Abstract
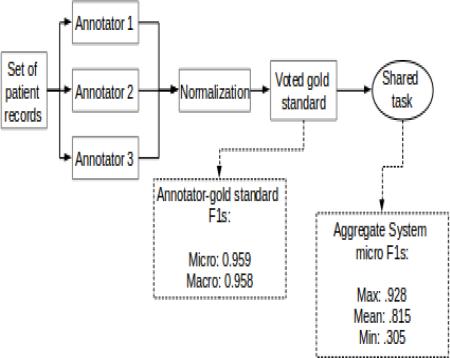
The 2014 i2b2/UTHealth natural language processing shared task featured a track focused on identifying risk factors for heart disease (specifically, Cardiac Artery Disease) in clinical narratives. For this track, we used a “light” annotation paradigm to annotate a set of 1,304 longitudinal medical records describing 296 patients for risk factors and the times they were present. We designed the annotation task for this track with the goal of balancing annotation load and time with quality, so as to generate a gold standard corpus that can benefit a clinically-relevant task. We applied light annotation procedures and determined the gold standard using majority voting. On average, the agreement of annotators with the gold standard was above 0.95, indicating high reliability. The resulting document-level annotations generated for each record in each longitudinal EMR in this corpus provide information that can support studies of progression of heart disease risk factors in the included patients over time. These annotations were used in the Risk Factor track of the 2014 i2b2/UTHealth shared task. Participating systems achieved a mean micro-averaged F1 measure of 0.815 and a maximum F1 measure of 0.928 for identifying these risk factors in patient records.
Graphical abstract
1. Introduction
While much information about a patient's medical history is stored in structured, easily searchable databases, still more information is contained within the narrative portions of the electronic medical records (EMRs). It is often necessary for clinicians to read through these narratives to gain a full perspective on a patient's history of a disease and other relevant factors. Yet reading through years of patient records is time-consuming, particularly when only certain pieces of information related to a particular medical question are sought.
Using natural language processing (NLP) to extract information about a specific clinical question was the focus for Track 2 of the 2014 i2b2/UTHealth (Informatics for Integrating Biology and the Bedside; University of Texas Health Science Center at Houston) NLP shared task. With the advice of practicing medical doctors and researchers, we developed an annotated corpus that answers the question “For each record in each patient's EMR, which heart disease risk factors were present before, during, and after the record's creation date?” We used this question as our starting point for enabling the use of EMRs in studying the clinical questions of “How do diabetic patients progress towards heart disease, specifically coronary artery disease? And how do diabetic patients with coronary artery disease differ from other diabetic patients who do not develop coronary artery disease?”
The development of coronary artery disease (CAD or “heart disease” for short) is complex, and many factors are involved in determining whether a patient is at risk. The World Health Organization defines “risk factors” as “any attribute, characteristic or exposure of an individual that increases the likelihood of developing a disease or injury” (WHO, 2014). Risk factors for heart disease include life-style and social factors such as smoking status and family medical history, as well as specific clinical conditions such as hypertension and hyperlipidemia. To understand the progression towards CAD in a patient, these risk factors are considered with their temporality and their time of onset.
In order to develop NLP systems that can extract disease-relevant information from narrative EMRs to help clinicians assess patients’ potential progression towards CAD over time, we built and de-identified a new corpus of longitudinal patient records. We annotated these records for heart disease risk factors and medical information that indicates the presence of these risk factors using a “light” annotation paradigm (Stubbs, 2013). This paradigm enabled us to annotate the corpus quickly and consistently.
This paper describes the Track 2 (also called the “Risk Factors Track”) corpus of the 2014 i2b2/UTHealth NLP Shared Task. Section 2 discusses related work, Section 3 provides an overview of the corpus, and Section 4 gives more in-depth information about the heart disease risk factors that we annotated. Section 5 discusses the annotation guidelines, Section 6 describes trial annotations, and Section 7 reviews the annotation procedures and provides statistics on the resulting corpus. Sections 8 and 9 close the paper with our discussions and conclusions.
2. Related work
Previous clinical NLP shared tasks have generally focused on identifying and extracting broad classes of information that can support multiple tasks. For example, the 2009 i2b2 shared task focused on identifying all medications mentioned in a corpus of 251 discharge summaries, along with related information: dosages, modes, frequencies, durations, reasons, and whether or not the information appeared in a list or narrative text (Uzuner et al., 2010). Other related tasks, such as the TREC Genomics shared tasks (Hersh and Vorhees, 2008) focused on biomedical corpora such as MEDLINE abstracts and journal articles; here we focus on corpora that use clinical narratives.
The 2007 Computational Medicine Challenge focused on assigning ICD-9-CM codes to a corpus of radiology reports (Pestian et al., 2007). The organizers used annotators from the coding staff of the Cincinnati Children's Hospital Medical Center and from two independent coding companies. They used a majority vote method to determine the gold standard, similar to the approach we describe in Section 7.3. Nearly 2,000 records were annotated for this project.
The 2010 i2b2/VA shared task (Uzuner et al., 2011) had three tracks: 1) Concept extraction, where systems had to identify patient medical problems, treatments, and tests; 2) Assertion classification, where identified concepts from the previous track were categorized as being present, absent, possible, conditional, hypothetical, etc.; 3) Relation classification, where relationships between the concepts were categorized into types. For example, medical problems could relate to tests in several ways including “test reveals medical problem”, “test conducted to investigate medical problem”, or “in the same sentence but the relationship is other/undefined”. 871 medical records were annotated for the 2010 shared task.
The 2012 temporal relations shared task (Sun et al., 2013) addressed temporal relations in clinical records. This shared task had two tracks: 1) identifying times and clinical events, and 2) identifying temporality and the temporal order of events. While the second track did limit the scope of possible relationships to those between times and events in the same sentence, main events in successive sentences, and two events where one has scope over the other, the task still encompasses many temporal relations and the resulting annotated corpus included 310 medical records.
The 2013 ShARe/CLEF eHealth Evaluation Lab (Suominen et al., 2013) featured three shared tasks: disorder identification and normalization; abbreviation identification and normalization; and information retrieval for answering medical questions. The first two tasks used clinical records from the ShARe corpus, while the third augmented those records with other sources of medical information.
Each of the aforementioned shared tasks focused on identifying and labeling all instances in medical documents of specific types of information: ICD-9-CM codes, medications, concepts, tests, events, and the relationships between them. Such exhaustive annotations often result in smaller annotated corpora, due to the complex nature of the annotations and the additional time and expense of creating the annotations. Additionally, while these annotations can identify all the types of, for example, medications in a document, they cannot necessarily support a diagnosis of a particular disease.
In order to find a way to minimize the time and expense of clinical annotation, our goal for the 2014 i2b2 shared task was to focus on a particular clinical question, and develop a light annotation model that made use of previous annotation efforts without annotating every example of a clinical concept, medication, etc. in the medical records. We focused on document-level annotations that were specific to CAD and its risk factors and indicators. Light annotation tasks require less effort than the more traditional exhaustive annotations, as they do not require that all instances of a particular item be annotated; instead, they require only that annotations be sufficient to support a claim. This is discussed further in Section 5.
Two previous i2b2 shared tasks used annotation guidelines that are relatively light: the 2006 challenge to identify patient smoking status placed patients into five categories: Past Smoker, Current Smoker, Smoker, Non-Smoker, and Unknown (Uzuner et al., 2007). We used these same basic categories for the Risk Factors track. Similarly, the 2008 challenge placed patients into four different classes for obesity and co-morbid diseases: present, absent, questionable, and unmentioned (Uzuner, 2009). Obesity is a risk factor for CAD, and as we describe below, we used broad, document-level classifications for this shared task, similar to those used for the 2006 and 2008 challenges.
