Abstract
The 2014 i2b2/UTHealth natural language processing shared task featured a track focused on the de-identification of longitudinal medical records. For this track, we de-identified a set of 1,304 longitudinal medical records describing 296 patients. This corpus was de-identified under a broad interpretation of the HIPAA guidelines using double-annotation followed by arbitration, rounds of sanity checking, and proof reading. The average token-based F1 measure for the annotators compared to the gold standard was 0.927. The resulting annotations were used both to de-identify the data and to set the gold standard for the de-identification track of the 2014 i2b2/UTHealth shared task. All annotated private health information were replaced with realistic surrogates automatically and then read over and corrected manually. The resulting corpus is the first of its kind made available for de-identification research. This corpus was first used for the 2014 i2b2/UTHealth shared task, during which the systems achieved a mean F-measure of 0.872 and a maximum F-measure of 0.964 using entity-based micro-averaged evaluations.
Graphical abstract
1. Introduction
Clinical narratives (i.e., free text records of patients’ health and medical history) provide information to researchers that cannot be found in structured medical records, such as family history, reasoning behind prescribed treatments, and details of the patient’s health. These clinical narratives are therefore an important resource for medical applications such as decision support (Demner-Fushman et al., 2009; Wagholikar et al., 2012) and cohort selection (Carroll et al., 2012; Weng et al; 2011). However, clinical narratives also contain information that identifies patients, such as their names, home addresses, and phone numbers. The Health Insurance Portability Accountability Act (HIPAA) requires that all information that identifies a patient be removed from these records before sharing the records outside of the clinical setting in which they were produced. The process of determining and removing patient-identifying information from medical records is called de-identification, also called anonymization. Often, removal of the patient-identifying information requires replacements with realistic placeholders, which we refer to as surrogates, also called pseudonyms. The replacement process is called surrogate generation.
HIPAA refers to patient-identifying information as Protected Health Information (PHI), and defines 18 categories of PHI as they relate to “the [patients] or of relatives, employers, or household members of the [patients]” (45 CFR 164.514). These categories are shown in Table 1.
Table 1.
18 HIPAA PHI categories (45 CFR 164.514)
|
|
The 2014 Informatics for Integrating Biology and the Bedside (i2b2) and the University of Texas Health Science Center at Houston (UTHealth) natural language processing (NLP) shared task featured a track focused on the de-identification of longitudinal medical records. Longitudinal medical records represent multiple time points in the care of a patient, making references to past records as appropriate; their de-identification needs to pay attention to indirect identifiers that can collectively reveal the identities of the patients, even when none of those indirect identifiers would be sufficient to reveal the identity of the patient on their own. For example, the description of a patient’s injuries as “resulting from Superstorm Sandy” would not be covered under the HIPAA guidelines, but they indirectly provide both a location and a year for that medical record. This information, paired with other hints about the patient’s identity, such as profession and number of children, could lead to the patient’s identity.
However, there are some rewards to mitigate the increased risks. Automated systems can take advantage of the repeated information: a name identified in one record as PHI can be searched for in other records in order to boost accuracy. Additionally, longitudinal records contain significantly more medical information about a patient, and they allow researchers to study a patient’s health over time. We selected the 2014 de-identification corpus in order to support research into the progression of Cardiac Artery Disease in diabetic patients, a different track for the 2014 i2b2/UTHealth shared task (Stubbs et al, this issue 2015b).
In addition to paying attention to the longitudinal aspects of the corpus, the preparation of the corpus for the shared task was guided by the following goals:
Given the intended widespread distribution of the corpus, we needed to apply a risk-averse interpretation of the HIPAA guidelines,
Given the intended use of the corpus for automatic system development, we needed to maintain the semantics and integrity of the data so that systems developed on these data could be useful on authentic data,
We needed to have sufficient representation of PHI categories, both in type and in quantity, so that machine learning based systems could learn automatically from available samples,
We needed to have granular PHI categories to maximize the usability of the data for research on any subsets of the PHI, and
We needed to replace the authentic PHI with realistic surrogates to maintain readability.
Given our goals, we developed annotation guidelines which we applied to the 2014 i2b2/UTHealth shared task corpus for manual de-identification of 1,304 longitudinal clinical narratives, generating gold standard annotations that researchers can use for automatic de-identification system development. We replaced the authentic PHI with realistic surrogates using a combination of automated systems and hand curation.
This paper describes the manual de-identification and the automatic surrogate generation processes applied to the 2014 de-identification shared task data, as well as the annotation guidelines generated for this shared task. Institutional review boards of MIT, Partners HealthCare, and SUNY Albany approved this study, and Partners HealthCare approved the de-identification methods described in Section 5.
2. Related work
Due to the strict regulations surrounding the release of medical records, very few clinical narrative data sets are currently available for de-identification research. The 2006 i2b2 NLP shared task had a de-identification track, and the corpus consisted of 889 hospital discharge summaries, which in total contained 19,498 PHI (Uzuner et al., 2007). This corpus is available on i2b2.org/NLP for researchers who sign a data use agreement (DUA). PhysioNet (Goldberger et al., 2000) includes a de-identification dataset created by Neamatullah et al., (2008), which is available at http://www.physionet.org/physiotools/deid/ with appropriate log ins and a DUA. The PhysioNet dataset contains 2,434 nursing notes and 1,779 instances of PHI.
Deleger et al., (2014) recently created a corpus of 3,503 de-identified medical records of 22 different types, including discharge summaries, progress notes, and referrals. In all, their corpus contains 30,815 instances of PHI and is available upon request.
All three of the above corpora, and the 2014 i2b2/UTHealth corpus described here, have the PHI replaced with realistic surrogates, making them suitable for NLP research into automated de-identification. All of the corpora follow HIPAA guidelines as a base for the PHI annotations, the annotations generally only have minor differences. For example, the corpus from Delager et al. (2014) conflates patient and doctor names into a single “name” category, while the other corpora maintain a distinction between patients and doctors. However, the 2014 i2b2/UTHealth de-identification corpus described in this paper is the only one that provides longitudinal data for patients, and it includes additional PHI categories, which we describe in Section 4.
