Abstract
Hippocampal sclerosis of aging (HS-Aging) is a common brain disease in older adults with a clinical course that is similar to Alzheimer’s disease. Four single-nucleotide polymorphisms (SNPs) have previously shown association with HS-Aging. The present study investigated structural brain changes associated with these SNPs using surface-based analysis. Participants from the Alzheimer’s Disease Neuroimaging Initiative cohort (ADNI; n = 1,239), with both MRI scans and genotype data, were used to assess the association between brain atrophy and previously identified HS-Aging risk SNPs in the following genes: GRN, TMEM106B, ABCC9, and KCNMB2 (minor allele frequency for each is >30%). A fifth SNP (near the ABCC9 gene) was evaluated in post-hoc analysis. The GRN risk SNP (rs5848_T) was associated with a pattern of atrophy in the dorsomedial frontal lobes bilaterally, remarkable since GRN is a risk factor for frontotemporal dementia. The ABCC9 risk SNP (rs704180_A) was associated with multifocal atrophy whereas a SNP (rs7488080_A) nearby (~50 kb upstream) ABCC9 was associated with atrophy in the right entorhinal cortex. Neither TMEM106B (rs1990622_T), KCNMB2 (rs9637454_A), nor any of the non-risk alleles were associated with brain atrophy. When all four previously identified HS-Aging risk SNPs were summed into a polygenic risk score, there was a pattern of associated multifocal brain atrophy in a predominately frontal pattern. We conclude that common SNPs previously linked to HS-Aging pathology were associated with a distinct pattern of anterior cortical atrophy. Genetic variation associated with HS-Aging pathology may represent a non-Alzheimer’s disease contribution to atrophy outside of the hippocampus in older adults.
Keywords: Arteriolosclerosis, dementia, KATP, progranulin, rs5848, rs704180, rs1990622, rs9637454, SUR2, TDP-43
INTRODUCTION
There is accumulating evidence of a common brain disease that mimics Alzheimer’s disease (AD) clinically, and which has been classified using the term “hippocampal sclerosis” based on pathologic observations [1–11]. This disease affects up to 25% of the “oldest-old” [1, 12–14] and is associated with substantial disease-specific cognitive impairment [7–9, 13–17]. We apply the term “hippocampal sclerosis of aging” (HS-Aging) [1], to differentiate this disease from other conditions referred to as “hippocampal sclerosis”, because the latter designation refers to a pathologic phenomenon observed in many different brain disorders [13, 18–20]. HS-Aging is distinguished by the advanced age of the affected individuals, by the usual lack of either seizure disorder or frontotemporal dementia symptoms clinically, and by the presence of hippocampal TDP-43 pathology at autopsy [1, 2, 21–24]. HS-Aging is generally misdiagnosed in live individuals as AD [9, 16]; put another way, a relatively large proportion of what is considered “Alzheimer’s disease” clinically is actually HS-Aging. Despite rapid progress from multiple research centers, much remains to be learned about HS-Aging pathoetiology.
One promising approach for better understanding HS-Aging is through the study of genetics and specifically genotype-phenotype correlations. HS-Aging pathology has been linked with single-nucleotide polymorphisms (SNPs) within or near four different genes: GRN, TMEM106B, ABCC9, and KCNMB2. Two of the putative HS-Aging risk SNPs were previously associated with risk for frontotemporal lobar degeneration (FTLD), namely rs5848 (GRN) and rs1990622 (near TMEM106B) [25–28]. Genome-wide association studies (GWAS) implicated the other two SNPs—rs704180 (ABCC9) and rs9637454 (KCNMB2)—in genes that encode potassium channel regulators [29, 30]. All four SNPs are relatively common, with minor allele frequencies of ~30%–50% in most populations. In terms of the disease phenotype, the study of multiple large cohorts indicated that HS-Aging, although diagnosed at autopsy according to the signal feature of hippocampal sclerosis, also affects brain areas outside of the hippocampus, including the frontal neocortex [31–33].
Collectively, these prior findings indicate that multiple genes can cause or exacerbate a disease (which we refer to currently as HS-Aging) that affects brain regions outside of the hippocampus. More specifically, we hypothesized that HS-Aging risk SNPs in older adults would be associated with brain structural variance detectable with brain neuroimaging, both within and outside the temporal lobe. To test this hypothesis, we evaluated data including magnetic resonance imaging (MRI) and genetic information, from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. These analyses indicated that the HS-Aging risk gene variants are associated with relatively widespread brain atrophy in older adults.
MATERIALS AND METHODS
Subjects
All individuals included in this report were participants in the ADNI cohort and aged 55–90 years at the time of scan. The ADNI initial phase (ADNI-1) was launched in 2003 to test whether serial MRIs, other biological markers, and clinical and neuropsychological assessment could be combined to measure the progression of mild cognitive impairment (MCI) and early AD. The ADNI-1 participants were recruited from 59 sites across the U.S. and Canada and included approximately 200 cognitively normal older individuals (HC), 400 patients diagnosed with MCI, and 200 patients diagnosed with early probable AD aged 55–90 years. ADNI-1 has been extended in subsequent phases (ADNI-GO and ADNI-2) for follow-up of existing participants and additional new enrollments. Inclusion and exclusion criteria, clinical and neuroimaging protocols, and other information about ADNI have been published previously [34] and can be found at http://www.adni-info.org. Demographic information, raw scan data, APOE and whole-genome genotyping data, neuropsychological test scores, and diagnostic information are publicly available from the ADNI data repository (http://www.loni.usc.edu/ADNI/). Written informed consent was obtained at the time of enrollment and/or genetic sample collection and protocols were approved by each participating study and sites’ Institutional Review Board. We only included non-Hispanic Caucasians in order to limit the impact of population stratification on association analysis (removing 140 participants).
