Abstract
Biomaterials are extensively used to restore damaged tissues, in the forms of implants (e.g. tissue engineered scaffolds) or biomedical devices (e.g. pacemakers). Once in contact with the physiological environment, nanostructured biomaterials undergo modifications as a result of endogenous proteins binding to their surface. The formation of this macromolecular coating complex, known as ‘protein corona’, onto the surface of nanoparticles and its effect on cell-particle interactions are currently under intense investigation. In striking contrast, protein corona constructs within nanostructured porous tissue engineering scaffolds remain poorly characterized. As organismal systems are highly dynamic, it is conceivable that the formation of distinct protein corona on implanted scaffolds might itself modulate cell-extracellular matrix interactions. Here, we report that corona complexes formed onto the fibrils of engineered collagen scaffolds display specific, distinct, and reproducible compositions that are a signature of the tissue microenvironment as well as being indicative of the subject's health condition. Protein corona formed on collagen matrices modulated cellular secretome in a context-specific manner ex-vivo, demonstrating their role in regulating scaffold-cellular interactions. Together, these findings underscore the importance of custom-designing personalized nanostructured biomaterials, according to the biological milieu and disease state. We propose the use of protein corona as in situ biosensor of temporal and local biomarkers.
Keywords: protein corona, tissue engineering scaffold, collagen, personalized nanostructured biomaterials, in situ biosensor
1. Introduction
The surface of nanostructured biomaterials (e.g. nano particles, tubes, and capsules) is covered by various types of macromolecules (e.g. proteins), upon their entrance to the biological fluids.[1–3] The absorption of these biomolecules onto biomaterial surfaces might confer them a new ‘biological identity’.[1,4–6] Therefore, in reality what the cells ‘see’ when interacting with implanted constructs is the protein coated nanostructures (so called ‘protein corona’) on their surface, rather than the pristine materials.[3,7] These protein constructs consist of hard corona (the irreversibly-bounded proteins) and soft corona (a dynamic exchange of proteins between the biomaterial surface and media).[8] Accordingly, once administered in vivo, the biological fate of biomaterials could be defined by the type and amount of the associated proteins in the composition of the protein corona.[2,9] For instance, negatively charged, poly(acrylic acid)-conjugated, gold nanoparticles can bind to fibrinogen and change its conformation. This new surface can promote interaction of nanoparticles with the integrin receptor and, consequently, release inflammatory cytokines.[9] The composition, conformation, and the quantity of the proteins constituting the corona medium are strongly dependent on factors including physicochemical properties of biomaterials,[10] incubation temperature[11,12] and time,[13] protein source,[4] and protein concentration.[14]
Although the formation of protein corona at the surface of various nano biomaterials has been investigated,[1,10] there is yet no report on the development of such protein constructs onto nanostructured porous scaffolds and their physiological implications in diverse tissue engineering applications. In addition to serving as a mechanical support, an optimal tissue engineering scaffold also provides an ‘informative’ biomimetic environment for the cells to direct them towards the targeted tissue regeneration pathway(s).[15–18] Whereas a significant amount of research has been conducted on the communications between various cells and scaffolding systems,[19–22] little is known about how the numerous macromolecules (proteins) existing in the biologic media may interact with the implanted biomaterial and its potential effects on the scaffold function. Thus, evaluation of protein corona, formed onto the scaffolds in vivo, may offer a greater understanding of the host tissue response to the implanted biomaterials.
In this study we examined the formation of the protein corona and cell interactions with protein corona, using an in vitro-reconstituted, type I collagen gel scaffold both in vitro and in vivo (Figure 1). The composition and the structure of the protein corona were probed under various physiologic conditions, including grafting onto the heart and subcutaneous muscle tissues in several murine disease models.
Figure 1.
Schematic illustration of the patch-corona study; A: Plastic compression of highly hydrated collagen gels was used to generate a dense fibrillar scaffold structure (i.e. collagen patch).[38,44] The panel on the right shows scanning electron microscopy of the collagen patch fibrillar ultrastructure. B: Collagen scaffolds were incubated in different biological environments including i: fetal bovine serum, and C57 mouse serum and plasma, in vitro, ii: onto the sham (healthy) or myocardial infarcted (MI) heart tissue in C57 mouse, in vivo, and iii: onto the subcutaneous muscle tissue in healthy or MI C57 mouse, in vivo. C: two hours post incubation under each condition (i-iii), patches were removed and the protein coronas formed onto the scaffold were analyzed using SEM, SDS-PAGE, LC-MS/MS, and Luminex (human cytokines) immunoassay.