3. Corpus
For this study, we selected and de-identified a new corpus of longitudinal medical records for 296 diabetic patients. Each longitudinal medical record contains many individual records, corresponding to different doctor—patient interactions. Each patient in this corpus belongs to one of three equally represented cohorts: 1) patients who have a diagnosis of CAD in their first selected record, 2) patients who do not have a diagnosis of CAD in their first selected record but get a diagnosis of CAD in their later selected records, and 3) patients who do not have a diagnosis of CAD in any of their records. Assuming that the diagnoses given in the records represent reality, the first group represents patients who already have CAD from the beginning of their records; the second group represents patients who do not have CAD in the beginning but progress towards it and develop it later; and the third group represents patients who have not yet (or ever) developed CAD. In total the corpus contains 1,304 narrative medical records, representing 2-5 records per patient. For training and testing purposes we used a 60/40 split: 790 records in the training set and 514 records in the test set. Each patient is assigned either to the training set or the test set. The full description of the corpus construction process for the 2014 i2b2/UTHealth shared task is in Kumar et al. (this issue).
4. Task definition and description
As described in the introduction, our goal for Track 2 of the 2014 i2b2/UTHealth NLP shared task was to pose a clinically significant question that could be answered through the use of NLP. After consulting with domain experts, we chose to focus on the progression of heart disease risk factors for diabetic patients. More specifically, our goal was to identify, for each record of each patient, which risk factor indicators were present before, during, and after the record's creation date, i.e., the time at which that record was kept. This goal allowed us to create a shared task that both has immediate relevance to medical researchers and also incorporates aspects of previous i2b2 NLP challenges.
For patients with diabetes, there is a set of risk factors that increase their risk of developing heart disease, specifically CAD. These are (NDIC, 2014):
hyperlipidemia/hypercholesterolemia,
hypertension,
obesity,
a family history of premature CAD, and
being a smoker.
Medical records may discuss medical conditions such as hyperlipidemia, hypertension, and obesity in various ways, including explicit references to a diagnosis or through indirect information such as clinical measurements, tests, and treatments that indicate either a finding of or an action taken in response to them. We refer to these explicit and indirect information as “indicators” of risk factors. Risk factors such as family history and smoking do not have indicators; rather they are categorized as “present” or “not present” for family history, and the patient's smoking status is categorized into one of five classifications (Uzuner et al., 2007).
Table 1 shows the risk factors in the left column and the indicators that correlate with the relevant risk factors in the right column. The bolded words in the right column mark the types of indicators, e.g., mention indicators (for explicit references to a diagnosis), test indicators (for medical exams that are indicative of the diagnosis), etc. For family history and smoking, the left column shows the relevant categories. Additionally, for heart disease risk factors, it is clinically important to know if the patients are taking certain classes of medications. Some medication classes are indicators for multiple risk factors (e.g., ACE inhibitors and beta-blockers suggest the possibility of both CAD and hypertension). Because of this, rather than associate each medication directly with a risk factor, the medication classes are treated as their own category. At the suggestion of our domain experts, we also included diabetes and CAD in the list of risk factors in order to provide a fuller view of the patient's medical status.
Table 1.
Indicators of CAD risk factors in diabetic patients
| Risk factor | Indicators |
|---|---|
| Diabetes | • Mention: A diagnosis of Type 1 or Type 2 diabetes • Test: An A1c test value of over 6.5 or 2 fasting blood glucose measurements of over 126 |
| CAD | • Mention: A diagnosis of CAD • Event: An event indicative of CAD (MI, STEMI, NSTEMI, revascularization procedures, cardiac arrest, ischemic cardiomyopathy) • Test: Test results: exercise or pharmacologic stress test showing ischemia, or abnormal cardiac catheterization showing coronary stenoses • Symptom: Chest pain consistent with angina |
| Hyperlipidemia/Hypercholesterolemia | • Mention: A diagnosis of Hyperlipidemia or Hypercholesterolemia • High cholesterol: Total cholesterol of over 240 • High LDL: LDL measurement of over 100mg/dL |
| Hypertension | • Mention: A diagnosis of hypertension • High blood pressure: BP measurement of over 140/90 mm/hg |
| Obesity | • Mention: A description of the patient as being obese • High body mass index (BMI): BMI over 30 • Large waist circumference: Waist circumference measurement of: ⎛ men: 40 inches or more ⎛ women: 35 inches or more |
| Family history of premature CAD | • Categories: ⎛ Present: Patient has a first-degree relative (parents, siblings, or children) who was diagnosed prematurely (younger than 55 for male relatives, younger than 65 for female relatives) with CAD ⎛ Not present: no positive mention of a family history of CAD |
| Smoking | • Categories: ⎛ Current: currently smokes or has smoked within the past year, ⎛ Past: quit over a year ago, ⎛ Ever: smoked at some point but their current status is unknown, ⎛ Never: never smoked, ⎛ Unknown: smoking status is not discussed |
| Medications | • ACE inhibitor, amylin, anti-diabetes, ARB, aspirin, beta blocker, calcium channel blocker, diuretic, DPP4 inhibitors, ezetimibe, fibrate, GLP1 agonist, insulin, Meglitinide, metformin, niacin, nitrate, obesity medications, statin, sulfonylurea, thiazolidinedione, thienopyridine, and drug combinations including these |
We used the indicators to track not just the existence of the risk factors, but also as a possible measure of the severity of the diagnoses. For example, a person who is diagnosed with hyperlipidemia in the document in addition to having high cholesterol may be more at risk for heart disease than someone with a diagnosis but whose cholesterol is being kept at healthy levels through diet or medication.
To link the identified indicators of risk factors with the patient's medical timeline, we marked each indicator with one or more temporal categories: before the document creation time (DCT), during the DCT, or after the DCT. These broad categories allowed the corpus to show progression (or lack thereof) of the diseases over the course of each patient's longitudinal records. For example, if a record did not contain a diagnosis of hyperlipidemia, but the next one implied that the condition had been previously established, one would be able to infer that the hyperlipidemia had probably been discovered sometime between those two records.
We posed the 2014 i2b2/UTHealth shared task as a document-level classification task. Given a set of patient records, for each patient record in a longitudinal set, we asked the participants to determine the risk factor indicators that are present in the record and the time of those indicators with respect to the DCT of the record. A single instance of an indicator in a file was sufficient evidence to support the presence of both the indicator and its related risk factor in the file.
5. Light Annotation Guidelines
The clinical question of identifying risk factor indicators and their times of existence would have been extremely complex and time-consuming if we had decided to exhaustively annotate every disease, medication, measurement, test, and temporal relation in every document by implementing all of the i2b2 annotation guidelines from previous shared tasks. Such an annotation project would have been time-consuming, expensive, and would have resulted in a much smaller corpus for the shared task.
Rather than annotate a small number of documents exhaustively, we chose to create a light annotation task, which lessened the burden on the annotators and enabled us to annotate the entire corpus (1,304 records) in a reasonable time. A light annotation implements the following principles: it employs expert annotators; it generates few, document-level, evidenced-based annotations; it uses guidelines that support best current medical practices; and its representation is extensible so that it can be augmented as necessary.