Genotyping and imputation
Genotyping was performed using the Illumina Human610-Quad BeadChip for the ADNI-1 participants, and the Illumina HumanOmni Express BeadChip and the Illumina Omni2.5M BeadChip for participants initially enrolled in ADNI-GO or ADNI-2. APOE genotyping was separately obtained using standard methods to yield the APOE ɛ4 allele defining SNPs (rs429358, rs7412). rs5848 (GRN) was genotyped only in the Illumina HumanOmni Express BeadChip and other SNPs (rs704180, rs7488080, rs1990622, and rs9637454) were not genotyped in any genotyping platforms. The un-genotyped SNPs were imputed separately in each phase as the ADNI cohort used different genotyping platforms. Before the imputation, we performed standard sample and SNP quality control procedures as described previously [35]. Furthermore, in order to prevent spurious association due to population stratification, we selected only non-Hispanic Caucasian participants that clustered with HapMap CEU or TSI populations using multidimensional scaling analysis (http://www.hapmap.org). Imputation was performed using MACH and minimac in a two-stage procedure as described previously [36]. The pilot 1 data of the 1000 Genomes Project were used as a reference panels for inferring missing genotypes. Minimac produced the posterior probabilities of the imputed genotypes at un-genotyped marker loci for each individual. In order to assess the quality of imputation, an r2 value equal to 0.30 was imposed as the threshold to accept the imputed genotypes.
Imaging processing
T1-weighted brain MRI scans at baseline were acquired using a sagittal 3D MP-RAGE sequence following the ADNI MRI protocol [37]. As detailed in previous studies [38], a widely employed automated MRI analysis technique was used to process MRI scans: FreeSurfer V5.1 software (http://surfer.nmr.mgh.harvard.edu/). FreeSurfer was used to process and extract brain-wide target MRI imaging phenotypes (region volume and cortical thickness) by automated segmentation and parcellation. The cortical surface was reconstructed to measure thickness at each vertex on the surface. The cortical thickness was calculated by taking the Euclidean distance between the grey/white boundary and the grey/cerebrospinal fluid (CSF) boundary at each vertex on surface. For surface-based comparison of the cortical thickness, all individual cortical surfaces were registered to a common surface template, which was an average created from all cognitively normal control subjects. The cortical thickness was smoothed with 10 mm FWHM Gaussian kernel to improve the signal-to-noise ratio and statistical power. The common surface template is a triangle mesh consisting of 327,684 vertices with a cortical thickness value at each vertex on surface. In order to assess relative size of significant clusters on surface, we identified the number of vertices belonging to the significant clusters in a 100 vertices unit.
Imaging genetics analysis
The SurfStat software package (http://www.math.mcgill.ca/keith/surfstat/) was employed to perform surface-based analysis using a general linear model (GLM) approach. GLMs were developed using age at scan, gender, years of education, intracranial volume (ICV), diagnosis at baseline, MRI field strength, and SNP as independent variables. Following analysis of cortical thickness to examine the association between candidate SNPs and localizable cortical thickness measures, correction for multiple comparisons was performed using the random field theory (RFT) correction method at a 0.05 level of significance.
RESULTS
The main research question was whether candidate SNPs previously associated with HS-Aging pathology are associated with brain atrophy detectable by structural MRI in older adults from the ADNI cohort. Detailed information about the SNPs, stratified by demographics and other parameters, is presented in Table 1. In the 1,239 included participants, the average age at scan was 73.8, average educational level was 15.9, percentage of participants with APOE ɛ4 allele was 46.0%, and percentage of participants that were female was 42.7%. The genes (SNPs) studied here were GRN (rs5848), TMEM106B (rs1990622), ABCC9 (rs704180), and KCNMB2 (rs9637454). The assumed models of mode of inheritance were derived from the published literature [29, 39]. We did not observe any associations with the non-risk alleles of these SNPs at the same statistical threshold (data not shown). For the SNPs showing an association (p < 0.05) with cortical thickness variance (following correction for multiple comparisons), tabular data about localization of atrophy and corrected p-values are presented in Table 2.
Table 1.
Genes and SNPs assessed in the current study, with additional information related to genotype, demographics, clinical diagnosis, and the MRI field strength (putative risk genotype shown in the leftward columns).
| Gene/SNP–> |
GRN/rs5848
|
ABCC9/rs704180
|
ABCC9/rs7488080
|
TMEM106B/rs19906722
|
KCNMB2/rs9637454
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype–> | TT/TC | CC | AA | GG/GA | AA/AG | GG | TT/CT | CC | AA | GG/AG |
| N | 668 | 571 | 335 | 904 | 135 | 1,104 | 1,022 | 217 | 89 | 1,150 |
| Sex (M/F) | 384/284 | 326/245 | 189/146 | 521/383 | 75/60 | 635/469 | 594/428 | 116/101 | 48/41 | 662/488 |
| APOE ɛ4 status (presence/absence) | 291/377 | 284/287 | 170/165 | 405/499 | 63/72 | 512/592 | 472/550 | 103/114 | 46/43 | 529/621 |
| Age (Mean ± SD) | 73.92 ± 7.29 | 73.75 ± 6.97 | 73.72 ± 7.41 | 73.89 ± 7.05 | 73.34 ± 7.47 | 73.90 ± 7.11 | 73.95 ± 7.17 | 73.33 ± 7.02 | 73.62 ± 7.70 | 73.86 ± 7.10 |
| Diagnosis (HC/EMCI/LMCI/AD) | 195/116/245/112 | 151/102/207/111 | 93/66/121/55 | 253/152/331/168 | 34/30/49/22 | 312/188/403/201 | 286/187/364/185 | 60/31/88/38 | 24/17/35/13 | 322/201/417/210 |
| Magnetic field strength (1.5T/3T) | 392/276 | 353/218 | 178/157 | 567/337 | 79/56 | 666/438 | 603/419 | 142/75 | 54/35 | 691/459 |
Table 2.