2. Results and Discussion
Upon their entrance to the body, the surface of biomaterials will be covered by a ‘protein corona’ consisting of a wide variety of biomolecules.[1,4,5] The way cells interact with the scaffold biomaterial strongly depends on the patch-biologic media interactions.[23,24] This study examined the formation of the protein corona onto collagen matrices in various protein environments (Figure 1). We tested the hypothesis that adsorption of various macromolecules, secreted/existing in the local biologic microenvironment, onto the patch biomaterial can be used to analyze the cardiac secretome in normal tissue and during injury, such as myocardial infarction.
2.1. Gel Electrophoresis Analysis of the Hard Protein Corona Formed on Collagen Scaffolds in Vitro
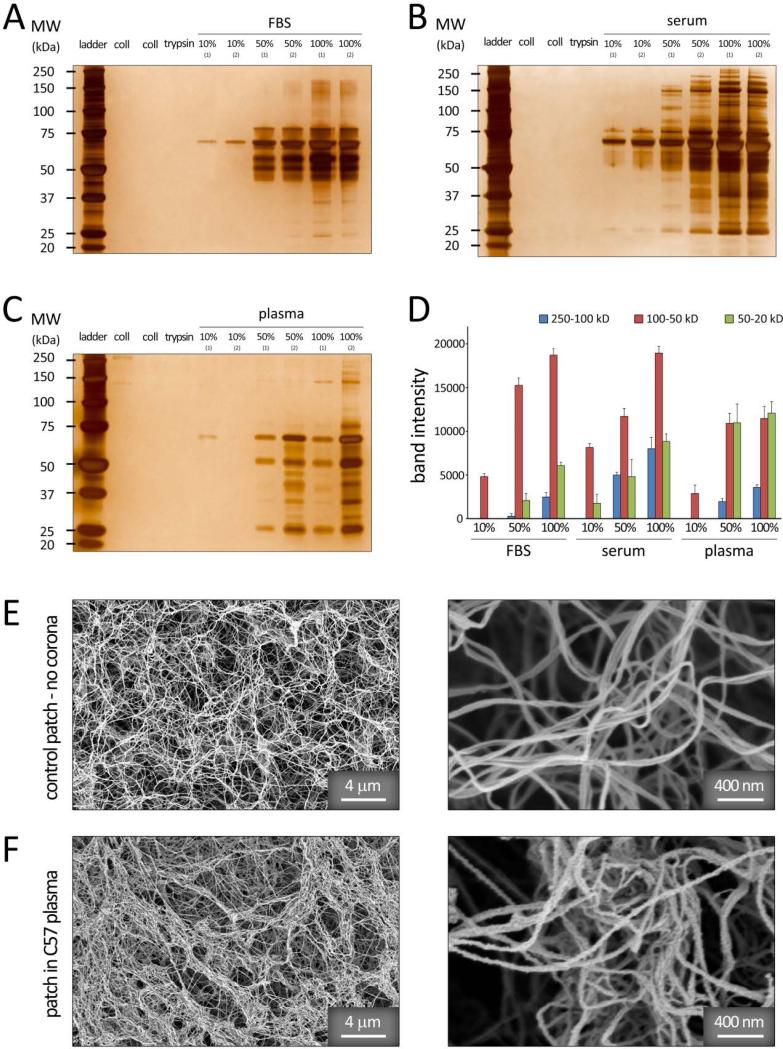
Patch incubation in various in vitro protein media (FBS, and C57 mouse serum and plasma) demonstrated significant differences in the composition and band intensities of protein corona adsorbed on the fibrous scaffold under varying conditions (Figure 2A-C). Semi-quantitative densitometry was utilized to further assess the relative amounts of proteins classified by molecular weight (20-50, 50-100, and 100-250 kDa) (Figure 2D), demonstrating significant variations in the protein pattern of scaffold hard coronas. The most striking observation was the association of low molecular weight proteins (Mw<50 kDa) in the corona composition of C57 plasma samples (with protein concentrations of 50% and 100%) (Figure 2D). The coagulation proteins (e.g., fibrinogen-based proteins) can be responsible for the observed difference of protein profiles from plasma and serum sources. These results were in agreement with our previous findings, demonstrating the variation of protein coronas formed on the surface of superparamagnetic iron oxide nanoparticles incubated in plasma versus serum.[4]
Figure 2.