Research into the annotation process for PHI has led to some interesting findings. South et al. (2014) performed an experiment to determine if pre-annotating a corpus using automated de-identification software had a substantial effect on the quality of the PHI annotations or the time it took human annotators to check the PHI when compared to their performance on un-annotated documents. They found that the pre-annotations did not, in fact, improve inter-annotator agreement or significantly decrease the amount of time that it took the annotators to complete the task.
Additionally, in a preliminary study to the de-identification process described in this paper, we performed an experiment to determine whether PHI annotation is more accurate when done in parallel (i.e., two annotators working separately on each document) or in series (one annotator reviews the document, then the second reviews the first one’s work and checks for un-annotated PHI). We found that the annotation process used had no effect on the quality of the annotations (Stubbs and Uzuner, 2014).
3. Corpus
The corpus selected for this project consisted of the longitudinal records for 301 patients, with 2–5 records selected per patient. The records came from Partners Healthcare, and were selected for use in Track 2 of the i2b2/UTHealth shared task: identification of risk factors for Cardiac Artery Disease in diabetic patients (Stubbs et al., this issue, 2015b). The records for each patient represent a snapshot of the patient’s health at different points in time, not the patient’s full medical history. The complete details of the corpus selection process are described in Kumar et al. (this issue). Over the course of the de-identification annotation, we had to remove some records from the corpus as they were incompatible with our surrogate generation software due to improper character encodings. The final corpus contains sets of longitudinal records for 296 patients, a total of 1,304 individual records, with 805,118 whitespace-separated tokens; an average of 617.4 tokens per file.
4. Annotation guidelines
Our annotation guidelines were guided by our project goals (see Introduction). Because we intended to release this corpus to a wide audience, we adhered to a risk-averse interpretation of the HIPAA guidelines. Essentially, we expanded the definition of category 18 to include other information that is indirectly related to patients and that could be used, either on their own or in combination, to identify patients. These indirect identifiers include information about hospitals, doctors and nurses, and patient’s professions. We also annotated all parts of dates, including years, as well as all locations, including states and countries.
Another PHI category guided by our risk-aversion was ages. HIPAA only considers ages above 89 to be PHI; however, we included all ages in our de-identification annotation guidelines. We used this approach for two reasons. First, in the case of a person’s age being over 90, all the other ages in the other records needed to be adjusted accordingly, so that the person’s age could not be calculated from information in other documents. Annotating all the ages allowed us to easily modify all the ages in a person’s records. This approach did make minor changes to the medical accuracy, protecting patient identities was more important. Second, carrying out this more expansive de-identification also remedied any issues we would have faced with respect to sample sizes of age PHI. Our data contains very few mentions of ages over 89 in the medical records. Marking all ages rather than only those above 89 allowed us to create a larger volume of age PHI which could serve as training data for systems that would try to address automatic de-identification.
Risk aversion was a key consideration in our annotation guidelines. We also kept in mind that ultimately this corpus would be used for NLP research, and therefore needed to accurately represent real clinical records. Therefore, the PHI in this corpus needed to preserve their semantics through the de-identification and surrogate generation processes. This goal led us to define PHI categories that were very fine-grained. Figure 1 shows the categories and sub-categories of PHI in our annotation guidelines.
Figure 1.
Categories and sub-categories in the i2b2 de-identification annotation (a version of this figure also appears in Stubbs and Uzuner, 2014a)
The fine-grained categories addressed two additional goals: First, we could be expansive in marking any potential PHI, as long as we maintained the semantics of the type of PHI, and adjusted the scope of the task as necessary down the road. For example, for the purposes of scrubbing the data, we wanted to adhere to our risk-averse strategy and mark all potential PHI; however, for the de-identification shared task evaluation, we could select a subset of the PHI and focus on categories that HIPAA cared about. Second, other users of the corpus down the road could focus on specific PHI categories for their research given their de-identification goals. For example, as Figure 1 shows, for the NAME category, we maintained the distinctions between PATIENT and DOCTOR names because some de-identification projects do not include the names of the hospital workers in PHI. Here, DOCTOR is used as an umbrella term for all hospital staff, including nurses, pharmacists, receptionists, and so on.
A third goal of the fine-grained PHI categories related to the surrogate generation process. By enabling us to gather some PHI together for uniform treatment while maintaining their fine-grained categories, we were able to simplify the surrogate generation process. For example, we were able to gather subsets of HIPAA categories 4–17 under CONTACTs and IDs. Most of these HIPAA categories are simply alphanumeric strings and are treated similarly for surrogate generation. The availability of fine-grained PHI categories enables for this step to be carried out efficiently, simplifying the surrogate generation process without any loss of semantics.
Similarly, fine-grained categories of LOCATIONs allowed us to maintain semantic details about the authentic PHI so we could more easily generate appropriate surrogates. For example, rather than give “Uganda”, “New York”, “Seattle”, “23 Fruit Street”, and “the East Coast” all the same label of LOCATION, we marked each one with a sub-category of LOCATION, specifically LOCATION:COUNTRY, LOCATION:STATE, LOCATION:CITY, LOCATION:STREET or LOCATION:OTHER. These fine-grained categories made it straightforward for surrogate generation to replace each PHI with a surrogate of the same semantic type.
We used the OTHER category (as opposed to LOCATION:OTHER) as a catch-all for information that could not be classified as any other PHI, but that could still potentially provide information about the patient, such as “is excited to see the Red Sox play a home game in the World Series next week”. The guidelines for the 2014 de-identification shared task reflected these goals. The full guidelines, including lists of generic department names and specific instructions regarding what temporal expressions and parts of phrases to annotate, can be found in Appendix A.
5. Annotation procedure
5.1 De-identification
We applied the 2014 de-identification shared task guidelines to longitudinal medical records of 301 patients. Each patient’s longitudinal records in our corpus were annotated for PHI by two independent annotators working in parallel. In total, we had 6 annotators, and we randomly assigned each set of patient records to two different annotators. Following that, we used multiple checks in order to ensure no PHI could be leaked, as we describe below. Delager et al. (2014) also used a “double annotation” method; Neamatullah et al. (2008) used three independent annotators, and Uzuner et al. (2006) used serial annotation. During the annotation process, the authors performed a study to determine if annotation in parallel or serial worked better for capturing all PHI in a record; the results of this study showed that neither method was more effective than the other (Stubbs and Uzuner, 2015).