Gene variants associated with MRI detected brain atrophy: anatomic localization, size, and corrected p values of significant clusters
| Gene (SNP) | Cluster | Anatomic localization | Cluster size* | Corrected p-value |
|---|---|---|---|---|
| GRN (rs5848) | 1 | Left frontal lobe | 33.4 | 4.0 × 10−5 |
| 2 | Right frontal lobe | 19.2 | 0.020 | |
| ABCC9 (rs7488080) | 1 | Right frontal and temporal lobes | 45.9 | 1.91 × 10−5 |
| 2 | Left parietal lobe | 31.8 | 6.42 ×10−4 | |
| 3 | Right temporal lobe including entorhinal cortex | 33.9 | 7.60 × 10−4 | |
| 4 | Left frontal and parietal lobes | 26.7 | 0.013 | |
| 5 | Right temporal lobe | 19.8 | 0.020 | |
| ABCC9 (rs704180) | 1 | Right frontal lobe | 61.6 | 7.13 × 10−8 |
| 2 | Right temporal and occipital lobes | 38.3 | 2.50 × 10−5 | |
| 3 | Left occipital lobe | 19.4 | 0.032 |
See text for calculation method.
None of the four SNPs previously identified to be associated with HS-Aging pathology showed associations with medial temporal lobe atrophy in the ADNI cohort. Two SNPs (rs5848 and rs1990622) previously linked to risk for both FTLD with TDP43-positive inclusions (FTLD-TDP) and HS-Aging showed different results. For GRN/rs5848 (Fig. 1; dominant model), the atrophy significantly associated with the risk allele was localized to the medial dorsal frontal cortex, roughly corresponding to Brodmann Area 9. Participants having at least one risk allele have decreased cortical thickness compared to those with no risk allele. As was the case for all SNPs tested, the non-risk SNPs did not show any significant spatial clusters (data not shown). For TMEM106B/rs1990622 (a dominant model was used), there was no area of MRI-detected atrophy (data not shown).
Fig. 1.
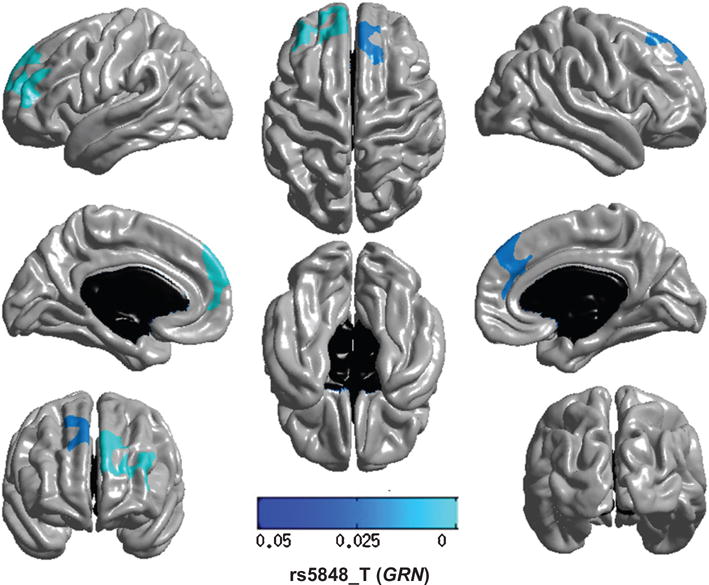
Regional brain atrophy associated with variation in GRN/rs5858. GRN encodes progranulin, a potent growth factor that plays important roles in wound healing and angiogenesis. Note that the GRN/rs5848_T allele is associated with atrophy that localizes on both sides to the dorsal and medial frontal neocortex. This is remarkable since GRN is a risk gene for frontotemporal dementia. The multi-tone blue tab indicates p value.
SNPs in two other genes were assessed that previously were linked to HS-Aging pathology: ABCC9 and KCNMB2. For ABCC9/rs704180, (Fig. 2; recessive model), the atrophy was multifocal with the largest area showing association to the SNP appearing in the right frontal lobe (Table 2). By contrast, for KCNMB2/rs9637454 (recessive model), we found no evidence of atrophy associated with the risk allele (data not shown).
Fig. 2.
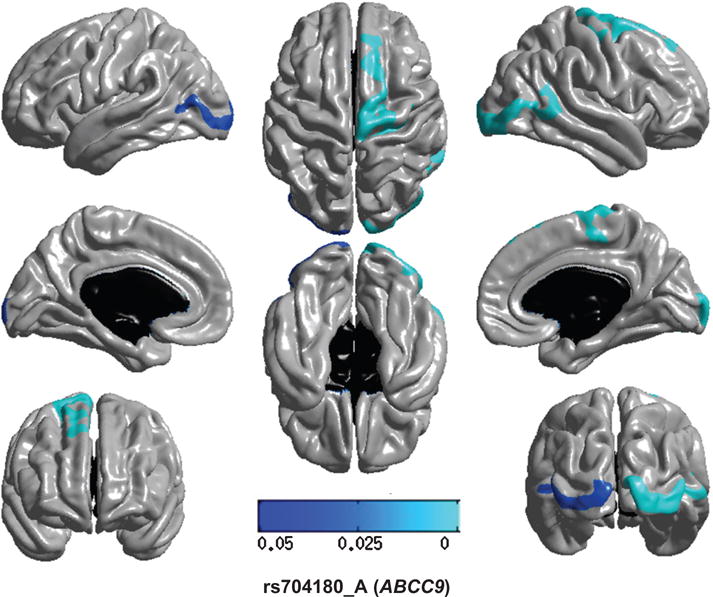
Regional brain atrophy associated with variation in ABCC9/rs704180. ABCC9 encodes SUR2 (sulfonylurea receptor 2), a protein that acts as a metabolic sensor important in brain’s responses to hypoxia and other stressors. The regions of the brain where atrophy is associated with the presence of the ABCC9/rs704180 risk allele were scattered throughout the brain, mostly outside of the temporal lobe. The multi-tone blue tab indicates p value.