1D SDS–PAGE hard-corona protein profile for collagen patches incubated with various concentrations of (A) FBS, (B) C57 mouse serum, and (C) C57 mouse plasma proteins. The molecular weights (KDa) of the proteins in the standard ladder are reported on the left for reference. D: Histogram demonstrating the total band intensity of proteins recovered from various corona patches in (A-C). E-F: SEM images of the collagen patch before (E) and after incubation in vitro (in C57 plasma, F) confirming the adsorption of various protein macromolecules onto the collagen fibrils.
Incubation in 10% plasma/serum resulted in relatively smaller amount and compositional range of associated proteins in the corona structure, when compared with those formed in 50% and 100% plasma/serum (Figure 2A-D). Higher plasma/serum concentrations yield greater amount of proteins with relatively higher affinity to the biomaterial surface, while the exposed surface remains constant. This results in formation of a thicker, multi-layer protein corona at elevated plasma/serum concentrations, which in turn, leads to entrapment of a number of excess proteins within the multi-layer structure.[4]
Scanning electron microscopy (SEM) demonstrated the adsorption of various biologic molecules onto the patch and its effect on the fibrillar structure of collagen in vitro (jagged fibrillar surface) (Figure 2E-F).
2.2. Mass Spectrometry – Heatmap Analysis of the Hard Protein Corona Formed on Collagen Scaffolds in Vitro
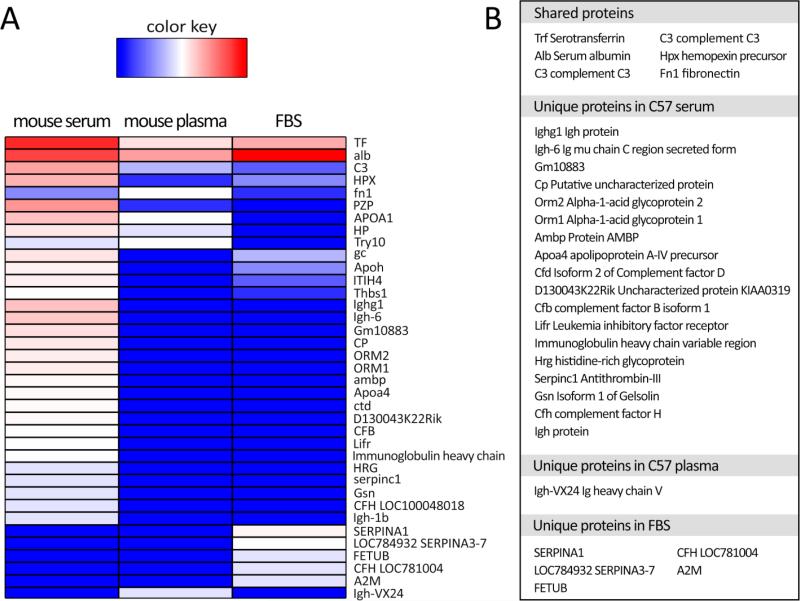
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to identify the common and unique proteins adsorbed onto the collagen matrices in vitro (Figure 3). Their abundances were plotted as a heatmap (Figure 3A) and the shared and unique proteins from each media were identified (Figure 3B). As expected, highly abundant blood proteins were common to all conditions, such as serotransferrin, serum albumin, complement C3, and hemopexin (beta-1B-glycoprotein). However, serum yielded more unique corona proteins (e.g. Ighg1, Orm1 and 2, Ambp, and Apoa proteins) than those found in plasma, suggesting that factors in the plasma prevented the attachment of serum proteins to the scaffold. Greater number of unique proteins in serum versus only one in plasma can be related to the fact that serum is considered as plasma without clotting factors, which suggests that proteins may be able to attach to the scaffold in serum condition more readily than the plasma environment.
Figure 3.
A: Heatmap data visualization of spectral counts of the associated proteins in the corona compositions of various in vitro conditions (C57 mouse serum, C57 muse plasma, and FBS), obtained by mass spectrometry. B: Table lists the proteins shared by C57 serum, plasma and FBS, and proteins unique to each condition.