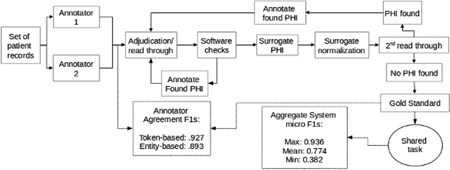
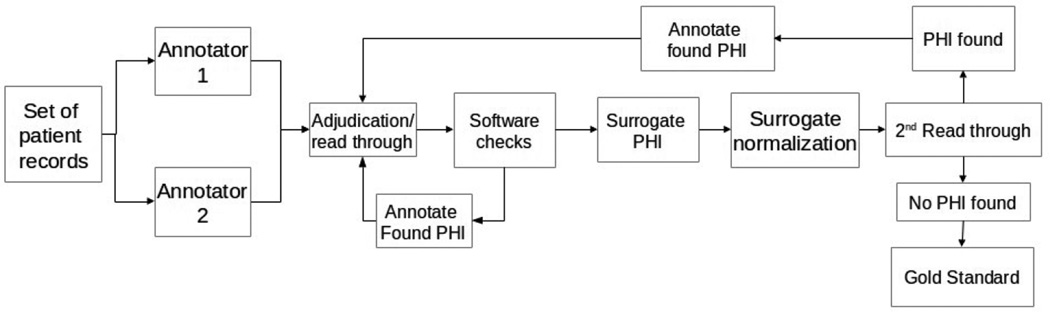
For annotation software, we used the Multi-purpose Annotation Environment (MAE; Stubbs, 2011). Figure 2 shows the entire annotation pipeline.
Figure 2.
Annotation pipeline for de-identification
For the purposes of de-identification annotation, we merged all the records for each patient into a single file. This allowed the annotators to use a new “annotate all” feature in MAE that enabled them to automatically carry forward any existing annotations to the rest of the file, e.g., marking “Kai Yamamoto” as DOCTOR and then applying “annotate all” automatically marked all occurrences of “Kai Yamamoto” as DOCTOR. This feature greatly sped up annotations, as PATIENT and DOCTOR names and DATEs are often repeated multiple times across patient records.
After double annotation, one of the authors (AS) adjudicated the discrepancies between the annotators, and also read the records for any missed PHI. We used the Multi-document Adjudication Interface (MAI) for adjudication (Stubbs, 2011). MAI loads the files of both annotators, and shows where they agree and disagree. This display allows the adjudicator to easily resolve conflicts, and also to check unannotated text for missed PHI. The output of MAI is a file with the gold standard annotations.
After adjudication, we used a script to check the records for PHI text that was annotated in one part of the records but not in another. AS added to the data any new annotations that resulted from these checks, and re-ran the script until no further missed PHI were found. Then we proceeded to surrogate generation.
5.2 Surrogate generation and final PHI check
Before we could make the medical records available to 2014 shared task participants, we needed to obfuscate all the PHI by replacing them with realistic surrogates. We built automatic surrogate generation software to perform the replacement. A full description of the surrogate generation process and its complexities are found in Stubbs et al. (anticipated 2015); we summarize the process in the remainder of this section.
Treating all of a patient’s records as a single file made it easy for us to maintain continuity between records during surrogate generation, so that all names in a patient’s longitudinal medical record were replaced consistently with the same surrogate, the dates were all offset by the same amount and intervals were preserved, etc.
We generated surrogate DATEs through date-shifting. For each document, we shifted all of the DATEs forward by the same random number of years, months, and days.
For NAMEs, for each new document we first randomly mapped each letter in the alphabet to another letter, and pre-decided that all NAMEs starting with, for example, letter A would be replaced by a surrogate that started with G as a way of simplifying initial generation for NAMEs. Then, we paid attention to maintain gender information and to replace NAMEs with NAMEs of the appropriate gender by selecting from lists generated from census data. For example, assuming a mapping from A to G, and from F to D, “Angie Ferrerro” became “Grace Dollard”. In order to preserve coreference information, all occurrences of an authentic NAME were replaced by the same surrogate. The surrogates mimic the surface form of the authentic PHI, mapping “A. Ferrerro”, “Mrs Ferrerro”, “Angie” to “G. Dollard”, “Mrs Dollard” and “Grace”, respectively.
We followed the same procedure for generating surrogates for LOCATIONs and PROFESSIONs, although without the alphabetic mappings. We used a pre-compiled list of surrogates, from which we selected as appropriate, while preserving coreference information.
We handled hospital DEPARTMENTs differently from other LOCATIONs: instead of replacing them with random surrogate department names, we matched specific department names to more generic ones. For example, the “Department for radiology, imaging, and oncology” became “Radiology”. If such a generic name did not exist, we changed the DEPARTMENT to “Internal Medicine”.
We left AGEs unchanged, unless they were 90 or over, in which case they were changed to “90”. In these cases, the ages in the other records were adjusted down in order to keep continuity between records.
We modified all numbers, including PHONEs, FAXes, and all sub-categories of IDs by randomly selecting new strings of digits/letters of the same length and format.
Any other PHI, such as EMAILs, URLs, and the OTHER category were initially replaced by strings of random characters; the adjudicator then modified those as necessary to make them more realistic.
The adjudicator modified two other types of surrogates by hand: ambiguous dates such as 02/03, which could potentially be February 3rd, March 2nd, or February of 2003; and nicknames, or misspellings of NAMEs and all other PHI.
After surrogate generation, the author AS again read through each patient’s set of longitudinal records and ensured that all PHI were changed. She also normalized surrogate PHI, for example, by checking that the appropriate article (“a” or “an”) appeared in front of a surrogate, and modifying the surrogates as necessary (e.g., “Cambodian” might be replaced by the surrogate “China”, which the adjudicator changed to “Chinese” to match the surrounding text). AS also re-created misspellings and other errors in the surrogate data: for example, if a patient named “Marissa” was identified as “Marrisa” in a different record, the surrogates in the de-identified data might have been changed to “Sara” and “Sarrah”.