In a post-hoc analyses, we also evaluated two SNPs that were intergenic, between ABCC9 and CMAS on chromosome 12p (Fig. 3). A prior study showed that rs10743430 is associated with entorhinal thinning [40], a neuroimaging phenotype that could be a proxy for HS-Aging pathology. We identified another SNP (rs7488080) that is closer to ABCC9 on chromosome 12p than rs10743430, and tested its association with brain atrophy in the ADNI cohort. These two SNPs are in strong linkage disequilibrium (LD) with each other (r2 = 0.87, D′= 0.99). We confirmed that in this sample (larger than in the prior study, but not an independent sample since Furney et al. [40] analyzed ADNI-1; see Discussion below), both rs10743430 and rs7488080 were associated with right entorhinal thinning.
Fig. 3.
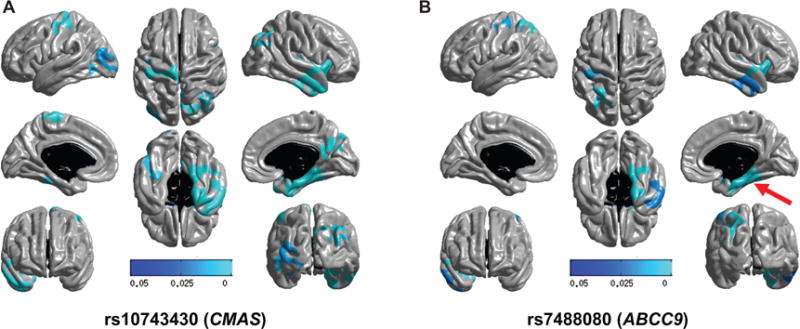
A prior study [40] identified an intergenic SNP between genes ABCC9 and CMAS on human chromosome 12p that is strongly associated with entorhinal cortical thinning in the ADNI and AddNeuroMed datasets (p < 1e−6). Here are shown results from that SNP ([A]; rs10743430; dominant model) and also a nearby SNP ([B]; rs7488080; dominant model) that is in close linkage disequilibrium (r2 = 0.87, D′ = 0.99). Note that for both SNPs, the risk allele is associated with right-sided entorhinal cortex atrophy (red arrow, panel B), as well as atrophy in other regions. The multi-tone blue tab indicates p value.
When the APOE ɛ4 status was added as a covariate, the results were not changed (Supplementary Material).
We investigated whether the four common variants (rs5848, rs1990622, rs704180, and rs9637454) previously associated with risk of HS-Aging may also collectively underlie brain atrophy using cumulative genetic risk scores or polygenic risk scores to model the aggregate effect. Note that the intergenic SNPs described above were not included in this analysis. A polygenic score for each subject was constructed by summing the number of risk alleles across the four risk SNPs without considering their effect sizes. Figure 4(A) showed the distribution of the polygenic risk scores for each diagnosis group. The association analysis between polygenic risk scores and cortical thickness identified significant regional differences in thickness including multiple, predominately frontal, neocortical areas.
Fig. 4.
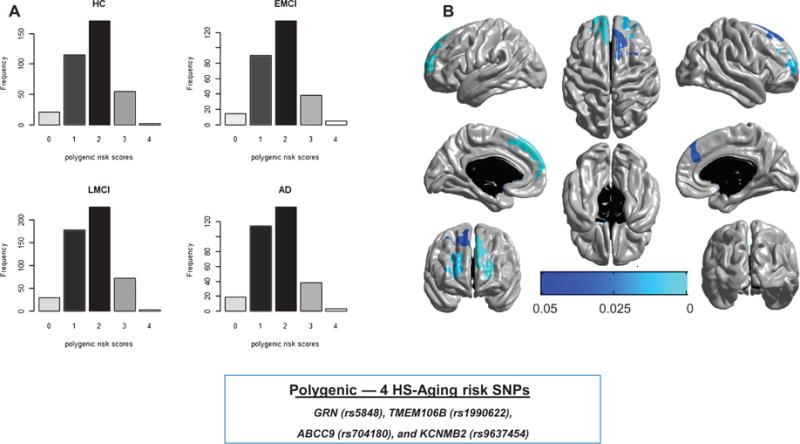
Distribution (A) of polygenic risk scores for each diagnosis group—cognitively normal control (HC), early MCI (EMCI), late MCI (LMCI), and Alzheimer’s disease (AD) by clinical impression—and regional brain atrophy (B) associated with polygenic risk scores. The polygenic risk scores were used to investigate the possibility of overlapping genetic factors. Those scores reflect a combined effect of four risk alleles for HS-Aging. The polygenic risk scores are associated with cortical thickness in multifocal brain atrophy including multiple areas of the neocortex, predominately in the frontal lobes. The multi-tone blue tab indicates p value.
DISCUSSION
Here we report that gene variants previously associated with risk of HS-Aging pathology are also associated with brain atrophy outside the temporal lobe. The GRN risk locus (rs5848) was associated with atrophy in the frontal cortex bilaterally, whereas the other risk locus also associated with FTLD, rs1990622 (TMEM106B), was not associated with cortical atrophy in this sample. ABCC9 gene variants (rs704180 and rs7488080) were associated with more multifocal, or generalized, atrophy. The only evaluated SNP that showed association with medial temporal structural changes (entorhinal cortex) in this dataset was rs7488080 which is ~50 kb upstream the ABCC9 gene. When the risk alleles were combined, they were related to a frontal cortex predominant pattern of brain atrophy. These data indicate that gene variants associated with HS-Aging pathology are also associated with vulnerability to neurodegeneration outside the hippocampal formation proper.