2.3. Gel Electrophoresis Analysis of the Hard Protein Corona Formed on Collagen Scaffolds in Vivo
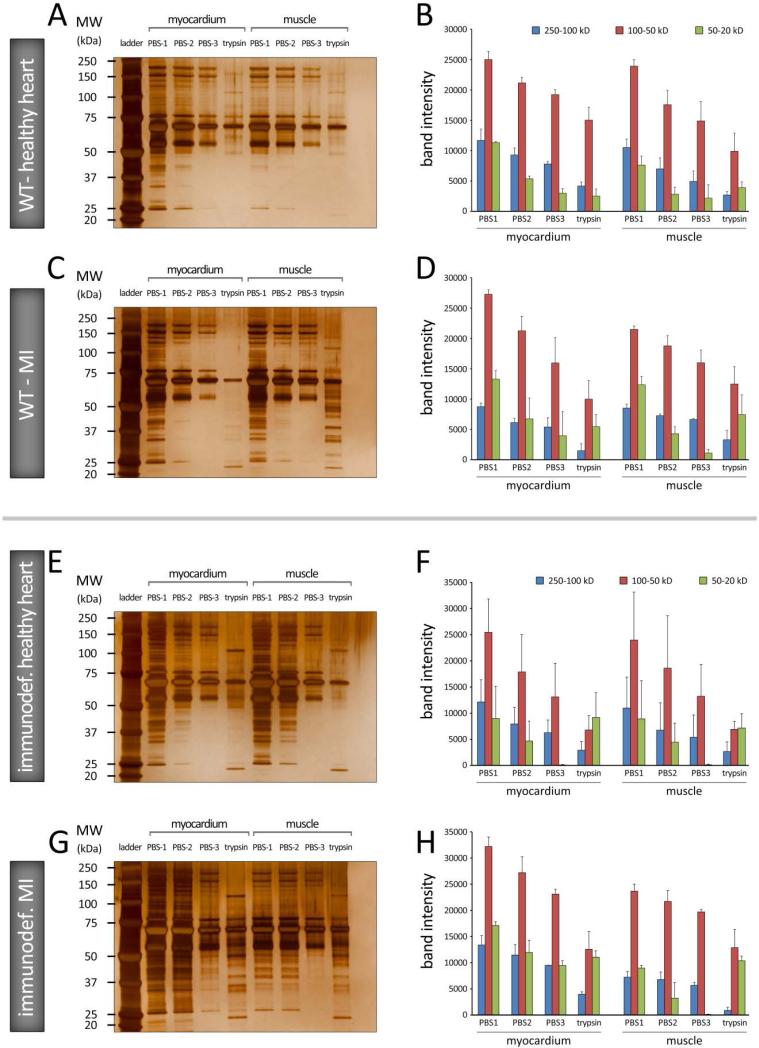
Protein profiles of the collagen patches were also evaluated in vivo, in wild type and immunodeficient (SCID) mice, and in control (sham) and after myocardial infarction (MI). Patches were inserted in two locations: on the epicardial surface of the heart and in peripheral (subcutaneous) muscle. SDS-PAGE analysis of the extracted patches identified patterns of protein complexes within the fibrillar constructs which were dependent both on the tissue type and physiologic conditions (Figure 4). Proteins with mid-range molecular weight (red bars, 50-100 kD) exhibited the greatest band intensities in all conditions. Band intensities declined continuously by subsequent rinses (from PBS1 to 3) (Figure 4B).
Figure 4.
A-D: 1D SDS–PAGE hard-corona protein profile for collagen patches incubated in vivo, in WT C57 BL/6J mice, under healthy heart (A and B) and myocardial infarction (MI) conditions (C and D) conditions. E-H: 1D SDS–PAGE hard-corona protein profile for patches incubated in immunodeficient C57 (SCID Beige) mice, under healthy heart (E and F) and MI (G and H) conditions. Each gel image shows the protein corona information of the patches grafted on myocardial (left) and subcutaneous muscle (right) tissues. The molecular weights (KDa) of the proteins in the standard ladder are reported on the left for reference. B, D, F, and H: Histograms demonstrating the total band intensity of proteins recovered from various corona patches in A, C, E, and G, respectively.