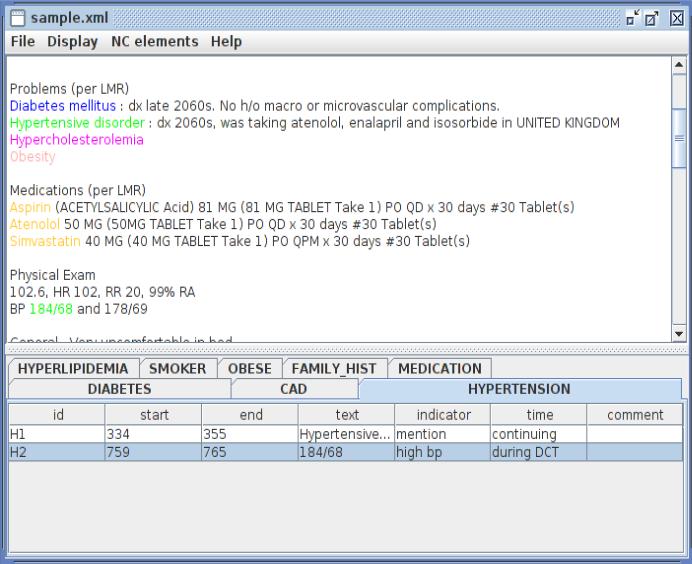
We implemented these principles by asking the annotators to mark only one positive piece of evidence for each risk factor indicator at each time it was present relative to the DCT. This guideline saved time, as it is common in this corpus for a record to describe a patient as having “DM”, show “diabetes mellitus” in a list of medical problems (as in Figure 1), and repeatedly describe the patient as being “diabetic”. Each annotator could use different piece of evidence to support an annotation. This allowed the annotators to skip vague information (e.g., “high BMI“) and focus on evidence that did not require judgment calls.
Figure 1.
Risk factor annotation in MAE
To simplify the temporal aspect of the annotations, we asked the annotators to categorize the temporal relationship between the indicator and the DCT into three broad categories: before DCT, during DCT, and after DCT. For indicators that most often referred to an ongoing condition (such as a pre-existing diagnosis of CAD), annotators could use the time value “continuing” as shorthand for all three temporal relations. The use of broad temporal categories is similar to the use of narrative containers (Pustejovsky and Stubbs, 2011), a practice that other researchers have found useful when annotating medical records (Miller et al., 2013). Our temporal anchoring of risk factors builds on the 2012 i2b2 challenge on temporal relations (Sun et al., 2014) and simplifies those relations for a light annotation task. The smoking status and family history risk factors did not have a temporal attribute.
Figure 1 shows a sample annotation in MAE (Multi-purpose Annotation Environment) (Stubbs, 2011). Notice that only one instance of “atenolol” is annotated. The unannotated mention only provides information about a past prescription, while the annotated instance provides evidence that the prescription existed before, during, and after the DCT, as the prescription is not indicated as being no longer necessary. There are two hypertension indicators: one is a diagnosis of the disease, labeled “mention” with a temporal annotation of “continuing”, and the other is a blood pressure measurement, labeled “high b.p.” and “during DCT”. Both of these annotations support the presence of hypertension in the patient; however, they provide different information about the time component, which requires both to be included.
Requiring annotators to annotate a piece of evidence in support of their document-level judgments kept the annotations objective. This resulted in high inter-annotator agreement and made the resulting gold standard a more feasible machine learning project. Asking the annotators to only mark a single positive example of each indicator/time combination made the annotation process rapid and less of a burden for the annotators. For more detail about how the annotations were applied to the records, please see the guidelines (Appendix A).
6. Annotation trials
Light annotation tasks result in rapid annotation and larger data sets, and they are easy for medical professionals to learn. However, at the same time, they generate data sets that contain no negative evidence (i.e., “patient not at risk for hypertension”) and do not directly link positive evidence to specific document-level annotations. Before deciding on the light annotation task described in Section 5, we experimented with three variations: “very light”, “moderately light”, and “exhaustive” annotation variations.
It should be noted that at the time we experimented with these variations, each condition (diabetes, CAD, hypertension, hyperlipidemia, and obesity) had a “medication” indicator that listed classes of medications indicative of it. Our experiments showed that some medications overlapped between conditions, which led us to move medications to their own category. This section reflects the fact that medications were indicators for conditions; for the sake of simplicity we limit the relevant medications in our examples to insulin (diabetes), eprosartan (hypertension), and simvastatin (hyperlipidemia).
Trial 1: Very light annotation
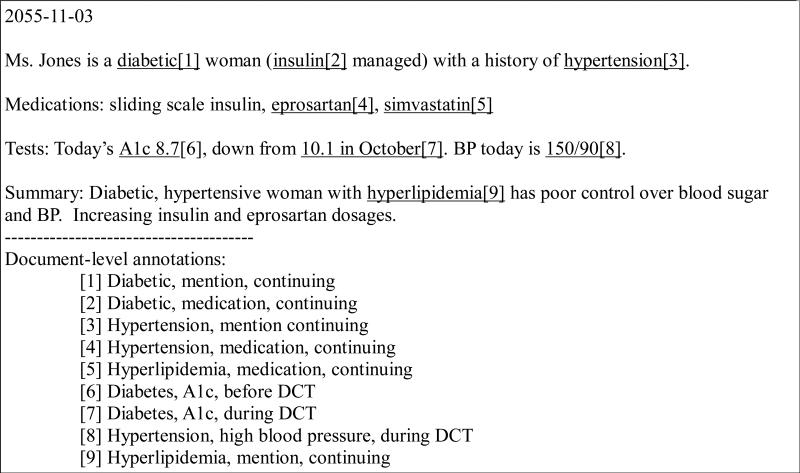
In this variation, the risk factors (i.e., diabetes, hyperlipidemia, hypertension, CAD, obesity, family history, and smoking status) were the primary unit of annotation. Any indicator associated with a risk factor could count as evidence for it, and we instructed annotators to mark only one indicator per risk factor per record. The annotators could mark multiple indicators only if the additional indicators provided new information about when the risk factor was seen in the patient (i.e., before, during, or after the DCT). Figure 2 shows an example, where textual evidence is underlined, and the bracketed number corresponds to the resulting document-level annotation, shown at the bottom.
Figure 2.
Example of Trial 1: very light annotation
As Figure 2 shows, in this variation annotators focused only on the risk factors, and it did not matter which indicator they used to support the assertion of the presence of a disease. It would have been equally valid to use the direct mention of “hyperlipidemia” as evidence of hyperlipidemia, or the mention of “eprosartan” as evidence of hypertension. The A1c measurement only provides information that is valid during the DCT, it could have been annotated for that purpose, but the mention “diabetic” provides more temporal information and deems annotation of A1c unnecessary.
Trial 2: Moderately light annotation
This is the variation adopted for the Risk Factors track. Details are in Section 5. The indicators are the main focus of this variation, but we still only require one annotation of each indicator per time per record. Figure 3 shows the same text from Figure 2, but with Trial 2's output.
Figure 3.
Example of Trial 2: moderately light annotation
As Figure 3 shows, Trial 2 led to many more annotations than Trial 1, and included much more information with regard to the risk factors and their times. Both A1c measurements are included in this annotation, as they relate to different times in the patient's medical history (i.e., before and during the DCT). Only one of the three mentions of insulin was annotated, as all three mentions would have led to the same document-level annotation (i.e., “Diabetic, medication, continuing”).
Trial 3: Exhaustive annotation
In this variation, we asked the annotators to mark every occurrence of every indicator, even if those occurrences were repetitive and redundant. This level of annotation meant that the annotators would mark every instance of “diabetes”, “diabetic”, “DM+”, and so on, even though subsequent annotations would not add to the information about the patient's medical history. Figure 4 shows output of Trial 3 on the same text as Figures 2 and 3.
Figure 4.
Example of Trial 3: exhaustive annotation
Figure 4 shows that Trial 3 resulted in even more annotations, many of which are redundant at the document level. Had this trial's guidelines been deemed most useful for our project, the gold standard would have used phrase-level rather than document-level annotations.