Finally, we performed an additional read-through of the text for any missed PHI. This step was shared between one of the authors (OU) and two medical professionals (one MD and one medical assistant).
If authentic PHI were found at any stage, we went back to the file that contained the authentic PHI, annotated the missed PHI there, and started the surrogate generation process over. Only after all these steps were complete did we deem the files ready for release.
In order to illustrate the surrogate generation process, Figure 2 shows an example of a fabricated clinical narrative before and after surrogate generation, using a simplified XML representation for readability. Here we show the PHI annotations in-line to simplify the presentation. The “before” segment is based on a segment of de-identified text with surrogate PHI from the i2b2/UTHealth corpus. The “after” segment shows a second run of surrogate generation on the same text, using the “before” segment as input. As Figure 2 shows, surrogate generation maintained the relationships between the DATEs in the file and preserved coreference so that all occurrences of a PHI were replaced by the same surrogate. Surrogate generation provided random numbers for the ID (marked by IDNUM), MEDICAL RECORD NUMBER (marked by MEDICALRECORD), and the numerical part of the USERNAME while still keeping consistent initials.
Before data release for the 2014 shared task, we unmarked some of the PHI categories, i.e., ROOM, DEPARTMENT, OTHER, from the gold standard. These PHI were included in our de-identification process as an extra precaution against PHI leak; however, ROOMs proved to be rare in the corpus, and the DEPARTMENTs were all made generic as such they would be easy to identify. Similarly, everything labeled as OTHER was re-written to not contain anything identifiable, making it a useless tag for a de-identification challenge. Therefore, we decided not to include those three tags in the de-identification shared task that would be addressed by the 2014 participants.
Finally, after generating the surrogate PHI we performed a final read through of the records one more time to look for any missed PHI. Found PHI would be annotated in the original medical records, and we would repeat the software check, surrogate generation, and read-throughs until we were confident that all PHI were removed.
5.3 Annotator backgrounds Five MIT undergraduates and one MIT senior researcher provided the double annotations for this project. None of the annotators have medical training, though the senior research is a member of the Clinical Decision Making Group. Prior to gaining access to the data, the annotators underwent training and obtained certification in how to treat documents with authentic PHI. Additionally, all the annotation was done remotely over secure connections to Partners servers; at no point did the data containing authentic PHI leave the Partners’ network. Prior to this project, none of the undergraduate annotators had experience de-identifying medical records.
Author AS acted as the adjudicator. AS is also certified for access to medical records, though prior to this project had not de-identified medical records. The final read-through was performed by author OU, who has worked on de-identification projects before (Uzuner et al., 2007; Uzuner, 2009), and two medical professionals with no prior de-identification experience.
5.4 Annotation time
In total, the annotators annotated 602 longitudinal medical records (double annotation for 301 patients) in approximately 310 hours. This amounts to an average of around 30 minutes per set of patient records. Adjudication took two months of part time effort.
The final read-through, took place after the individual records were split apart, and took an average of 3 minutes per record (roughly 15 minutes per set of patient records).
6. Annotation quality
In order to ensure the removal of all PHI from the medical records, we implemented multiple checks, including both human and software, as shown in Figure 2. The initial annotations captured the majority of PHI, as we show by comparing the annotations to the gold standard prior to surrogate generation.
We measured agreement using precision (Eq. 1), recall (Eq. 2), and F-measure (F1) (Eq. 3) at the macro and micro levels. For macro evaluation, the scores for each document are calculated and then averaged across the corpus. For mirco evaluation, the scores for each annotation is calculated across the corpus as a whole. We determined inter-annotator agreement (IAA) by comparing both sets of annotators to the adjudicated gold standard and averaging the results. We used two methods to generate agreement scores. First, we calculated an entity-based (sometimes referred to as “instance-based” or “instance-level” (Uzuner et al., 2007)) inter-annotation agreement, which looks at whether the system output matches the gold standard in PHI sub-category, and the start and end attributes for every PHI entity identified in the gold standard.
| Eq. 1 |
| Eq. 2 |
| Eq. 3 |
We also calculated IAA using a token-based evaluation, which looks at the tag associated with each individual whitespace-separated token rather than PHI entities in the corpus. The token-based measurements allow for “John” and “Smith” (two separate PHI) to match “John Smith” (a single PHI), if their PHI sub-categories match. The numbers in Table 2 show the entity- and token-based agreement scores at the macro and micro levels.
Table 2.
Micro- and macro-averaged P, R, and F1 for entity- and token-based inter-annotator agreement (IAA)
| Granularity | Micro Precision |
Micro Recall |
Micro F1 | Macro Precision |
Macro Recall |
Macro F1 |
|---|---|---|---|---|---|---|
| Entity-based evaluation | 0.904 | 0.887 | 0.895 | 0.902 | 0.886 | 0.892 |
| Token-based evaluation | 0.939 | 0.920 | 0.930 | 0.939 | 0.921 | 0.928 |
Overall, the agreement levels are quite high. Table 10 in Appendix C shows the IAA scores by PHI category. The categories that proved most difficult were generally the ones that required some knowledge of medical records to label correctly. For example, the ID category by itself has an F1 average of 0.867, but the annotators often had difficulty determining the different between, for example, a BIOID and a MEDICALRECORD. Similarly, low scores in the LOCATION subcategories stem in part from the annotators not being sure if “Springfield Medical Center: Colorado Hospital” should be tagged as ORGANIZATION, DEPARTMENT, HOSPITAL, or a combination of those. Another source of error in LOCATION was DEPARTMENTs: the annotation guidelines (see Appendix A) specified that annotators should mark only “unique” department names, but that if they were unsure they should mark the name anyway. This led to some annotators annotating all of the department names they encountered, which lowered the agreement overall. Finally, many disagreements stemmed from phrases that appeared to be PHI, but were not. For example, a common phrase, “2/2”, was often marked as a DATE. However, “2/2” appears in contexts such as “toe amputation 2/2 diabetes”, where it stands for “secondary to”. Similarly in “patient transferred from OSH”, “OSH” stands for “outside hospital” rather than the abbreviation of a specific hospital name. However, all of these problems were resolved during adjudication.