2.4. Mass Spectrometry – Heatmap Analysis of the Hard Protein Corona Formed on Collagen Scaffolds in Vivo
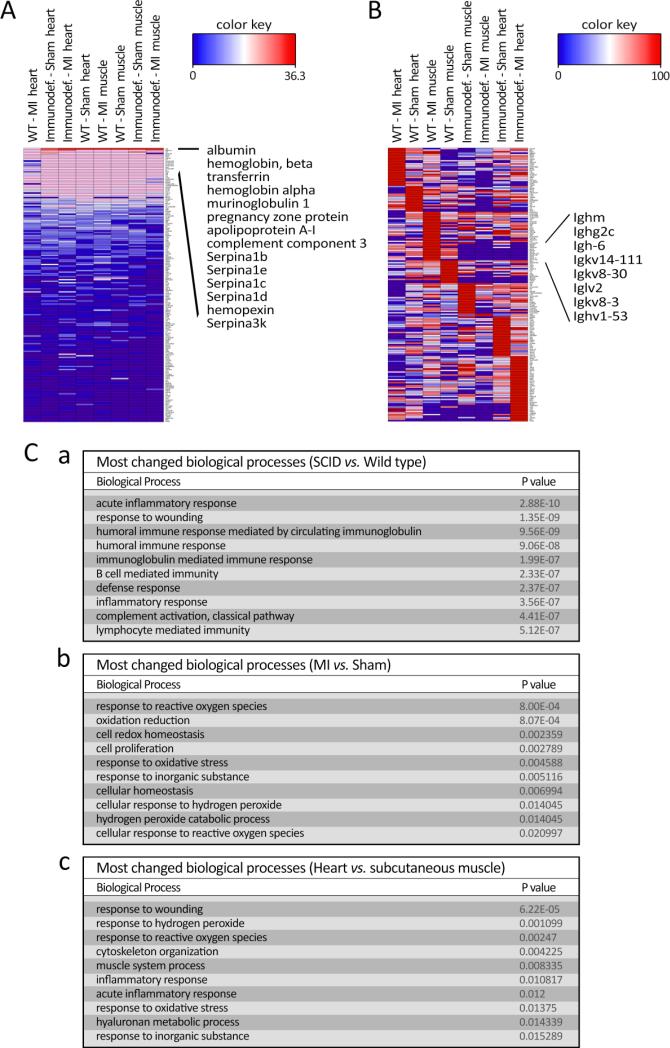
Heatmap visualization of spectral counts of proteins detected by LC-MS/MS was generated by Hierarchical Clustering using total protein spectral count with global normalization (Figure 5A). The common serum proteins (serum albumin, serotransferrin, serpin family proteins) and abundant blood cell proteins (hemoglobins) accounted for the majority of the detected proteins and the overall pattern of detection was similar between the four different conditions. These housekeeping proteins were subsequently subtracted by normalization of the individual protein content in each condition, to highlight the differential detection of proteins (Figure 5B). Individual protein spectral counts are listed in Supplementary Tables S1-20. The majority of the proteins negatively regulated in the SCID condition were components of immunoglobins that are not produced in SCID mice. The absence of immunoglobin-affiliated proteins in the corona structure in the immunodeficient mice suggests that the collagen scaffold representatively captured its surrounding proteins and the corona faithfully recapitulated the in vivo environment that it was embedded into. This may occur due to the significant reduction in the concentration of the immunoglobin proteins in the immunodeficient mouse and their consequent affinity changes (reduction) to the surface of nanostructured scaffold [25,26].
Figure 5.
Heatmap data visualization of spectral counts of the associated proteins in corona composition obtained from different in vivo conditions: A: showing the total spec count ranking obtained by global normalization; and B: demonstrating expression ranking of normalized individual proteins which associated in corona composition of each in vivo condition. C: tables listing P values of top 10 significantly changed biological processes between (a) SCID vs. wild type, (b) MI vs. sham, and (c) heart vs. subcutaneous muscle, ranked by P value.
