH NLM 2U54LM008748 PI: Isaac Kohane, by NIH NLM 5R13LM011411 PI: Ozlem Uzuner, and by NIH NIGMS 5R01GM102282, PI: Hua Xu.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Informatics for Integrating Biology and the Bedside
University of Texas Health Science Center at Houston
Conflict of Interest Statement
This statement affirms that Vishesh Kumar, Amber Stubbs, Stanley Shaw, Ozlem Uzuner, the authors of the paper “Creation of a new longitudinal corpus of clinical narratives” do not have any conflicts of interest relating to this paper or its results.
Works Cited
- Hersh William, Buckley Chris, Leone TJ, Hickam David. OHSUMED: an interactive retrieval evaluation and new large test collection for research. In: Bruce Croft W, van Rijsbergen CJ, editors. Proceedings of the 17th annual international ACM SIGIR conference on Research and development in information retrieval (SIGIR '94) Springer-Verlag New York, Inc.; New York, NY, USA: 1994. pp. 192–201. [Google Scholar]
- Yeh Alexander, Hirschman Lynette, Morgan Alexander. Background and overview for KDD Cup 2002 task 1: information extraction from biomedical articles. SIGKDD Explor. Newsl. 2002 2002 Dec;4(2):87–89. DOI=10.1145/772862.772873 http://doi.acm.org/10.1145/772862.772873. [Google Scholar]
- Hersh William, Voorhees Ellen. TREC genomics special issue overview. Information Retrieval. 2008;12:1–15. [Google Scholar]
- Chapman WW, Nadkarni PM, Hirschman L, D'Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. Journal of the American Medical Informatics Association. 2011;18(5):540–543. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford GD, Scott DJ, Villarroel M. User Guide and Documentation for the MIMIC II Database 2012, database version 2.6. available online: https://mimic.physionet.org/UserGuide/UserGuide.html.
- Suominen Hanna, Salanterä Sanna, Velupillai Sumithra, Chapman Wendy W., Savova Guergana, Elhadad Noemie, Pradhan Sameer, South Brett R., Mowery Danielle L., Jones Gareth J. F., Leveling Johannes, Kelly Liadh, Goeuriot Lorraine, Martinez David, Zuccon Guido. Forner Pamela, Müller Henning, Paredes Roberto, Rosso Paolo, Stein Benno., editors. Overview of the ShARe/CLEF eHealth Evaluation Lab 2013. Chapter in Information Access Evaluation. Multilinguality, Multimodality, and Visualization. 2013:212–231. Volume 8138 of the series Lecture Notes in Computer Science. [Google Scholar]
- Kelly Liadh, Goeuriot Lorraine, Suominen Hanna, Schreck Tobias, Leroy Gondy, Mowery Danielle L., Velupillai Sumithra, Chapman Wendy W., Martinez David, Zuccon Guido, Palotti João, Toms Elaine. Kanoulas Evangelos, Lupu Mihai, Clough Paul, Sanderson Mark, Hall Mark, Hanbury Allan., editors. Overview of the ShARe/CLEF eHealth Evaluation Lab 2014. Chapter in Information Access Evaluation. Multilinguality, Multimodality, and Interaction. 2014:172–191. Volume 8685 of the series Lecture Notes in Computer Science. [Google Scholar]
- Styler William, Bethard Steven, Finan Sean, Palmer Martha, Pradhan Sameer, de Groen Piet, Erickson Brad, Miller Timothy, Chen Lin, Savova Guergana K., Pustejovsky James. Temporal annotations in the clinical domain. Transactions of the Association for Computational Linguistics. 2014 [PMC free article] [PubMed] [Google Scholar]
- Deleger L, Lingren T, Ni Y, Kaiser M, Stoutenborough L, Marsolo K, Kouril M, Molnar K, Solti I. Preparing an annotated gold standard corpus to share with extramural investigators for de-identification research. 2014. J Biomed Inform. 2014 Aug;50:173–83. doi: 10.1016/j.jbi.2014.01.014. doi: 10.1016/j.jbi.2014.01.014. Epub 2014 Feb 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees Ellen M., Hersh William. The Twenty-First Text REtrieval Conference (TREC 2012) Proceedings. NIST Special Publication; Overview of the TREC 2012 Medical Records Track. SP 500-298. http://trec.nist.gov/pubs/trec21/t21.proceedings.html. [Google Scholar]
- Uzuner Ö, Luo Y, P Szolovits. Evaluating the State-of-the-Art in Automatic De-identification. Journal of the Medical Informatics Association. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. doi:10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Özlem, Goldstein Ira, Luo Yuan, Kohane Isaac. Identifying patient smoking status from medical discharge records. Journal of the American Medical Informatics Association. 2008;15:14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö. Focus on i2b2 Obesity NLP Challenge: Viewpoint Paper: Recognizing Obesity and Comorbidities in Sparse Data. Journal of the Medical Informatics Association. 2009;16:4, 561–570. doi: 10.1197/jamia.M3115. doi:10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö , Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010;17:514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. Journal of the American Medical Informatics Association. 2012;19(5):786–791. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Rumshisky A, Uzuner O. Evaluating Temporal Relations in Clinical Text: 2012 i2b2 Challenge. J Am Med Inform Assoc. 2013 2013 Sep-Oct;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A, Uzuner Ö, Kotfila C, Goldstein I, Szolovitz P. Challenges in Synthesizing Replacements for PHI in Narrative EMRs. In: Gkoulalas-Divanis Aris, Loukides Grigorios., editors. Chapter in Medical Data Privacy Handbook. Springer. Anticipated publication; 2015. [Google Scholar]
- Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biber Douglas. Representativeness in Corpus Design. Literary and Linguistic Computing. 1993;8:243–257. [Google Scholar]
- Ferraro Jeffrey P, Daumé Hal, III, DuVall Scott L, Chapman Wendy W, Harkema Henk, Haug Peter J. Improving performance of natural language processing part-of-speech tagging on clinical narratives through domain adaptation. J Am Med Inform Assoc. Sep. 2013;20(5):931–939. doi: 10.1136/amiajnl-2012-001453. doi: 10.1136/amiajnl-2012-001453 PMCID: PMC3756264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs Amber, Uzuner Ozlem. Annotating Risk Factors for Heart Disease in Clinical Narratives for Diabetic Patients. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2015.05.009. Supplement: 2014 i2b2 Natural Language Processing Challenge in Clinical Data. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian Dariush, Benjamin Emelia J., Go Alan S., Arnett Donna K., Blaha Michael J., Cushman Mary, de Ferranti Sarah, Després Jean-Pierre, Fullerton Heather J., Howard Virginia J., Huffman Mark D., Judd Suzanne E., Kissela Brett M., Lackland Daniel T., Lichtman Judith H., Lisabeth Lynda D., Liu Simin, Mackey Rachel H., Matchar David B., McGuire Darren K., Mohler Emile R., Moy Claudia S., Muntner Paul, Mussolino Michael E., Nasir Khurram, Neumar Robert W., Nichol Graham, Palaniappan Latha, Pandey Dilip K., Reeves Mathew J., Rodriguez Carlos J., Sorlie Paul D., Stein Joel, Towfighi Amytis, Turan Tanya N., Virani Salim S., Willey Joshua Z., Woo Daniel, Yeh Robert W., Turner Melanie B. Heart Disease and Stroke Statistics—2015 Update: A Report From the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. Published online before print December 17, 2014, doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- Amber tubbs, Uzuner Ozlem. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. J Biomed Inform. 2015 Aug 28;:S1532–0464(15)00182-3. doi: 10.1016/j.jbi.2015.07.020. doi: 10.1016/j.jbi.2015.07.020. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, An Y, Hu X. Bioinformatics and Biomedicine (BIBM), 2014 IEEE International Conference on. IEEE; Nov, 2014. A matching framework for modeling symptom and medication relationships from clinical notes. pp. 515–520. [Google Scholar]
- Fan JW, Yang EW, Jiang M, Prasad R, Loomis RM, Zisook DS, Denny JC, Xu H, Huang Y. Syntactic parsing of clinical text: guideline and corpus development with handling ill-formed sentences. J Am Med Inform Assoc. 2013 Nov-Dec;20(6):1168–77. doi: 10.1136/amiajnl-2013-001810. doi: 10.1136/amiajnl-2013-001810. Epub 2013 Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejan CA, Vanderwende L, Xia F, Yetisgen-Yildiz M. Assertion modeling and its role in clinical phenotype identification. J Biomed Inform. 2013 Feb;46(1):68–74. doi: 10.1016/j.jbi.2012.09.001. doi: 10.1016/j.jbi.2012.09.001. Epub 2012 Sep 21. [DOI] [PubMed] [Google Scholar]
- Grouin Cyril. Presentation at the Seventh i2b2 Shared Task and Workshop: Challenges in Natural Language Processing for Clinical Data. Washington DC.: Nov 14, 2014. Identification of medication side effects in clinical records: an experiment based on the 2014 i2b2/UTHealth corpus. [Google Scholar]
- Ling Yuan, Jiang Xingpeng, An Yuan, Hu Xiaohua. Presentation at the Seventh i2b2 Shared Task and Workshop: Challenges in Natural Language Processing for Clinical Data. Washington DC.: Nov 14, 2014. Data Exploration and Visualization of Risk Factors for Heart Disease from Medical Documents Using Non-Negative Matrix Factorization (NMF). [Google Scholar]
- Jonnagaddala Jitendra, Liaw Siaw-Teng, Ray Pradeep, Kumar Manish, Chang Nai-Wen, Dai Hong-Jie. Coronary artery disease risk assessment from unstructured electronic health records using text mining. Journal of Biomedical Informatics. 2015 doi: 10.1016/j.jbi.2015.08.003. Supplement: 2014 i2b2 Natural Language Processing Challenge in Clinical Data. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivade Chaitanya, Hebert Courtney, Lopetegui Marcelo, de Marneffe Marie-Catherine, Fosler-Lussier Eric, Lai Albert M. Textual Inference for Eligibility Criteria Resolution in Clinical Trials. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2015.09.008. Supplement: 2014 i2b2 Natural Language Processing Challenge in Clinical Data. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes Solomon John, Nielsen Rodney. Predicting Changes in Systolic Blood Pressure Using Longitudinal Patient Records. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2015.06.024. Supplement: 2014 i2b2 Natural Language Processing Challenge in Clinical Data. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Chih-Wei, Chang Nai-Wen, Chang Yung-Chun, Dai Hong-Jie. Lecture Notes in Computer Science Volume 8916. Springer; 2014. Section Heading Recognition in Electronic Health Records Using Conditional Random Fields. Technologies and Applications of Artificial Intelligence. pp. 47–55. [Google Scholar]
- Zweigenbaum P, Grouin C. Reformatting Clinical Records Based on Global Layout Statistics.. Proc of SMBM, 2014; Aveiro, Portugal. October 6th-7th, 2014. [Google Scholar]