After adjudication and the read through, the software checks occasionally revealed missed PHI. In many cases, a patient or doctor name might be found in the middle of a paragraph describing medical histories, or a date would appear in the middle of an unformatted table, where it would be easy to miss. Very little PHI made it through the adjudication, scripts, and surrogate generation checks. The second read-through step only revealed six instances of PHI that were missed during the previous steps, and these were all minor PHI, such as “spring” or an age under 90. Even though no major PHI made it through to the second read through, the step did prevent a small number of potential leaks.
7. Distribution of tags in the corpus
Table 3 shows the distribution of different PHI categories in the final version of the corpus, and the split between the training data (790 files) and the testing data (514 files).
Table 3.
PHI distributions in the i2b2/UTHealth 2014 de-identification corpus
| PHI category | # in training data | # in test data | Total # in corpus |
|---|---|---|---|
| NAME: PATIENT | 1316 | 879 | 2195 |
| NAME: DOCTOR | 2885 | 1912 | 4,797 |
| NAME: USERNAME | 264 | 92 | 356 |
| PROFESSION | 234 | 179 | 413 |
| LOCATION: HOSPITAL | 1,437 | 875 | 2,312 |
| LOCATION: ORGANIZATION | 124 | 82 | 206 |
| LOCATION: STREET | 216 | 136 | 352 |
| LOCATION: CITY | 394 | 260 | 654 |
| LOCATION: STATE | 314 | 190 | 504 |
| LOCATION: COUNTRY | 66 | 117 | 183 |
| LOCATION: ZIP CODE | 212 | 140 | 352 |
| LOCATION: OTHER | 4 | 13 | 17 |
| AGE | 1,233 | 764 | 1,997 |
| DATE | 7,507 | 4,980 | 12,487 |
| CONTACT: PHONE | 309 | 215 | 524 |
| CONTACT: FAX | 8 | 2 | 10 |
| CONTACT: EMAIL | 4 | 1 | 5 |
| CONTACT: URL | 2 | 0 | 2 |
| CONTACT: IPADDRESS ID: SSN | 0 0 | 0 0 | 0 0 |
| ID: MEDICAL RECORD | 611 | 422 | 1033 |
| ID: HEALTH PLAN | 1 | 0 | 1 |
| ID: ACCOUNT | 0 | 0 | 0 |
| ID: LICENSE | 0 | 0 | 0 |
| ID: VEHICLE | 0 | 0 | 0 |
| ID: DEVICE | 7 | 8 | 15 |
| ID: BIO ID | 1 | 0 | 1 |
| ID: ID NUMBER | 261 | 195 | 456 |
| Total # of tags | 17,410 | 11,462 | 28,872 |
| Average PHI per file | 22.03 | 22.3 | 22.14 |
Overall, the tags are distributed relatively evenly between the training and testing data. DATEs are the most prevalent form of PHI, with NAMEs and LOCATIONs also appearing very frequently. Some tags did not appear at all: CONTACT: IPADDRESS, ID: SSN, ID: ACCOUNT, ID: LICENSE, and ID: VEHICLE. Some PHI categories included in the annotation guidelines were not included in the final gold standard (LOCATION: ROOM, LOCATION: DEPARTMENT, OTHER), as we described in Section 5.2.
8. 2014 i2b2/UTHealth De-identification Shared Task
We utilized the generated data for the 2014 i2b2/UTHealth de-identification shared task. The full analysis of the different systems and their rankings can be found in Stubbs, Kofila, and Uzuner (this issue); here we present the overview statistics of the system results. We used two subsets of the annotated PHI for the challenge evaluation. One set contained all of the categories shown in Table 3; the other contained only those categories identified by HIPAA. The HIPAA-identified categories are: NAME:PATIENT, AGE, LOCATION:CITY, LOCATION:STREET, LOCATION:ZIP, LOCATION:ORGANIZATION, DATE, CONTACT:PHONE, CONTACT:FAX, CONTACT:EMAIL, ID:SSN, ID:MEDICALRECORD, ID:HEALTHPLAN, ID:ACCOUNT, ID:LICENSE, ID:VEHICLE, ID:DEVICE, ID:BIOID, and ID:IDNUM.
Overall, we received 22 submissions from 10 teams. We calculated the aggregate precision, recall, and f-measure (F1) for all submissions using both entity-based and token-based evaluations. Micro score calculations evaluate all the tags in the corpus as a single set, while macro score calculations evaluate all the tags in each document, then average across the corpus.
Table 4 shows the aggregate statistics for all submitted systems when evaluated on matching all of the PHI categories using the entity-based evaluation metric. The macro scores are comparable to those of the inter-annotator agreement scores in Table 2. The maximum system scores for precision, recall, and F1 are slightly higher than those for IAA by 0.062, 0.030, and 0.047, respectively. This is likely due in part to the fact that the DEPARTMENT tag, which lowered inter-annotator agreement, was not included in the gold standard for the challenge.
Table 4.
Aggregate statistics for entity-based evaluation of all submissions -- all PHI categories
| Minimum | Mean | Median | Maximum | Std. deviation | |
|---|---|---|---|---|---|
| Micro Precision | 0.527 | 0.872 | 0.920 | 0.964 | 0.121 |
| Micro Recall | 0.242 | 0.717 | 0.794 | 0.909 | 0.212 |
| Micro F1 | 0.382 | 0.774 | 0.845 | 0.936 | 0.180 |
| Macro Precision | 0.566 | 0.872 | 0.921 | 0.965 | 0.113 |
| Macro Recall | 0.267 | 0.720 | 0.794 | 0.916 | 0.215 |
| Macro F1 | 0.411 | 0.777 | 0.845 | 0.940 | 0.179 |
Table 5 shows the entity-based evaluation for only the HIPAA-identified categories. The evaluations based only on HIPAA-identified PHI categories are marginally higher than the expanded set of categories we defined. This is possibly because other PHI datasets that participants could have used to train their systems do not contain PHI categories such as PROFESSION and LOCATION:HOSPITAL, which would make machine learning systems less likely to be able to detect them.