Grouping the protein counts by WT vs. SCID, heart vs. subcutaneous muscle, and MI vs. sham, produced a comparison of altered proteins in the designated groups (Supplementary Table S1) and the top altered biological processes generated through gene ontology (Figure 5C). Most highly changed biological processes in SCID vs. WT are related to the immune system, including all immunoglobin proteins, validating that the corona from the scaffold faithfully represented its interacting biologic environment. In the case of MI vs. sham our analysis demonstrated the involvement of reactive oxygen species and oxidative stress to be a main biological change during the first two hours after ischemic injury. In particular, Prdx family genes (Prdx1, 2, 6) were specifically enriched in the MI corona, suggesting the notable presence of these novel peroxidase enzymes in the infarct area. Cytoskeleton organization and muscle system process were among the top changes between heart and subcutaneous muscle, suggesting that tissue specific cells were also attracted to the scaffolds and hence, various intracellular proteins (e.g. Actn2, Actn4, and Tuba4a, Supplementary Table S1) abundantly existed in the corona (Figure 5C).
To probe the biosensing capability of the collagen patches, the most enriched proteins binding to scaffolds from in vivo data were defined (Supplementary Figure S1). Based on KEGG pathway analysis of significant proteins binding to collagen, we found that the complement and coagulation cascades were the most enriched pathways involved in binding to the collagen patch.
2.5. Assessment of Cell-Biomaterial Interactions in the Presence of Protein Corona
To assess how cells will react to the different protein coronas formed on the collagen patches, human umbilical vein endothelial cells (HUVECs) were cultured onto the matrices extracted after 2-hour in vivo implantation (Figure 6). Cells showed a normal (healthy) morphology when attached to the fibrillar corona-coated matrices (scanning electron microscopy, Figure 6A). The number of HUVECs attaching to the sham–collagen substrate was qualitatively greater than those attached to MI matrices. Cells cultured on MI versus healthy sham patches showed changes in morphology, with a relative increase in cell surface roughness (jagged cell membrane).
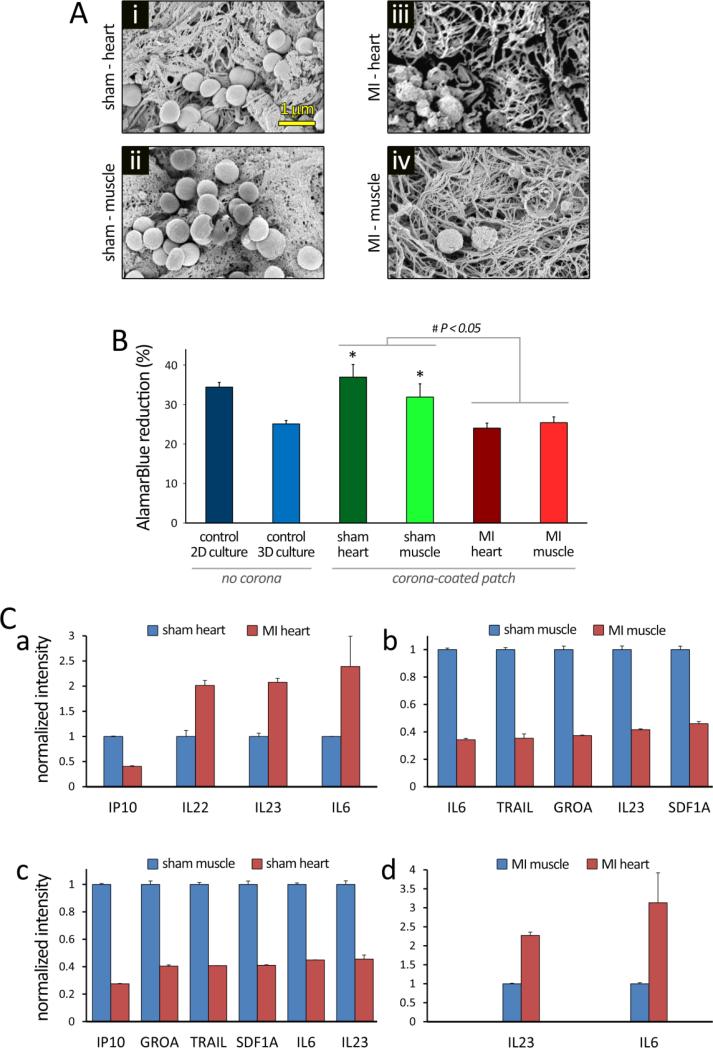
Figure 6.