There are limitations to this study. The ADNI dataset is enriched for individuals at risk for AD and the present study relates to a disease (HS-Aging) which is similar but not identical to AD clinically [16, 41], so, the study findings may best relate to the subset of individuals with HS-Aging related genotype that have an “AD-like” phenotype. From a technical standpoint, the medial and more variable brain regions can be challenging to assess confidently across individuals using structural MRI. Further, the average age of the included subjects at the time of their MRI scan was not extremely old (73.8 years) in comparison to the ages of the individuals with autopsy-proven HS-Aging (often over 85 years at death) [1, 2, 6, 7, 9, 12, 16, 17, 31, 42]. However, the current study also lacks some of the sources of bias that affect most autopsy studies (for examples, see [43, 44]). It should be noted that the observations made on the ADNI research subjects could provide important insights about the structural brain changes that occur earlier in the disease course.
An assumption underlying the current study is that neuropathologic classification is continuously evolving and fails to capture the full complexity of the aged human brain. The term “sclerosis” means “hardening” in Greek, and lacks a specific connotation in terms of molecular pathogenesis. Hippocampal sclerosis in aged individuals is diagnosed at autopsy, according to consensus-based criteria [45], when neuron loss and astrocytosis are observed in the hippocampal formation, out of proportion to AD-type plaques and tangles in the same structure. However, the actual anatomical localization of HS-Aging related pathologic changes are not confined to the hippocampal formation [1, 31, 32] and the applied nomenclature for the disease is heterogeneous [13, 19]. Even at academic centers specializing in neurodegenerative disease, HS-Aging tends to be misdiagnosed as AD clinically because of overlapping symptoms and imperfectly understood biomarkers [1, 16, 41]. Hence the disease nosology, and even the awareness of its existence, should be recognized as being currently in a state of rapid change.
As described above, genetic risk factors for HS-Aging have provided insights into potential disease mechanisms. For example, APOE gene variants are not associated with altered risk for HS-Aging [1, 12, 16, 41, 46] and including APOE genotype in our models did not alter our results in terms of the genotype associations. These findings provide strong support for the hypothesis that HS-Aging is a separate disease entity from AD.
In the present study we found that the pattern of brain atrophy associated with different candidate HS-Aging risk alleles was not the same for each gene, and that atrophy was localized to regions mostly outside of the hippocampus. For GRN (rs5848), the atrophy associated with the risk genotype was localized to the dorsal and (mostly) medial frontal cortex. There was a striking bilaterally symmetric pattern that seemed to correspond to or near Brodmann Area 9. This is intriguing because GRN mutations lead to a frontotemporal dementia (FTD, a clinical term) that is associated with a pathologic diagnosis of FTLD-TDP. Remarkably, there was no apparent atrophy signal related to rs5848 and the anterior temporal lobe such as occurs in FTLD-TDP. In addition to HS-Aging pathology, rs5848 has also been linked to multiple other brain disorders [47–49]. It is thus possible that the rs5848-associated structural changes that were elucidated in the current study were due to brain conditions other than HS-Aging.
Another gene that was associated with brain atrophy in the ADNI cohort and previously associated with the HS-Aging pathology is ABCC9. The risk SNP, rs704180, was also associated with brain atrophy in the ADNI cohort but the statistically significant atrophy (controlling for multiple comparisons) is outside of the hippocampal formation. A SNP near ABCC9 (rs7488080) was associated with entorhinal cortex atrophy in the ADNI cohort. This SNP is <50 kb upstream of the ABCC9 start site, the next closest gene is CMAS. Also noteworthy is that rs10743430, in strong linkage disequilibrium (R2 = 0.87) with rs7488080, but not rs704180, was previously found to be the second strongest SNP in association with the endophenotype of entorhinal thinning in the ADNI-1 and AddNeuroMed cohorts [40]. At the time of that prior study, only 939 participants were evaluated for entorhinal thickness whereas the current study included 1,239 participants. Notably, entorhinal cortical atrophy may be a strong proxy for HS-Aging in a MRI study since the entorhinal cortex appears involved even early in the disease process [50].
The KCNMB2 and TMEM106B variants, when assessed individually, did not show significant association signals with cerebral atrophy in the ADNI dataset. However, prior evidence suggests that risk of HS-Aging pathology is increased when risk alleles are present in combination [39]. We therefore tested for potential interactions and found that frontal cortical atrophy may be associated with a polygenic combination of HS-Aging risk SNPs.
The present results can be reconciled with prior studies to help refine our expectations about a prevalent and high-morbidity brain disease that mimics AD. A directly relevant recent study from Rush University found that in elderly individuals that died with eventually autopsy-proven HS, premortem neuroimaging showed extensive brain atrophy on MRI, especially in the frontal lobes [32]. We previously reported evidence for brain pathology outside the hippocampal formation (specifically in the frontal cortex) in patients with comorbid HS-Aging [31], and multiple lines of evidence point to a multi-domain cognitive deficit in patients affected by the disease [1, 16]. The hypothesis that HS-Aging is actually a diffuse or multifocal disease that is often comorbid with brain arteriolosclerosis pathology rather than one localized to medial temporal lobe structures, is quite credible given the prior reports [13, 16, 31, 33, 50–52]. We conclude that the present study adds additional evidence to support a role for genetic variation in brain pathologies, and to the appreciation that this is a multi-factorial condition that can impact brain areas well beyond the hippocampus.
Supplementary Material
Acknowledgments
We are profoundly grateful to the research participants and clinicians that enabled us to perform these studies.
This work was supported by the following National Institute of Health (NIH) grants: P30 AG028383, R01 AG038651, R01 AG042419, T32 AG 000242, R00 LM011384, R01 LM011360, P30 AG10133, and R01 AG19771.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (NIH Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the University of Southern California.
Samples from the National Cell Repository for AD (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the NIA, were used in this study.
Footnotes
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/16-0077).
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-160077.