Using 30 documents selected for development purposes, we assigned two annotators to each variation, then evaluated the inter-annotator agreement (IAA), time taken to create the annotations, and usability of the annotations for NLP purposes. We calculated IAA using the standard precision, recall, and f-measure equations; more details can be found in Stubbs et al., (this issue). Table 2 shows the average time and IAA scores for each variation. We calculated the f-measure by taking one annotator to be the “gold standard” and evaluating a second annotator against this gold standard.
Table 2.
Comparison of the trials
| Variation | Annotation time | F-measure |
|---|---|---|
| 1) Very light | 16 min/record | 0.84 |
| 2) Moderately light | 11 min/record | 0.36 |
| 3) Exhaustive | 14.5 min/record | 0.62 |
The low f-measure for Trial 2 surprised us, but further investigation revealed that one of the annotators assigned to that variation was not a medical professional, and was therefore unable to interpret many of the abbreviations in the records. When we repeated the variation with two registered nurses, the f-measure increased to 0.75.
These f-measures provided a reasonable starting place for refining the annotation guidelines. Given little difference in the time taken for each trial, the deciding factor was the usefulness of each trial's output to clinicians. After consulting with MDs involved in the i2b2/UTHealth shared task, we determined that Trial 1 would only inform a clinician that a patient is hypertensive, but the document-level annotation would not include information about individual indicators. By not including information about which indicators are present at what times, the clinician would form an incomplete picture of the patient's health. A lone diagnosis of hypertension does not provide information about whether the patient is taking related medication or consistently has high blood pressure, which are important factors when considering a course of treatment. On the other hand, Trial 2 would include all the clinically relevant information that a clinician would need, including for example, whether a hypertensive patient is still experiencing high blood pressure even while on a medication for lowering blood pressure. Finally, Trial 3 would contain the most information for an NLP system, but for a clinician it is only necessary to know about the diagnosis of diabetes and its accompanying indicators and their times, not the number of times the record repeated that same information. Therefore, we determined that Trial 2 hit the sweet spot for capturing sufficient clinically relevant information.
7. Annotation procedure
Once we determined that Trial 2 would be the most efficient and effective variation of the task, we incorporated the relevant instructions in the guidelines (see Appendix and Section 5).
7.1 Annotator expertise
To create the gold standard we hired 7 annotators, all of whom had medical training. We hired five registered nurses, one medical doctor, and one medical assistant.
7.2 Training
All of the annotators attended at least one online training session to go over the annotation guidelines and software. The annotators practiced on the development set, and sent us both their annotated documents and any questions about the guidelines. We then used the annotator's work to create a voted gold standard (see Section 7.4), then compared each annotator to that gold standard and averaged the precision, recall, and f-measure for all the annotators. We repeated this procedure until the average metrics were all approaching .90. The annotators’ work and feedback led to revisions of the guidelines, including the creation of the Medication category described in Section 6; the final version of the guidelines is found in the Appendix.
7.3 Triple annotation and voted gold standard
In order to create the gold standard, we used majority vote. We assigned each record to three different annotators, who all worked independently. During the annotation training described in 7.2, we determined that all the annotators were equally capable at performing the annotation task, and so we did not require that each file be annotated by the doctor, the medical assistant, and an RN. To make the annotation process as efficient as possible, we assigned annotations on a first-come, first-served basis: annotators with more time to work received annotation assignments more frequently. We normalized these annotations (see section on post-processing) and included in the gold standard any annotation that was supported by two or more annotators.
7.4 Post-processing
Before creating the gold standard, we took the following steps:
-
1)
We removed the offset and text information, corresponding to the span of textual evidence, from the document-level annotations. The spans of textual evidence were intended to keep the annotators objective; however, it did not matter if the annotators found the same exact evidence, as long as at least two of them generated the same combination of risk factor, indicator, and time.
-
2)
In cases where the annotators used the “continuing” option for the time attribute, we split each of those annotations into three, one for each of the “before”, “during”, and “after” values of the time attribute. As described in the Annotation Guidelines section, the “continuing” option was shorthand for three annotations with all the different time values represented.
After post-processing, we ran the voting algorithm to generate the gold standard. We also made available a version of the gold standard that contained the textual evidence with the document-level annotations, for those researchers who wanted to benefit from the evidence for system development.
7.5 Annotation quality and statistics
We calculated the final IAA in terms of precision, recall, and f-measure by comparing each annotator against the gold standard. Table 3 shows the average macro- and micro-averaged scores for all the annotators.
Table 3.
Average macro- and micro-averaged precision, recall, and f-measure scores for annotators compared to the gold standard
| Macro-averaged | Micro-average | |
|---|---|---|
| Precision | 0.957 | 0.958 |
| Recall | 0.959 | 0.960 |
| F-measure | 0.958 | 0.959 |
Inter-annotator agreement was quite high. In the vast majority of cases the annotators agreed upon when the medical record indicated various risk factor indicators were present in the patient history. Additionally, the annotation was done quickly: on average each annotator took only 10 minutes per record, which was faster than all of the trials we performed, but is also substantially faster than other exhaustive type of annotations. For example, the de-identification annotation for this same corpus took, on average, 30 minutes per document (Stubbs, and Uzuner, this issue)
The following tables show the distribution of the different risk factors, indicators, and times over the entire corpus of medical records. Each table represents a single risk factor, and each row shows a single indicator for that risk factor (see Table 1 in Section 4). For example, Table 4 shows the distribution of CAD indicators in the corpus, focusing on mentions (i.e., “patient with history of CAD”), events (occurrences that indicate CAD, such as myocardial infarctions), symptoms (such as chest pain consistent with angina), and test results (such as stress tests that reveal ischemia), etc. The columns then show the temporal labels assigned to each indicator split between the training and testing data. As we mentioned in Section 7.4, many “before”, “during”, and “after” annotations resulted from splitting apart the “continuing” judgments from the annotators. “Mentions” and “medications” were most frequently marked as “continuing” and therefore, show similar numbers across all three temporal categories.
Table 4.
Distribution of CAD indicators
| Indicator | Before DCT | During DCT | After DCT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Total | Train | Test | Total | Train | Test | Total | |
| Mention of CAD | 260 | 170 | 430 | 261 | 175 | 436 | 259 | 171 | 430 |
| Event indicative of CAD | 224 | 129 | 353 | 20 | 9 | 29 | 2 | 1 | 3 |
| Symptoms indicative of CAD | 54 | 41 | 95 | 24 | 26 | 50 | 3 | 3 | 6 |
| Test result indicating CAD | 66 | 53 | 119 | 13 | 6 | 19 | 0 | 0 | 0 |
Table 4 shows that “mention” was the most common indicator for CAD by far, with 430, 436, and 430 records containing before, during, and after annotations respectively. Only “event, before DCT” annotations came close to having the same representation. The presence of more mentions than any other indicator continues with all of the other condition categories (see tables for diabetes, hypertension, hyperlipidemia, obesity). This skewed distribution does make it more difficult to train machine learning algorithms to identify the less well-represented indicators, but represents the realities of the data.
Even for diagnoses that exist in a patient, some records may not indicate them. Table 5 shows the distribution of indicators of diabetes. Every single patient in this corpus is diabetic, yet the fact that they are currently diabetic (“mention, during DCT”) is only explicitly present in 880 records, which is 67% of the corpus.
Table 5.