Evaluation of cells interaction with protein corona-patch following 24 hours of culture in vitro. Human umbilical vein endothelial cells (HUVECs) were plated onto collagen patches that were previously incubated in different in vivo conditions for 2 hours. HUVECs were also cultured on 2D substrate, and on 3D collagen patches, without protein corona, as controls. A: SEM images show HUVECs seeded onto the collagen patches incubated in i) Sham-heart, ii) Sham-muscle, iii) MI-heart, and iv) MI-muscle tissues. Remarkable differences were observed in the patch fibrillar ultrastructure, protein corona formation, and cell number/attachment. B: AlamarBlue assay demonstrated significantly different HUVEC viability/metabolism under varying conditions (P<0.05). *: significantly different compared to the 3D culture control (P<0.05). The differences in the means (± SEM) were tested using ANOVA to check for statistical significance (P < 0.05). C: the 63-plex Luminex Immunoassay of human cytokines detected significant differences in the production of multiple cytokines in the supernatant of HUVECs, after 24 hours of culture in vitro. Patch without cells was used to establish background signal in the assay; values smaller or not significantly greater than background were considered as negative. In all positively detected cytokines, those with levels significantly changed more than 2 folds between conditions are summarized in panels a-d.
To determine the effect of protein corona–coated matrices on cell metabolism, AlamarBlue™ reduction was determined following 24 hrs of culture (Figure 6B). In comparison to the 2D control culture and sham-incubated patches, HUVECs cultured on either 3D patches without corona or MI – incubated patches exhibited significantly reduced metabolic activity (P < 0.05). We further analyzed the pattern of cytokine production (Luminex Immunoassay) of the HUVEC cells after incubation for 24 hrs with control patch (with no protein corona) and different corona coated-patches (incubated in sham heart, sham muscle, MI heart, and MI muscle) (Figure 6C and Supplementary Figure S2). Considerable substrate-dependent differences were observed in the release of positively-detected cytokines, including those related to the inflammatory response (e.g., Interleukin-6 (IL6), IL8, monocyte chemotactic protein 1 (MCP1), and tumor necrosis factor beta (TNFB)), cell adhesion properties (e.g., vascular cell adhesion protein 1 (VCAM1)), and fibrinolysis (e.g., plasminogen activator inhibitor-1 (PAI1)). HUVECs cultured on the patches extracted from MI heart released greater levels of IL22, IL23, and IL6, in comparison with those from sham heart (Figure 6C, panel a). In contrast to the heart tissue, sham muscle induced greater levels of cytokine release in comparison to MI muscle (Figure 6C, panel b).
Among the pool of proteins in the plasma, it is well recognized that unfolded fibrinogen has a crucial role in activation of the integrin receptor (Mac-1 pathway), through its C-terminus of the γ chain.7 Our LC-MS/MS results confirmed that there are significant variations in fibrinogen concentration in the corona compositions of different patches (Supplementary Tables S1-20 and Figure S2). Therefore, one can expect that the observed changes in the inflammatory cytokines could be occurring due to the different decoration of fibrinogen in the corona composition of the patches.
The monocyte chemotactic protein 1 (MCP1) is another inflammatory cytokine that recruits immune cells (e.g., monocytes, T cells, and dendritic cells) into injured tissue[27]. Interestingly, the secretion of MCP1 from HUVEC cells after interaction with various corona coated-patches was significantly higher than the control patch, indicating the effective and accurate role of the corona coated-patch in reflecting the injured tissue microenvironment. In addition, patches obtained from MI heart tissue (with a great level of tissue damage), had the greatest amount of MCP1 compared to the other tissues. It has been shown that the amount of plasminogen activator inhibitor-1 (PAI1) cytokine, an inhibitor of fibrinolysis (the process that degrades blood clots[28]), is significantly increased after MI.[29] Accordingly, the greatest amount of secreted PAI1 we observed was in the supernatant of HUVEC cells interacted with MI heart tissue patch corona.
The cardiac secretome has recently generated substantial interest as new high-throughput methods for detecting small changes in secreted proteins have been developed.[30–33] The secretome can be utilized to detect new signaling pathways associated with cardiac pathology and therefore new drug targets, as well as to search for novel biomarkers,[34,35] which could then potentially be detected in the plasma. However, current techniques used in animal models such as pericardial fluid analysis[36] or plasma sampling have significant limitations. Our results demonstrate the use of a nanostructured collagen-based scaffold as an in situ biosensor in the studies of various disease biomarker discovery, to directly detect – and collect – the diverse macromolecules that are present (even transiently) in the tissue microenvironment. In terms of its potential clinical application, catheter-based techniques can be used to deliver and apply the biosensor patch system to the target tissue/organ.