References
- 1.Nelson PT, Schmitt FA, Lin Y, Abner EL, Jicha GA, Patel E, Thomason PC, Neltner JH, Smith CD, Santacruz KS, Sonnen JA, Poon LW, Gearing M, Green RC, Woodard JL, Van Eldik LJ, Kryscio RJ. Hippocampal sclerosis in advanced age: Clinical and pathological features. Brain. 2011;134:1506–1518. doi: 10.1093/brain/awr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zarow C, Sitzer TE, Chui HC. Understanding hippocampal sclerosis in the elderly: Epidemiology, characterization, and diagnostic issues. Curr Neurol Neurosci Rep. 2008;8:363–370. doi: 10.1007/s11910-008-0057-3. [DOI] [PubMed] [Google Scholar]
- 3.Attems J, Jellinger KA. Hippocampal sclerosis in Alzheimer disease and other dementias. Neurology. 2006;66:775. doi: 10.1212/01.wnl.0000200959.50898.26. [DOI] [PubMed] [Google Scholar]
- 4.Corey-Bloom J, Sabbagh MN, Bondi MW, Hansen L, Alford MF, Masliah E, Thal LJ. Hippocampal sclerosis contributes to dementia in the elderly. Neurology. 1997;48:154–160. doi: 10.1212/wnl.48.1.154. [DOI] [PubMed] [Google Scholar]
- 5.Davidson YS, Raby S, Foulds PG, Robinson A, Thompson JC, Sikkink S, Yusuf I, Amin H, DuPlessis D, Troakes C, Al-Sarraj S, Sloan C, Esiri MM, Prasher VP, Allsop D, Neary D, Pickering-Brown SM, Snowden JS, Mann DM. TDP-43 pathological changes in early onset familial and sporadic Alzheimer’s disease, late onset Alzheimer’s disease and Down’s syndrome: Association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 2011;122:703–713. doi: 10.1007/s00401-011-0879-y. [DOI] [PubMed] [Google Scholar]
- 6.Dickson DW, Davies P, Bevona C, Van Hoeven KH, Factor SM, Grober E, Aronson MK, Crystal HA. Hippocampal sclerosis: A common pathological feature of dementia in very old (> or = 80 years of age) humans. Acta Neuropathol. 1994;88:212–221. doi: 10.1007/BF00293396. [DOI] [PubMed] [Google Scholar]
- 7.Jellinger KA. Hippocampal sclerosis: A common pathological feature of dementia in very old humans. Acta Neuropathol. 1994;88:599. doi: 10.1007/BF00296500. [DOI] [PubMed] [Google Scholar]
- 8.Leverenz JB, Lipton AM. Clinical aspects of hippocampal sclerosis. Handb Clin Neurol. 2008;89:565–567. doi: 10.1016/S0072-9752(07)01252-3. [DOI] [PubMed] [Google Scholar]
- 9.Pao WC, Dickson DW, Crook JE, Finch NA, Rademakers R, Graff-Radford NR. Hippocampal sclerosis in the elderly: Genetic and pathologic findings, some mimicking Alzheimer disease clinically. Alzheimer Dis Assoc Disord. 2011;25:364–368. doi: 10.1097/WAD.0b013e31820f8f50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Probst A, Taylor KI, Tolnay M. Hippocampal sclerosis dementia: A reappraisal. Acta Neuropathol. 2007;114:335–345. doi: 10.1007/s00401-007-0262-1. [DOI] [PubMed] [Google Scholar]
- 11.Thom M, D’Arrigo C, Scaravilli F. Hippocampal sclerosis with hypertrophy of end folium pyramidal cells. Acta Neuropathol. 1999;98:107–110. doi: 10.1007/s004010051057. [DOI] [PubMed] [Google Scholar]
- 12.Leverenz JB, Agustin CM, Tsuang D, Peskind ER, Edland SD, Nochlin D, DiGiacomo L, Bowen JD, McCormick WC, Teri L, Raskind MA, Kukull WA, Larson EB. Clinical and neuropathological characteristics of hippocampal sclerosis: A community-based study. Arch Neurol. 2002;59:1099–1106. doi: 10.1001/archneur.59.7.1099. [DOI] [PubMed] [Google Scholar]
- 13.Nelson PT, Smith CD, Abner EL, Wilfred BJ, Wang WX, Neltner JH, Baker M, Fardo DW, Kryscio RJ, Scheff SW, Jicha GA, Jellinger KA, Van Eldik LJ, Schmitt FA. Hippocampal sclerosis of aging, a prevalent and high-morbidity brain disease. Acta Neuropathol. 2013;126:161–177. doi: 10.1007/s00401-013-1154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarow C, Weiner MW, Ellis WG, Chui HC. Prevalence, laterality, and comorbidity of hippocampal sclerosis in an autopsy sample. Brain Behav. 2012;2:435–442. doi: 10.1002/brb3.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson PT, Abner EL, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, Davis DG, Poduska JW, Patel E, Mendiondo MS, Markesbery WR. Modeling the association between 43 different clinical and pathological variables and the severity of cognitive impairment in a large autopsy cohort of elderly persons. Brain Pathol. 2010;20:66–79. doi: 10.1111/j.1750-3639.2008.00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenowitz WD, Monsell SE, Schmitt FA, Kukull WA, Nelson PT. Hippocampal sclerosis of aging is a key Alzheimer’s disease mimic: Clinical-pathologic correlations and comparisons with both Alzheimer’s disease and non-tauopathic frontotemporal lobar degeneration. J Alzheimers Dis. 2014;39:691–702. doi: 10.3233/JAD-131880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nag S, Yu L, Capuano AW, Wilson RS, Leurgans SE, Bennett DA, Schneider JA. Hippocampal sclerosis and TDP-43 pathology in aging and Alzheimer disease. Ann Neurol. 2015;77:942–952. doi: 10.1002/ana.24388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatanpaa KJ, Raisanen JM, Herndon E, Burns DK, Foong C, Habib AA, White CL. 3rd, Hippocampal sclerosis in dementia, epilepsy, and ischemic injury: Differential vulnerability of hippocampal subfields. J Neuropathol Exp Neurol. 2014;73:136–142. doi: 10.1097/OPX.0000000000000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutra JR, Cortes EP, Vonsattel JP. Update on hippocampal sclerosis. Curr Neurol Neurosci Rep. 2015;15:592. doi: 10.1007/s11910-015-0592-7. [DOI] [PubMed] [Google Scholar]