Distribution of Diabetes indicators
| Indicator | Before DCT | During DCT | After DCT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Total | Train | Test | Total | Train | Test | Total | |
| Mention of diabetes | 518 | 354 | 872 | 524 | 356 | 880 | 518 | 355 | 873 |
| High A1c measurement | 89 | 71 | 160 | 21 | 11 | 32 | 0 | 0 | 0 |
| High glucose measurement | 16 | 18 | 34 | 9 | 15 | 24 | 0 | 0 | 0 |
As Tables 6 and 7 show, for both hyperlipidemia and hypertension, as with CAD, mentions of the diseases in all time periods were far more common than high test results. In our corpus, hypertension was a more common problem than hyperlipidemia: 577 (44%) records contain “mention, during DCT” annotations of hyperlipidemia, compared to the 888 (68%) for hypertension.
Table 6.
Distribution of Hyperlipidemia indicators
| Indicator | Before DCT | During DCT | After DCT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Total | Train | Test | Total | Train | Test | Total | |
| Mention of hyperlipidemia/hypercholesterolimia | 340 | 237 | 577 | 340 | 237 | 577 | 340 | 237 | 577 |
| High LDL | 23 | 26 | 49 | 10 | 3 | 13 | 0 | 0 | 0 |
| High cholesterol | 8 | 9 | 17 | 1 | 2 | 3 | 0 | 0 | 0 |
Table 7.
Distribution of hypertension indicators
| Indicator | Before DCT | During DCT | After DCT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Total | Train | Test | Total | Train | Test | Total | |
| Mention of hypertension | 523 | 365 | 888 | 521 | 367 | 888 | 519 | 366 | 885 |
| High blood pressure | 41 | 20 | 61 | 322 | 175 | 497 | 0 | 0 | 0 |
Table 8 shows the distribution of obesity indicators. Descriptions of the patient as obese are by far the most common indicator: 234 (18%) records describe the patient as currently obese, compared to the 28 (2%) that contain current BMI scores. Waist circumferences are not mentioned at all. Overall, obesity indicators were sparse in this corpus.
Table 8.
Distribution of obesity indicators
| Indicator | Before DCT | During DCT | After DCT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Total | Train | Test | Total | Train | Test | Total | |
| Patient described as “obese” | 133 | 79 | 212 | 147 | 87 | 234 | 133 | 79 | 212 |
| High BMI | 3 | 2 | 5 | 15 | 13 | 28 | 2 | 2 | 4 |
| Large waist circumference | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Table 9 shows the distribution of CAD-related family history in the corpus. By far, most records either contained no mention of family history or contained a family history with no mention of CAD.
Table 9.
Distribution of CAD-related family history in corpus
| Training | Test | Total | |
|---|---|---|---|
| No mention of CAD-related family history | 768 | 495 | 1263 |
| Mention of CAD-related family history | 22 | 19 | 41 |
Table 10 shows that 614 (47%) of records do not discuss smoking status at all, but a majority do discuss whether or not the patient smokes or smoked in the past.
Table 10.
Distribution of smoking status tags
| Smoking status | Count | ||
|---|---|---|---|
| Training | Test | Total | |
| Current smoker | 58 | 33 | 91 |
| Ever smoked | 9 | 3 | 12 |
| Never smoked | 184 | 120 | 304 |
| Past smoker (quit over 1 year ago) | 149 | 113 | 262 |
| Unknown (no mention of smoking) | 371 | 243 | 614 |
Table 11 shows all the medication classes described in Table 1 and that appear in our corpus. Only three classes of combination drugs appeared in the corpus: ACE inhibitor + diuretics, ARBs + diuretics, and Sulfonylureas + metformin. Most classes of medications occurred relatively infrequently, with only 3 appearing “during DCT” in more than half the corpus (i.e., more than 652 records). The “during DCT” annotations indicate that the patient was taking the medication at the time of the visit, indicating that it was a necessary part of their medical health, and presumably being used to actively treat one of the risk factors we were interested in tracking. As we previously noted, if a person has high blood pressure and is taking a medication to treat hypertension, that can indicate that the hypertension is not under control and the patient is therefore at higher risk for CAD. The medications that more than half the patients were actively taking are aspirins (708), beta-blockers (753), and statins (693). 947 records (72.6%) contain patients who are taking metformin (289), insulin (349), sulfonylureas (239), thiazolidinediones (60), or drugs containing a combination of sulfonylureas and metformin (10) (i.e., medication classes associated with diabetes) during the DCT.
Table 11.
Distribution of medication tags
| Medication category | Before DCT | During DCT | After DCT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Total | Train | Test | Total | Train | Test | Total | |
| ACE inhibitor | 322 | 205 | 527 | 314 | 195 | 509 | 319 | 200 | 519 |
| ACE inhibitor + diuretic | 4 | 5 | 9 | 4 | 5 | 9 | 5 | 5 | 10 |
| Amylin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| anti diabetes | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 |
| ARB | 95 | 59 | 154 | 90 | 57 | 147 | 94 | 57 | 151 |
| ARB + diuretic | 3 | 8 | 11 | 3 | 7 | 10 | 3 | 5 | 8 |
| aspirin | 424 | 263 | 687 | 435 | 273 | 708 | 424 | 262 | 686 |
| Beta blocker | 469 | 276 | 745 | 472 | 281 | 753 | 470 | 278 | 748 |
| Calcium channel blocker | 186 | 129 | 315 | 178 | 128 | 306 | 181 | 128 | 309 |
| diuretic | 113 | 79 | 192 | 99 | 69 | 168 | 105 | 71 | 176 |
| DPP4 inhibitors | 1 | 2 | 3 | 0 | 2 | 2 | 0 | 2 | 2 |
| Ezetimibe | 12 | 11 | 23 | 12 | 11 | 23 | 12 | 14 | 26 |
| Fibrate | 22 | 31 | 53 | 20 | 29 | 49 | 22 | 30 | 52 |
| GLP1 agonists | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Insulin | 204 | 132 | 336 | 218 | 131 | 349 | 212 | 132 | 344 |
| Metformin | 187 | 125 | 312 | 176 | 113 | 289 | 181 | 118 | 299 |
| Niacin | 7 | 9 | 16 | 6 | 7 | 13 | 7 | 9 | 16 |
| Nitrate | 117 | 91 | 208 | 126 | 95 | 221 | 93 | 85 | 178 |
| Obesity meds | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Statin | 436 | 274 | 710 | 427 | 266 | 693 | 438 | 277 | 715 |
| Sulfonylureas | 154 | 93 | 247 | 150 | 89 | 239 | 152 | 91 | 243 |
| Sulfonylureas + metformin | 5 | 5 | 10 | 5 | 5 | 10 | 5 | 5 | 10 |
| Thiazolidinedione | 43 | 22 | 65 | 41 | 19 | 60 | 40 | 20 | 60 |
| Thienopyridine | 97 | 97 | 194 | 98 | 95 | 193 | 97 | 92 | 189 |
Overall, risk factor “mentions” and medications tend to appear before, during, and after the DCT, while specific events tend to have occurred in the past, with the exception of high blood pressure measurements which mostly take place during the DCT. For the most part, the distribution of indicators between training and testing data were roughly proportional to the 60/40 split of the corpus overall.
8. 2014 i2b2/UTHealth Risk Factors for CAD in Diabetic Patients Shared Task
We used the voted gold standard for the 2014 i2b2/UTHealth shared task Risk Factor track. Overall, we received 49 submissions from 20 teams for this track. We calculated the aggregate precision, recall, and f-measure at the micro- and macro-averaged levels for evaluating the submissions. To do this, we compared the document-level gold standard annotations to the document-level annotations generated by the systems. Table 12 shows the aggregate scores for all the submitted systems.
Table 12.