Table 5.
Aggregate statistics for entity-based evaluation of all submissions – HIPAA-identified PHI categories only
| Minimum | Mean | Median | Maximum | Std. deviation | |
|---|---|---|---|---|---|
| Micro Precision | 0.538 | 0.887 | 0.932 | 0.976 | 0.121 |
| Micro Recall | 0.135 | 0.752 | 0.816 | 0.939 | 0.222 |
| Micro F1 | 0.235 | 0.800 | 0.863 | 0.957 | 0.196 |
| Macro Precision | 0.602 | 0.879 | 0.927 | 0.975 | 0.116 |
| Macro Recall | 0.143 | 0.752 | 0.813 | 0.942 | 0.222 |
| Macro F1 | 0.235 | 0.801 | 0.860 | 0.958 | 0.193 |
Tables 6 and 7 show the token-based evaluations for all PHI (Table 6) and HIPAA-identified PHI (Table 7).
Table 6.
Aggregate statistics for token-based evaluation of all submissions – all PHI categories
| Minimum | Mean | Median | Maximum | Std. deviation | |
|---|---|---|---|---|---|
| Micro Precision | 0.716 | 0.927 | 0.953 | 0.982 | 0.073 |
| Micro Recall | 0.211 | 0.777 | 0.863 | 0.941 | 0.203 |
| Micro F1 | 0.344 | 0.832 | 0.907 | 0.961 | 0.164 |
| Macro Precision | 0.731 | 0.924 | 0.951 | 0.981 | 0.069 |
| Macro Recall | 0.244 | 0.772 | 0.856 | 0.940 | 0.203 |
| Macro F1 | 0.385 | 0.828 | 0.902 | 0.960 | 0.161 |
Table 7.
Aggregate statistics for token-based evaluation of all submissions -- HIPAA-identified PHI categories
| Minimum | Mean | Median | Maximum | Std. deviation | |
|---|---|---|---|---|---|
| Micro Precision | 0.744 | 0.941 | 0.967 | 0.989 | 0.068 |
| Micro Recall | 0.108 | 0.807 | 0.882 | 0.963 | 0.211 |
| Micro F1 | 0.193 | 0.853 | 0.922 | 0.976 | 0.184 |
| Macro Precision | 0.686 | 0.927 | 0.963 | 0.986 | 0.082 |
| Macro Recall | 0.121 | 0.800 | 0.872 | 0.960 | 0.208 |
| Macro F1 | 0.206 | 0.848 | 0.915 | 0.973 | 0.180 |
Comparing the macro scores in Table 6 to the token-based IAA scores in Table 2, we see that the maximum system performance is again higher than the annotator agreement in precision, recall and f-measure by 0.042, 0.019, and 0.033, respectively, and likely for the same reasons. For both annotators and system scores, the token-based evaluations are higher. Given that token-based evaluations allow for more flexibility in what is considered a “correct” annotation, this boost in scores is expected. Table 7 shows that the highest system evaluation scores are token-based and on only the HIPAA-identified PHI categories.
We performed one other evaluation on the system outputs, which does not have a correlated IAA score. This evaluation is a “relaxed” entity-based evaluation which allowed some leeway for systems to include trailing punctuation in their output. The results for this analysis are included in Appendix B.
9. Conclusions
This paper describes the corpus for the 2014 i2b2/UTHealth NLP shared task in medical record de-identification. This is the first longitudinal set of patient records annotated for de-identification and available for de-identification research. It consists of 1,304 records for 296 patients, and contains over 28,000 PHI. The data were made available for de-identification shared task in May 2014 and will be available online to the rest of the research community in November 2015 at http://i2b2.org/NLP/DataSets with a Data Use Agreement.
Figure 3.
Sample of clinical text before and after surrogate generation using simplified XML representation
Highlights.
De-identification shared task for longitudinal clinical records
Protected Health Information in records replaced with realistic surrogates
First corpus of its kind available for distribution
Used for Track 1 of the 2014 i2b2/UTHealth NLP Shared Task
Acknowledgments
This project was funded by NIH NLM 2U54LM008748, PI: Isaac Kohane, and by NIH NLM 5R13LM011411, PI Ozlem Uzuner. We would like to thank our de-identification annotators and medical professionals: Zachary Franco, Ye Eun Jeong, Bill Long, Kaitlin Mahar, Tony Ping, Kathleen Ririe, and Nick Uhlenhuth. Thanks also to Vishesh Kumar, Shawn Murphy, and Stanley Shaw, who advised us on HIPAA regulations and identifying PHI. We would also like to thank the reviewers, for their insightful comments and questions.
Appendix A: Annotation guidelines
De-Identification annotation task
Amber Stubbs and Ozlem Uzuner
Last updated: November 22, 2013
-------------------------------------
TASK DESCRIPTION
HIPAA requires that patient medical records have all identifying information removed in order to protect patient privacy. There are 18 categories of Protected Health Information (PHI) identifiers of the patient or of relatives, employers, or household members of the patient that must be removed in order for a file to be considered de-identified.
In order to de-identify the records, each file must have the PHI marked up so that it can be removed/replaced later. This will be done using a graphical interface, and all PHI will be given an XML tag indicating its category and type, where applicable.
For the purposes of this annotation project, the 18 categories have been expanded to include more specific identifiers, which have been grouped into 6 larger categories. These are:
NAME
patient
doctor
username
PROFESSION
LOCATION
room
department
hospital
organization
street
city
state
country
ZIP
other
AGE
DATE
CONTACT
telephone
fax
email
URL
IP address
IDs
SSN
record id
health plan/insurance id
account number
certificate/license number
car id
device id
biometric id
other id number
-------------------------------------
ANNOTATION NOTES
General
Only annotate the information that would need to be replaced when the file is re-identified.
When in doubt, annotate!
When tagging something that is PHI but it’s not obvious what to tag it as, think about what it should be replaced by, and whether that will make sense in the document
-
ORGANIZATION tags will be replaced with a company name, like Google. So try not to use that for medical facilities.