- 20.Rauramaa T, Pikkarainen M, Englund E, Ince PG, Jellinger K, Paetau A, Alafuzoff I. Consensus recommendations on pathologic changes in the hippocampus: A postmortem multicenter inter-rater study. J Neuropathol Exp Neurol. 2013;72:452–461. doi: 10.1097/NEN.0b013e318292492a. [DOI] [PubMed] [Google Scholar]
- 21.Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, Graff-Radford NR, Hutton ML, Dickson DW. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 23.Lee EB, Lee VM, Trojanowski JQ, Neumann M. TDP-43 immunoreactivity in anoxic, ischemic and neoplastic lesions of the central nervous system. Acta Neuropathol. 2008;115:305–311. doi: 10.1007/s00401-007-0331-5. [DOI] [PubMed] [Google Scholar]
- 24.Wilson RS, Yu L, Trojanowski JQ, Chen EY, Boyle PA, Bennett DA, Schneider JA. TDP-43 pathology, cognitive decline, and dementia in old age. JAMA Neurol. 2013;70:1418–1424. doi: 10.1001/jamaneurol.2013.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutherford NJ, Carrasquillo MM, Li M, Bisceglio G, Menke J, Josephs KA, Parisi JE, Petersen RC, Graff-Radford NR, Younkin SG, Dickson DW, Rademakers R. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology. 2012;79:717–718. doi: 10.1212/WNL.0b013e318264e3ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickson DW, Baker M, Rademakers R. Common variant in GRN is a genetic risk factor for hippocampal sclerosis in the elderly. Neurodegener Dis. 2010;7:170–174. doi: 10.1159/000289231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Langenhove T, van der Zee J, Van Broeckhoven C. The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann Med. 2012;44:817–828. doi: 10.3109/07853890.2012.665471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray ME, Cannon A, Graff-Radford NR, Liesinger AM, Rutherford NJ, Ross OA, Duara R, Carrasquillo MM, Rademakers R, Dickson DW. Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol. 2014;128:411–421. doi: 10.1007/s00401-014-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, Bennett DA, De Jager PL, Larson EB, Crane PK, Kamboh MI, Kofler JK, Mash DC, Duque L, Gilbert JR, Gwirtsman H, Buxbaum JD, Kramer P, Dickson DW, Farrer LA, Frosch MP, Ghetti B, Haines JL, Hyman BT, Kukull WA, Mayeux RP, Pericak-Vance MA, Schneider JA, Trojanowski JQ, Reiman EM, Alzheimer’s Disease Genetics Consortium (ADGC) Schellenberg GD, Montine TJ. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10:e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson PT, Estus S, Abner EL, Parikh I, Malik M, Neltner JH, Ighodaro E, Wang WX, Wilfred BR, Wang LS, Kukull WA, Nandakumar K, Farman ML, Poon WW, Corrada MM, Kawas CH, Cribbs DH, Bennett DA, Schneider JA, Larson EB, Crane PK, Valladares O, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, Scheff SW, Sonnen JA, Haines JL, Pericak-Vance MA, Mayeux R, Farrer LA, Van Eldik LJ, Horbinski C, Green RC, Gearing M, Poon LW, Kramer PL, Woltjer RL, Montine TJ, Partch AB, Rajic AJ, Richmire K, Monsell SE, Alzheimer’ Disease Genetic Consortium. Schellenberg GD, Fardo DW. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol. 2014;127:825–843. doi: 10.1007/s00401-014-1282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neltner JH, Abner EL, Baker S, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, Hammack E, Kukull WA, Brenowitz WD, Van Eldik LJ, Nelson PT. Arteriolosclerosis that affects multiple brain regions is linked to hippocampal sclerosis of ageing. Brain. 2014;137:255–267. doi: 10.1093/brain/awt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotrotsou A, Schneider JA, Bennett DA, Leurgans SE, Dawe RJ, Boyle PA, Golak T, Arfanakis K. Neuropathologic correlates of regional brain volumes in a community cohort of older adults. Neurobiol Aging. 2015;36:2798–2805. doi: 10.1016/j.neurobiolaging.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neltner JH, Abner EL, Jicha GA, Schmitt FA, Patel E, Poon LW, Marla G, Green RC, Davey A, Johnson MA, Jazwinski SM, Kim S, Davis D, Woodard JL, Kryscio RJ, Van Eldik LJ, Nelson PT. Brain pathologies in extreme old age. Neurobiol Aging. 2016;37:1–11. doi: 10.1016/j.neurobiolaging.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, Ramanan VK, Foroud TM, Faber KM, Sarwar N, Munsie LM, Hu X, Soares HD, Potkin SG, Thompson PM, Kauwe JS, Kaddurah-Daouk R, Green RC, Toga AW, Weiner MW, Alzheimer’s Disease Neuroimaging Initiative Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11:792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, Swaminathan S, Ramanan VK, Liu Y, Foroud T, Inlow MH, Siniard AL, Reiman RA, Aisen PS, Petersen RC, Green RC, Jack CR, Weiner MW, Baldwin CT, Lunetta K, Farrer LA, Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study. Furney SJ, Lovestone S, Simmons A, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, AddNeuroMed Consortium. McDonald BC, Farlow MR, Ghetti B, Indiana Memory and Aging Study. Huentelman MJ, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative (ADNI) Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–787. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nho K, Kim S, Risacher SL, Shen L, Corneveaux JJ, Swaminathan S, Lin H, Ramanan VK, Liu Y, Foroud TM, Inlow MH, Siniard AL, Reiman RA, Aisen PS, Petersen RC, Green RC, Jack CR, Jr, Weiner MW, Baldwin CT, Lunetta KL, Farrer LA, MIRAGE. (Multi-Institutional Research on Alzheimer Genetic Epidemiology) Study. Furney SJ, Lovestone S, Simmons A, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, AddNeuroMed Consortium. McDonald BC, Farlow MR, Ghetti B, Indiana Memory and Aging Study. Huentelman MJ, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative Protective variant for hippocampal atrophy identified by whole exome sequencing. Ann Neurol. 2015;77:547–552. doi: 10.1002/ana.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, Harvey DJ, Jack CR, Jr, Weiner MW, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative Longitudinal MRI atrophy biomarkers: Relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson PT, Wang WX, Partch AB, Monsell SE, Valladares O, Ellingson SR, Wilfred BR, Naj AC, Wang LS, Kukull WA, Fardo DW. Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J Neuropathol Exp Neurol. 2015;74:75–84. doi: 10.1097/NEN.0000000000000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, Hodges A, Powell J, Wahlund LO, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Spenger C, Lathrop M, Shen L, Kim S, Saykin AJ, Weiner MW, Lovestone S, Alzheimer’s Disease Neuroimaging Initiative. AddNeuroMed Consortium Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2011;16:1130–1138. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pao WC, Dickson DW, Crook JE, Finch NA, Rademakers R, Graff-Radford NR. Hippocampal sclerosis in the elderly: Genetic and pathologic findings, some mimicking Alzheimer disease clinically. Alzheimer Dis Assoc Disord. 2011;25:364–368. doi: 10.1097/WAD.0b013e31820f8f50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amador-Ortiz C, Dickson DW. Neuropathology of hippocampal sclerosis. Handb Clin Neurol. 2008;89:569–572. doi: 10.1016/S0072-9752(07)01253-5. [DOI] [PubMed] [Google Scholar]
- 43.Haneuse S, Schildcrout J, Crane P, Sonnen J, Breitner J, Larson E. Adjustment for selection bias in observational studies with application to the analysis of autopsy data. Neuroepidemiology. 2009;32:229–239. doi: 10.1159/000197389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuang D, Simpson KL, Li G, Barnhart RL, Edland SD, Bowen J, McCormick W, Teri L, Nochlin D, Larson EB, Thompson ML, Leverenz JB. Evaluation of selection bias in an incident-based dementia autopsy case series. Alzheimer Dis Assoc Disord. 2005;19:67–73. doi: 10.1097/01.wad.0000165507.67993.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Trojanowski JQ, Vinters HV, Hyman BT, National Institute on Aging; Alzheimer’s Association National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 2012;123:1–11. doi: 10.1007/s00401-011-0910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Troncoso JC, Kawas CH, Chang CK, Folstein MF, Hedreen JC. Lack of association of the apoE4 allele with hippocampal sclerosis dementia. Neurosci Lett. 1996;204:138–140. doi: 10.1016/0304-3940(96)12331-4. [DOI] [PubMed] [Google Scholar]
- 47.van Blitterswijk M, Mullen B, Wojtas A, Heckman MG, Diehl NN, Baker MC, DeJesus-Hernandez M, Brown PH, Murray ME, Hsiung GY, Stewart H, Karydas AM, Finger E, Kertesz A, Bigio EH, Weintraub S, Mesulam M, Hatanpaa KJ, White CL, 3rd, Neumann M, Strong MJ, Beach TG, Wszolek ZK, Lippa C, Caselli R, Petrucelli L, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Mackenzie IR, Seeley WW, Grinberg LT, Miller BL, Boylan KB, Graff-Radford NR, Boeve BF, Dickson DW, Rademakers R. Genetic modifiers in carriers of repeat expansions in the C9ORF72 gene. Mol Neurodegener. 2014;9:38. doi: 10.1186/1750-1326-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ, Josephs KA, Boeve BF, Knopman DS, Petersen RC, Parisi JE, Caselli RJ, Wszolek ZK, Uitti RJ, Feldman H, Hutton ML, Mackenzie IR, Graff-Radford NR, Dickson DW. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viswanathan J, Makinen P, Helisalmi S, Haapasalo A, Soininen H, Hiltunen M. An association study between granulin gene polymorphisms and Alzheimer’s disease in Finnish population. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:747–750. doi: 10.1002/ajmg.b.30889. [DOI] [PubMed] [Google Scholar]
- 50.Ighodaro ET, Jicha GA, Schmitt FA, Neltner JH, Abner EL, Kryscio RJ, Smith CD, Duplessis T, Anderson S, Patel E, Bachstetter A, Van Eldik LJ, Nelson PT. Hippocampal sclerosis of aging can be segmental: Two cases and review of the literature. J Neuropathol Exp Neurol. 2015;74:642–652. doi: 10.1097/NEN.0000000000000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson PT, Jicha GA, Wang WX, Ighodaro E, Artiushin S, Nichols CG, Fardo DW. ABCC9/SUR2 in the brain: Implications for hippocampal sclerosis of aging and a potential therapeutic target. Ageing Res Rev. 2015;24(Pt B):111–125. doi: 10.1016/j.arr.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ighodaro ET, Abner EL, Fardo DW, Lin AL, Katsumata Y, Schmitt FA, Kryscio RJ, Jicha GA, Neltner JH, Monsell SE, Kukull WA, Moser DK, Appiah F, Bachstetter AD, Van Eldik LJ, Alzheimer’s Disease Neuroimaging Initiative (ADNI) Nelson PT. Risk factors and global cognitive status related to brain arteriolosclerosis in elderly individuals. J Cereb Blood Flow Metab. 2016 Jan 6; doi: 10.1177/0271678X15621574. 2016. pii: 0271678X15621574 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