Aggregate statistics for all systems compared to the gold standard
| Min. | Mean | Median | Max | Std. Dev. | |
|---|---|---|---|---|---|
| Micro-averaged Precision | .455 | .808 | .852 | .913 | .119 |
| Micro-averaged Recall | .203 | .835 | .908 | .969 | .175 |
| Micro-averaged F-measure | .305 | .815 | .872 | .928 | .145 |
| Macro-averaged Precision | .455 | .800 | .849 | .914 | .121 |
| Macro-averaged Recall | .258 | .834 | .904 | .968 | .162 |
| Macro-averaged F-measure | .365 | .812 | .870 | .928 | .137 |
Table 12 shows that the maximum scores are slightly lower than the agreement achieved by the annotators, but only by relatively small margins. The annotator's micro- and macro-averaged f-measures were .959 and .958, respectively, with the top system scoring .928 for both, differences of only .031 and .03. In fact, the top system's recall (.968) was higher than the annotators’ at both the micro- (.960) and macro-averaged levels (.959). A full evaluation of the individual system scores can be found in Stubbs et al (this issue).
9. Discussion
We employed a light annotation paradigm to annotate a corpus of 1,304 longitudinal medical records for the i2b2/UTHealth shared task. We applied this paradigm to generate annotations that address a complex medical question in a reasonably short amount of time, and with high levels of inter-annotator agreement. These data were distributed to the i2b2/UTHealth shared task participants for system development. While one team did create additional annotations to train their systems (Roberts et al., this issue), most of them did not. All of the top ten systems showed micro-averaged f-measures over 0.87 (Stubbs et al., this issue), which indicates that the gold-standard contained sufficient information to train automated systems. However, the feedback at the workshop indicated that participants felt that it would have been easier to train systems if the extents of the textual evidences were standardized and if the annotations had contained negative examples of the risk factor indicators. In the future we will consider small augmentations to our light annotation paradigm that can improve annotations without placing excessive burdens on the annotators.
10. Conclusions
The corpus for the 2014 i2b2/UTHealth NLP challenge is a new resource for researchers in clinical NLP. We annotated this corpus by applying light annotation procedures and determined the gold standard using majority voting. The document-level annotations provided for each record in a longitudinal EMR provide information that can support studies of progression of heart disease risk factors in a patient over time. The annotation of risk factors, associated indicators, and their temporal placements enables the use of NLP to answer important, clinically-relevant questions using the information in patients’ narrative EMRs. The output from systems trained on this data could potentially be used to create visualizations of patient timelines, predict patient outcomes based on larger trends, and show comorbidities between different risk factors and their indicators. This data set is distributed under data use agreements from i2b2.org/NLP.
Highlights.
NLP task focused on identifying risk factors over time in clinical narratives
Corpus of 1,304 longitudinal medical records for 296 patients
“Light” annotation task for domain expert annotators
Gold standard created through voting
Corpus used for track 2 of 2014 i2b2/UTHealth NLP Shared Task
Acknowledgements
This project was funded by NIH NLM 2U54LM008748 PI: Isaac Kohane, by NIH NLM 5R13LM011411 PI: Ozlem Uzuner, and by NIH NIGMS 5R01GM102282, PI: Hua Xu.
We would like to thank Stanley Shaw and Vishesh Kumar for their help creating the annotation guidelines, and we would like to thank our annotators for their excellent work: Zachary Franco, Tara Liddiard, Aubrey Qualls Kathleen Ririe, Katie Ruri, Thao Nguyen, and Tracy Nguyen. We would also like to thank the JBI editor and reviewers for their constructive and thoughtful feedback.
Appendix A
Annotation guidelines:
Risk factors for Heart Disease in Diabetic Patients
Amber Stubbs, Ozlem Uzuner, Vishesh Kumar, Stanley Shaw April 1, 2014
1. Overview
This annotation task will create a set of medical documents that track the progression of heart disease in diabetic patients. Multiple records will be annotated for each patient, which will allow a general timeline to be created from the set. This project uses tags and attributes used to indicate the presence and progression of disease (diabetes, heart disease), associated risk factors (hypertension, hyperlipidemia, smoking status, obesity status, and family history), and the time they were present in the patient's medical history.
2. Tags
Each of the diseases and risk factors associated with this task has its own set of indicators that we will use to identify whether or not the disease or risk factor is present for that patient, and when it is present. This section discusses each tag that will be used in this annotation.
Every tag (except for those related to smoking status and family history) has an “indicator” attribute and a “time” attribute. The indicator attributes are described below; the time attribute is described in Section 3.
For each indicator, only text that matches the description of the indicator should be annotated as evidence that the risk factor is present. If an indicator is negated (“no history of diabetes”), that text should not be annotated.
Please note that medications now have their own tag—see section 2.8 for details.
2.1 Diabetes
Table 1.
indicators and descriptions for the Diabetes tag
| indicator | description |
|---|---|
| mention | a diagnosis of Type 1 or Type 2 diabetes, or a mention of a preexisting diagnosis |
| High A1c | An A1c test value of over 6.5 |
| High glucose | 2 fasting blood glucose measurements of over 126 |
Phrases that should be annotated for Diabetes:
Mention: patient has h/o DMII, diabetic ketoacidosis
Med-Sulfonylureas: Medications on admission: Glyburide, ...
A1c: 7/18: A1c: 7.3
Glucose: Example: (8:00am)glu: 145 [. . . ] (8:00pm)glu: 139
Phrases that should not be annotated for Diabetes:
Sister with h/o DMII
pre-diabetic
Will consider starting Glyburide at next visit
A1c: 6.2
Glucose after eating: 200
2.2 CAD
Table 2.
indicators and descriptions for the Diabetes tag
| indicator | description |
|---|---|
| mention | a diagnosis of CAD, or a mention of a history of CAD |
| event | – MI, STEMI, NSTEMI – revascularization procedures (bypass surgery, CABG, percutaneous) – cardiac arrest – ischemic cardiomyopathy |
| test result | – exercise or pharmacologic stress test showing ischemia – abnormal cardiac catheterization showing coronary stenoses (narrowing) |
| symptom | chest pain consistent with angina |
Phrases that should be annotated for CAD:
mention: PMH: significant for CAD
med-Thienopyridines Discharge medications: continue on Prasugrel;
med-Nitrates increase Nitrostat
event: s/p STEMI in 2004; CABG in 1999
test result: dolbutamine stress test revealing ischemia; cath. of LAD revealed 50% lesion
symptom: patient being treated for stable angina
Phrases that should not be annotated for CAD:
Ruled out for MI
Atrial fib.
Reason for admission: chest pain
Performed stress test; neg. for ischemia
2.3 Hyperlipidemia/Hypercholesterolemia
Table 3.
Indicators and descriptions of Hyperlipidemia/Hypercholesterolemia
| indicator | description |
|---|---|
| mention | a diagnosis of Hyperlipidemia or Hypercholesterolemia or a mention of the patient already having that condition |
| high cholesterol | total cholesterol of over 240 |
| high LDL | LDL measurement of over 100 mg/dL |
Phrases that should be annotated for Hyperlipidemia:
mention: I agree with his risk factor modification including control of his diabetes, hypertension, and hypercholesterolemia.
me-Statin: Current medications: Lipitor [...]
high cholesterol: The result of your latest chol. test is 250
high LDL: latest LDL: 135
Phrases that should not be annotated for Hyperlipidemia:
Allergies: Lipitor
At risk for high chol.