PROFESSION tags will be replaced with job names, like “lawyer”
“lawyer at Harvard” should be tagged as “PROFESSION at ORGANIZATION”
DEPARTMENT tags will be replaced with something like “internal medicine”
HOSPITAL tags will be replaced with things that sound like hospitals. Use this if there’s a name of a medical facility and you’re not sure if it should be a HOSPITAL or a DEPARTMENT, you should probably go with HOSPITAL. For example: I would tag “MGH Everett Family Center” as two hospitals: one tag for MGH, the other for “Everett Family Center”; that way MGH is consistently replaced in the document and “Everett Family Center” can be replaced with a different phrase
“Bigelow C” should be tagged as HOSPITAL(Bigelow) ROOM(C)
Names
Annotate initials at end of documents-- even ones that don’t seem to match any names
Titles (Dr., Mr., Ms., etc.) do not have to be annotated.
Information such as “M.D.”, “R.N.” do not have to be annotated
If a name is possessive (e.g., Sam’s) do not annotate the ‘s
-
the USERNAME tag should only be used for names that follow the Partners username standard: initials followed by numbers (i.e., arw4)
In “entered by gsmith”, the “gsmith” should be tagged as a Doctor, not a username
Profession
Any job that is mentioned that is not held by someone on the medical staff should be tagged
Dates
Any calendar date, including years, seasons, months, and holidays, should be annotated
Days of the week should also be annotated
Do not include time of day
If the phrase has ‘s (i.e., “in the ‘90’s), annotate “‘90’s”
Include annotations of seasons (“Fall ‘02”)
Include quote marks that stand in for years (“‘92”)
Locations
Hospital room numbers should be annotated as ROOM
-
Floors and suites can also be annotated as ROOM
“Floor 2, room 254” can all be one ROOM tag
-
Annotate state/country names as well as addresses and cities. Each part of an address will get its own tag. For example:
32 Vassar Street -- Street
Cambridge -- City
MA -- State
02142 -- ZIP
USA -- Country
The departments inside of hospitals should be annotated, but only if they are unique. There is a list of generic hospital units at the end of this file; if a department is not on that list, it should be annotated.
If in doubt, annotate
Generic locations like “hair salon” do not need to be annotated, but named organizations (i.e., “Harvard University”) do
Age
Annotate all ages, not just those over 90, including those for patient’s families if they are mentioned
Contact
Pager numbers should be annotated as phone numbers
IDs
When in doubt, call something a record ID
Doctor or nurse IDs should be annotated as “other id”
No need to label names of devices (for example: “25 mm Carpentier-Edwards magna valve”, “3.5mm by 32mm Taxus drug-eluting stent”, Angioseal”)
-------------------------------------
TASK PROCEDURE
Each file being de-identified will be reviewed by two annotators. Every piece of information that meets the criteria for PHI should be tagged using the appropriate annotation tag, and then the type of PHI should be indicated where appropriate.
-------------------------------------
GENERIC HOSPITAL DEPARTMENTS
Acute assessment unit
Cardiology
Coronary care unit / CCU
Critical care
Ear nose and throat (ENT)
Emergency department / ED
Emergency room / ER
Emergency ward / EW
Gastroenterology
General surgery
Geriatric intensive-care unit
Gynaecology
Haematology
Intensive care unit (ICU)
Internal medicine
Maternity
Medical records department
Neonatal unit
Neonatal intensive care unit (NICU)
Nephrology
Neurology
Obstetrics
Occupational therapy
Oncology
Operating room / OR
Ophthalmology
Orthopaedics
Pediatric intensive care unit (PICU)
Pharmacy
Physical therapy
Post-anesthesia care unit
Psychiatric Unit / Psychiatry
Radiology
Rheumatology
Surgery
Urgent care
Urology
Appendix B: Aggregate results for relaxed entity-based system evaluations
We included a third evaluation for the systems output: a “relaxed” entity-based evaluation in which the beginning offset for each tag had to match the gold standard, but the ending offset could be up to 2 characters larger than the gold standard. This helped take into account systems which may have included spaces or punctuation in their output. Tables 8 and 9 show the evaluation results for all PHI (Table 8) and HIPAA-identified PHI categories (Table 9). However, these results are only marginally better than the standard evaluations, showing that trailing spaces and punctuations did not impact the performance of most systems in a significant way.
Table 8.
Aggregate statistics for relaxed evaluation of all submissions – all PHI categories
| Minimum | Mean | Median | Maximum | Std. deviation | |
|---|---|---|---|---|---|
| Micro Precision | 0.538 | 0.877 | 0.924 | 0.966 | 0.118 |
| Micro Recall | 0.242 | 0.721 | 0.799 | 0.911 | 0.211 |
| Micro F1 | 0.383 | 0.779 | 0.851 | 0.938 | 0.178 |
| Macro Precision | 0.575 | 0.877 | 0.924 | 0.967 | 0.110 |
| Macro Recall | 0.267 | 0.724 | 0.798 | 0.917 | 0.214 |
| Macro F1 | 0.412 | 0.781 | 0.851 | 0.941 | 0.178 |
Table 9.
Aggregate statistics for relaxed evaluation of all submissions -- HIPAA-identified PHI categories only
| Minimum | Mean | Median | Maximum | Std. deviation | |
|---|---|---|---|---|---|
| Micro Precision | 0.544 | 0.893 | 0.936 | 0.978 | 0.120 |
| Micro Recall | 0.136 | 0.757 | 0.824 | 0.941 | 0.221 |
| Micro F1 | 0.236 | 0.805 | 0.870 | 0.959 | 0.195 |
| Macro Precision | 0.606 | 0.884 | 0.931 | 0.977 | 0.114 |
| Macro Recall | 0.143 | 0.756 | 0.820 | 0.944 | 0.221 |
| Macro F1 | 0.236 | 0.805 | 0.867 | 0.960 | 0.192 |
Appendix C: Entity-based micro IAA scores by PHI category
Table 10.