Will start Lipitor if test results not improved on next visit
LDL 90 mg/dL
Chol. good @ 190
2.4 Hypertension
Table 4.
indicators and descriptions for the Hypertension tag
| indicator | description |
|---|---|
| mention | a diagnosis of Hypertension or a mention of a pre-existing condition |
| high blood pressure | BP measurement of over 140/90 mm/hg (if either value is high, the patient has hypertension) |
Phrases that should be annotated for Hypertension:
mention: PMH: HTN
med-Beta blockers: His current medications include Toprol, and Glucophage
high blood pressure: at admit, bp 140/100
Phrases that should not be annotated for Hypertension:
At risk for HTN
bp 120/80
Allergies: Toprol
2.5 Obesity
Table 5.
indicators and descriptions for the Obesity tag
| indicator | description |
|---|---|
| mention | a description of the patient as being obese |
| BMI | BMI over 30 |
| waist circumference | Waist circumference measurement of: • men: 40 inches or more • women: 35 inches or more |
Phrases that should be annotated for Obesity:
mention: 57y/o obese white male
BMI: spoke to patient about lowering BMI (31.4 last August)
waist circumference: 42in waist meas.
Phrases that should not be annotated for Obesity:
- weight: 340
- ⎛ weight alone is not an indicator of obesity
- lost 150lbs over past year
- ⎛ weight/weight loss is not an indicator of obesity
2.6 Family History of premature CAD
The FAMILY_HIST tag has only an indicator attribute that has the values “present” and “not present”. This tag should only be marked as present if the patient is has a first-degree relative (parents, siblings, or children) who was diagnosed prematurely (younger than 55 for male relatives, younger than 65 for female relatives) with CAD. Family histories of diabetes or any of the other risk factors should not be annotated.
Phrases that should be annotated for Family History of CAD:
Family/social History: Father diagnosed w/ CAD at 49
Fam.hist. significant for premature CAD
Phrases that should not be annotated for Family History of CAD:
- Father w/ CAD, died at 82yrs
- ⎛ unknown when diagnosis occurred
- No known relatives with CAD
- ⎛ does not confirm CAD
- Both grandfathers prem. CAD
- ⎛ not first-degree relatives
2.7 Smoker
The SMOKER tag does not have indicator or time attributes. Instead, it has a status attribute that indicates whether the person is currently a smoker (CURRENT), used to smoke but quit over a year ago (PAST), smoked at some point but it is unclear if they are still smoking or have quit (EVER), has never smoked (NEVER), or if their smoking is not mentioned (UNKNOWN).
For the purposes of this research, a person is considered to be a current smoker if they have smoked at all within the past year. A patient is given “past” status if they quit smoking more than a year ago, “ever” status if it is unclear whether they have quit and “never” if the text states that they have never been a smoker. If no information is provided about their smoking status, they are to be labeled as “unknown”.
Phrases that should be annotated for Smoker:
CURRENT: Patient says trying to quit 1pack/day habit; quit 6mos ago
PAST: Social history: patient used to smoke, quit 10 yrs ago
- EVER: quit smoking; remote history of tobacco dependence
- ⎛ Unless we know how “remote” the history is, it has to be “ever”
NEVER: denies tobacco, alcohol, illicits
UNKNOWN: (no mention of smoking/tobacco use)
Phrases that should not be annotated for Smoker:
recreational marijuana
chewing tobacco
2.8 Medications
Due to overlaps between medications prescribed for different diseases/risk factors, MEDICATION is now its own tag. The list below contains all the different categories of medications that we are interested in for this task.
Please remember that you should annotated every category of medication that the patient is on—do not only annotate one CAD medication if the person is taking, for example, both aspirin and a beta blocker—annotate both.
While you do not need to indicate the risk factor/disease related to each medication, the correlations are provided here for reference:
Diabetes: Metformin, insulin, sulfonylureas, thiazolidinediones, GLP-1 agonists, Meglitinides, DPP-4 inhibitors, Amylin, anti-diabetes medications, combinations including these
CAD: Aspirin, Thienopyridines, beta blockers, ACE inhibitors, nitrates, calcium-channel blockers, combinations including these
Hyperlipidemia: statins, fibrates, niacins, ezetimibes, combinations including these
Hypertension: beta-blockers, ACE inhibitors ARBs, Thiazide diuretics, calcium-channel blockers, combinations including these
Obesity: orlistat (xenical) or Lorqess (Lorcaserin)
The medication tag has two “type” attributes for identifying what category of drug(s) a medication falls into. For combination drugs, indicate both categories. For drugs that are only of one type, the “type2” attribute can be left blank.
Phrases that should be annotated for medications:
ACE inhibitor: Current medications: Lisinopril
Thiazolidinediones & metformin: increase Avandamet
etc
Phrases that should not be annotated for medications:
 Consider starting Humalog if next A1c measurement too high
Consider starting Humalog if next A1c measurement too high Allergic to Simvastatin
Allergic to Simvastatin
3. The Time attribute
Every tag (except for SMOKER and FAMILY_HIST) has a time attribute that is used to show when the indicator for each medical problem is known to have existed. These reflect when the indicator occurred/was active in relation to the date the medical record was written. This document creation time (DCT) most often refers to an entire day, usually the admission date for a hospital stay, the day of an outpatient visit, or the date a report is written describing the events of a previous hospital stay or doctor's visit. Whichever of these is used, all indicators should be annotated in relation to the DCT. The possible values for the time attribute are:
 continuing
continuing before DCT
before DCT during DCT
during DCT after DCT
after DCT not mentioned
not mentioned
The rest of this section discusses when to use each possible attribute value.
3.1 The DCT
All the files in this dataset start with the line “Record date: “ followed by the calendar date that the record was written. This date is the Document Creation Time (DCT), and it is the date against with all the risk factors and indicators in the document are evaluated. Because the DCT is always a calendar date, you can think about it as a “container” for the events of the day—we aren't concerned with when in the container the events occurred, just whether they occurred inside it or overlap with it either before, after, or both.
In most records, the DCT is the date of the patient's visit to the doctor, or possibly the date of one day of a multi-stay visit. In either of those cases, simply evaluate the risk factors in the record in relation to the date of that record. In other words, if the record describes one day in a multi-day visit, don't try to extrapolate the events to the end of the patient's stay.
In some cases, the records are summary letters sent from a specialist to a PCP. In those cases, use the date of the letter as the DCT, rather than the date of the visit being described.
3.2 “Continuing” times
The “continuing” value for the time attribute is used to show that whatever the annotated indicator is, it was present before the DCT, during day of the DCT, and continued after the day of the DCT. In general, this attribute value will only be used for indicators that represent a state or condition that is unlikely to change, such as an existing diagnosis (and corresponding mention) of diabetes, CAD, hypertension, etc. This attribute may also be used if a medication is mentioned in the context of “continue/increase XXX med. following discharge”.
Correct examples of “continuing” times:
CAD, mention: h/o CAD
Diabetes, mention: diabetes diagnosed 2004
Hyptertension, mention: PMH significant for HTN
Hyperlipidemia, mention: current problems: Hyperlipidemia, GERD, Arthritis.
- Obesity, mention: Obese white female, 45yo
- ⎛ (note that this doesn't mean the person is obese their whole life, just in the time surrounding that DCT)
Medication: Discharge instructions: continue on Glucophage
CAD, symptom: patient being treated for ongoing stable angina.
Medication: continue full-dose aspirin and plavix
Incorrect “continuing” times:
Diabetes, mention: This visit confirms diabetes
Obese, mention: weight still around 260
CAD/HTN, mention: at risk for CAD/HTN
any measurement such as blood pressure, cholesterol, LDL, blood glucose, A1c, etc
any CAD-related event or test
Note that “continuing” can be used for medications, events, etc on the Incorrect list only if “continuing” is being used to indicate that those things are not mentioned in the text (see Section 4.1 for more information).
3.2.1 “Continuing” medications
Previous versions of this guideline suggested that medications should not, in general, be considered continuing unless specifically stated. However, this assessment was incorrect. A more correct interpretation (provided by our MD consultants) is that if a medication is mentioned in a list, whether it be “medications on admission” or “medications”, or just in a list with no title, the assumption has to be that the patient was on them before and during the DCT, and unless a medication is mentioned specifically as being discontinued, will be on them afterwards as well.
This assumption applies to write-ups of inpatient and outpatient visits, as well as to summary letters written from a specialist to a PCP. In the case of the summary letters, the DCT is still the date that the document was written, not the date of the visit (which might not even be stated in the document itself).
3.3 “Before DCT” times
This attribute value is used to indicate that the indicator can only be stated to be present prior to the date of the record. This will most often apply to:
lab values from previous tests
events or symptoms that occurred prior to admission
Correct “before admit” annotations:
Hyperlipidemia, high LDL: lab values from previous visit: LDL: 135, [...]
CAD, event: s/p CABG 2004
Incorrect “before admit” annotations:
- CAD, test result: An exercise Myoview performed in 01/1999 revealed no evidence of inducible ischemia
- ⎛ If the test did reveal ischemia, then this would be a valid “before admit” annotation
3.4 “During DCT” times
The “during DCT” value for the time attribute indicates that a risk factor indicator occurred the day of the date on the record. This will generally apply to:
tests or surgeries performed during the day the document was written
medications prescribed during that day
symptoms occurring that day (that are not indicated as being ongoing problems)
Note that if a patient is diagnosed with a condition such as CAD, diabetes, etc. for the first time during a visit, those mentions should be annotated as both “during DCT” and “after DCT” (see next section), as those conditions will be ongoing from that time.
Correct “during DCT” annotations:
Hypercholestorolimia, high cholestorol: today's lab values: Chol. 247 ...
Hypertension, medication: started on Lisinopril
CAD, symptom: patient developed chest pain during stay
Diabetes, mention: patient confirmed as diabetic, will be trained in managing blood sugar
Incorrect “during DCT” annotations:
Events or test results that are from a previous visit
Events, tests, medications, etc, that are being considered or scheduled, but whose results are not in the record
3.5 “After DCT” times
The “after DCT” value for the time attribute indicates that the risk factor indicator applies to the days after the date of the record. This will mostly be used for:
newly prescribed medications (in conjunction with a “during DCT” annotation)
ongoing diagnoses that were discovered during the day
Correct “after discharge” annotations:
Hyptertension, medication: Discharge medications: Diovan [...]
Diabetes, mention: patient confirmed as diabetic, will be trained in managing blood sugar
CAD, medication: patient continuing aspirin regimen begun this visit...
Incorrect “after discharge” annotations:
Any medication: that is only being suggested, recommend talking to GP about starting (medication); will consider starting (medication) next visit
Any mention or indicator that is mentioned as a possibility, that is being recommended, or that is suggested, but that is not definitely performed
4. Annotation Procedure
The procedure for annotating explicit and implicit events is that every risk factor/indicator/time combination mentioned in the document should be addressed in the annotations.
4.2 Completed annotations
When an annotation is completed, all of the risk factors with multiple indicators (Diabetes, Hypertension, Hyperlipidemia, Obese) should, at minimum, have tag(s) for each indicator that is present in the text.
Note that it's fine to annotate multiple mentions of the same indicator if you want to, as long as all the indicators mentioned in the text have at least one tag.
5. Annotation software
Annotation will be done using MAE (Multi-purpose Annotation Environment). The newest test version is available at your account on the UAlbany server, packaged with the test annotation files and these instructions. On most systems you can run the .jar file by double-clicking, loading the DTD, then loading the file you want to annotate. Best practice is to run the software from the command line (see the included user guide).
6. A note on HIPAA/PHI
While the files we are sharing with you have been de-identified and changed to protect the identities of the patients and doctors, we ask that you not share these files or send them to anyone. If you know someone who wants access to the files, please direct them to Amber Stubbs (astubbs@albany.edu). If you find information in the files that seems to be original PHI, please contact Amber immediately.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Works Cited
- Kumar Vishesh, Stubbs Amber, Shaw Stanley, Uzuner Ozlem. Creation of a new longitudinal corpus of clinical narratives. doi: 10.1016/j.jbi.2015.09.018. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller Timothy, Bethard Steven, Dligach Dmitriy, Pradhan Sameer, Lin Chen, Savova Guergana. Proceedings of the 2013 Workshop on Biomedical Natural Language Processing. Association for Computational Linguistics; Sofia, Bulgaria: 2013. Discovering Temporal Narrative Containers in Clinical Text. pp. 18–26. [Google Scholar]
- NDIC (National Diabetes Information Clearinghouse) [February 19, 2014];Diabetes, Heart Disease, and Stroke. http://diabetes.niddk.nih.gov/dm/pubs/stroke/index.aspx.
- Pestian John P., Brew Christopher, Matykiewicz Paweł, Hovermale DJ, Johnson Neil, Bretonnel Cohen K, Duch Włodzisław. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing (BioNLP '07) Association for Computational Linguistics; Stroudsburg, PA, USA: 2007. A shared task involving multi-label classification of clinical free text. pp. 97–104. [Google Scholar]
- Pustejovsky James, Stubbs Amber. 2011 Proceedings of the Linguistic Annotation Workshop V, Association of Computational Linguistics. Portland, Oregon: Jul 23-24, 2011. Increasing Informativeness in Temporal Annotation. [Google Scholar]
- Stubbs Amber. 2011 Proceedings of the Linguistic Annotation Workshop V. Association of Computational Linguistics; Portland, Oregon: Jul 23-24, 2011. MAE and MAI: Lightweight Annotation and Adjudication Tools. [Google Scholar]
- Stubbs Amber. Doctoral Dissertation. Brandeis University; Feb, 2013. A Methodology for Using Professional Knowledge in Corpus Annotation. [Google Scholar]
- Stubbs Amber, Kotfila Christopher, Xu Hua, Uzuner Ozlem. Identifying risk factors for heart disease over time: Overview of the 2014 i2b2/UTHealth shared task Track 2. doi: 10.1016/j.jbi.2015.07.001. this special issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs Amber, Uzuner Ozlem. De-identifying longitudinal medical records. (This special issue) [Google Scholar]
- Suominen Hanna, Salanterä Sanna, Velupillai Sumithra, Chapman Wendy W., Savova Guergana, Elhadad Noemie, Pradhan Sameer, South Brett R., Mowery Danielle L., Jones Gareth J.F., Leveling Johannes, Kelly Liadh, Goeuriot Lorraine, Martinez David, Zuccon Guido. Multilinguality, Multimodality, and Visualization. Springer; Berlin Heidelberg: 2013. Overview of the ShARe/CLEF eHealth Evaluation Lab 2013. Information Access Evaluation. pp. 212–231. [Google Scholar]
- Sun W, Rumshisky A, Uzuner O. Evaluating Temporal Relations in Clinical Text: 2012 i2b2 Challenge Overview. Journal of the American Medical Informatics Association. 2013 doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Özlem. Recognizing Obesity and Comorbidities in Sparse Data. Journal of the American Medical Informatics Association. 2009 Jul-Aug;16(4):561–570. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Özlem, Goldstein Ira, Luo Yuan, Kohane Isaac. Identifying patient smoking status from medical discharge records. Journal of the American Medical Informatics Association. 2007;15:14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö , Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010;17:514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization) [2014];Health Topics: Risk Factors. http://www.who.int/topics/risk_factors/en/.