Entity-based micro-averaged IAA by PHI category
| PHI category | Micro precision | Micro Recall | Micro F1 |
|---|---|---|---|
| NAME | 0.971 | 0.944 | 0.957 |
| NAME: PATIENT | 0.958 | 0.934 | 0.946 |
| NAME: DOCTOR | 0.966 | 0.928 | 0.947 |
| NAME: USERNAME | 0.858 | 0.923 | 0.889 |
| PROFESSION | 0.756 | 0.678 | 0.714 |
| LOCATION | 0.838 | 0.862 | 0.850 |
| LOCATION: ROOM | 0.521 | 0.629 | 0.570 |
| LOCATION: DEPARTMENT | 0.320 | 0.497 | 0.389 |
| LOCATION: HOSPITAL | 0.866 | 0.808 | 0.836 |
| LOCATION: ORGANIZATION | 0.406 | 0.520 | 0.454 |
| LOCATION: STREET | 0.938 | 0.930 | 0.934 |
| LOCATION: CITY | 0.943 | 0.933 | 0.938 |
| LOCATION: STATE | 0.961 | 0.966 | 0.964 |
| LOCATION: COUNTRY | 0.822 | 0.774 | 0.797 |
| LOCATION: ZIP CODE | 0.995 | 0.985 | 0.990 |
| LOCATION: OTHER | 0.15035 | 0.41176 | 0.22 |
| AGE | 0.96937 | 0.94101 | 0.95498 |
| DATE | 0.96413 | 0.93437 | 0.94902 |
| CONTACT | 0.95068 | 0.92745 | 0.93891 |
| CONTACT: PHONE | 0.9439 | 0.92857 | 0.93616 |
| CONTACT: FAX | 1.0 | 0.625 | 0.7619 |
| CONTACT: EMAIL | 1.0 | 1.0 | 1.0 |
| CONTACT: URL | 0 | 0 | 0 |
| CONTACT: IPADDRESS | n/a | n/a | n/a |
| ID | 0.8644 | 0.87335 | 0.86882 |
| ID: SSN | n/a | n/a | n/a |
| ID: MEDICAL RECORD | 0.88386 | 0.87354 | 0.87837 |
| ID: HEALTH PLAN | 0.1 | 0.5 | 0.16667 |
| ID: ACCOUNT | n/a | n/a | n/a |
| ID: LICENSE | n/a | n/a | n/a |
| ID: VEHICLE | n/a | n/a | n/a |
| ID: DEVICE | 0.5035 | 0.4 | 0.44505 |
| ID: BIO ID | 0.01667 | 0.5 | 0.03226 |
| ID: ID NUMBER | 0.557 | 0.52922 | 0.54043 |
| OTHER | 0 | 0 | 0 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, Pacheco JA, Boomershine CS, Lasko TA, Xu H, Karlson EW, Perez RG, Gainer VS, Murphy SN, Ruderman EM, Pope RM, Plenge RM, Ngo Kho A, Liao KP, Denny JC. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Informatics Association. 2012 Jun;19(e1):e162–e169. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleger L, Lingren T, Ni Y, Kaiser M, Stoutenborough L, Marsolo K, Kouril M, Molnar K, Solti I. Preparing an annotated gold standard corpus to share with extramural investigators for de-identification research. Journal of Biomedical Informatics. 2014 doi: 10.1016/j.jbi.2014.01.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demner-Fushman D, Chapman WW, McDonald CJ. What can Natural Language Processing do for Clinical Decision Support? Journal of Biomedical Informatics. 2009 Oct;42(5):760–772. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. PhysioBank, PhysioToolkit, and Physionet: Components of a New Research Resource for Complex Physiologic Signals. Circulation. 2000 Jun 13;101(23):e215–e220. doi: 10.1161/01.cir.101.23.e215. [Circulation Electronic Pages; http://circ.ahajournals.org/cgi/content/full/101/23/e215] [DOI] [PubMed] [Google Scholar]
- Kumar V, Stubbs A, Shaw S, Uzuner Ö. Creation of a new longitudinal corpus of clinical narratives. doi: 10.1016/j.jbi.2015.09.018. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neamatullah I, Douglass M, Lehman LH, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD. Automated De-Identification of Free-Text Medical Records. BMC Medical Informatics and Decision Making. 2008;8:32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South BR, Mowery D, Suo Y, Leng J, Ferrandez O, Meystre SM, Chapman WW. Evaluating the effects of machine pre-annotation and an interactive annotation interface on manual de-identification of clinical text. Journal of Biomedical Informations in press. 2014 May; doi: 10.1016/j.jbi.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A. MAE and MAI: Lightweight Annotation and Adjudication Tools; 2011 Proceedings of the Linguistic Annotation Workshop V, Association of Computational Linguistics, Portland, Oregon; 2011. Jul 23–24, [Google Scholar]
- Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. This issue. 2015a doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A, Kotfila C, Uzuner Ö. Identifying risk factors for heart disease over time: Overview of 2014 i2b2/UTHealth shared task Track 2. This issue. 2015b doi: 10.1016/j.jbi.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A, Uzuner Ö. De-identification of Medical Records Through Annotation. In: Nancy Ide and James Pustejovsky, editor. Chapter in Handbook of Linguistic Annotation. Springer; 2015. Submitted, anticipated publication. [Google Scholar]
- Stubbs A, Uzuner Ö, Kotfila C, Goldstein I, Szolovitz P. Challenges in Synthesizing Replacements for PHI in Narrative EMRs. In: Gkoulalas-Divanis Aris, Loukides Grigorios., editors. Chapter in Medical Data Privacy Handbook. Springer; 2015. Anticipated publication. [Google Scholar]
- Uzuner Ö. “Focus on i2b2 Obesity NLP Challenge: Viewpoint Paper: Recognizing Obesity and Comorbidities in Sparse Data”. Journal of the Medical Informatics Association. 2009;16(4):561–570. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö, Luo Y, Szolovits P. “Evaluating the State-of-the-Art in Automatic De-identification”. Journal of the Medical Informatics Association. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagholikar KB, MacLaughlin KL, Henry MR, Greenes RA, Hankey RA, Liu H, Chaudhry R. Clinical decision support with automated text processing for cervical cancer screening. Journal of the American Medical Informatics Association. 2012 doi: 10.1136/amiajnl-2012-000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association. 2011 Dec;18(Suppl 1):i116–i124. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]