3. Conclusion
In summary, we demonstrated the formation of complex protein shells onto fibrillar tissue engineering constructs when they were introduced into various biological environments. The protein corona composition was shown to be a function of tissue type and health condition. Moreover, the interaction between corona-coated collagen matrices with endothelial cells demonstrated a significant effect on cell morphology, viability and metabolism, as well as the cytokine profile released by cells. Evaluating the potential of such fibrillar scaffold systems to serve as a local biosensor/collector would further enable the study of the expression of various emerging biomarkers for different diseases, at the cellular and tissue levels.
4. Experimental Section
Preparation of collagen patch
Hydrated collagen gels were fabricated by adding 1.1 ml of 1X DMEM (Sigma, MO, US) to 0.9 ml of sterile, rat tail-derived, type I collagen solution (in acetic acid, 3.84 mg/ml, Millipore, MA, US) and neutralizing by 0.1 N NaOH in a drop-wise manner.[16,37–39] For the entire patch preparation process, collagen solution was kept on the ice (4°C) to prevent premature polymerization of the collagen.[40] The collagen solution (0.9 ml) was subsequently cast into the wells of 24-well plates (d=15.6) and placed in a tissue culture incubator for 30 mins at 37°C for polymerization. Thereupon, highly hydrated collagen gels with poor mechanical integrity were produced.[41–43] To improve the hydrogels physical stability, hydrated collagen gels underwent plastic compression (as previously described[37,41,43]). Briefly, cast collagen gels were removed from the well (used as mold) and transferred to a porous substrate comprising (bottom to top) paper layers, a steel mesh, two polymer meshes, and two fabric meshes (Figure 1A). By applying a static compressive stress of 1400 N/m2 to the scaffold, for 5 minutes, the excess fluid was expelled out of the construct, yielding a dense biomaterial (Figure 1B) with improved biomechanical properties.[16,38,44] Compressed collagen sheets (used as collagen patch) were then transferred to and rinsed in PBS (using the fabric mesh) before incubating in various media.
Extraction of protein corona in vitro
Collagen patches were incubated at different concentrations (10%, 50%, and 100%) of various protein sources including FBS, C57-serum, and C57-plasma. Scaffolds were immersed in 1ml of protein solution (in 24 well plates) and incubated for 2 hrs in the tissue culture incubator (37°C - 5% CO2). Following the incubation in protein source, the supernatants were completely removed and the patches were extensively washed with PBS (3 times, 5 mins each, on the shaker), in order to dislodge loosely attached proteins (aka, ‘soft corona’) from the 3D structure of the patch. Subsequently, to remove the more strongly attached proteins (aka, ‘hard corona’) from the collagen constructs, patches were immersed in 1ml of trypsin (0.05%, HyClone 1X, SH40003.12) for 5 mins and the supernatants were collected. The obtained solutions were used for gel electrophoresis and mass spectroscopy analysis.
Mouse model of myocardial infarction – patch implantation
All procedures involving animal use, housing, and surgeries were approved by the Stanford Institutional Animal Care and Use Committee (IACUC). Animal care and interventions were provided in accordance with the Laboratory Animal Welfare Act. Male, 10-13 weeks old, wild type (C57BL/6J) and severe combined immunodeficiency (SCID Beige) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and studied in four different groups: I) healthy myocardium, II) myocardial infarction (MI), III) immunodeficient mouse with healthy myocardium, and IV) immunodeficient mouse with MI. In each group, the patch was grafted onto both the epicardium and the extra-thoracic subcutaneous muscle (Figure 1B, n=4 per group). Mice were anesthetized utilizing an isoflurane inhalational chamber (2%). This was followed by endotracheal intubation using a 20-gauge angiocatheter (Becton, Dickinson Inc., Sandy, Utah) that was connected to a small animal volume-control ventilator (Harvard Apparatus, Holliston, MA). A left thoracotomy was performed via the fourth intercostal space and the lungs retracted to expose the heart. For the mice with MI, once the pericardium was opened, a 7-0 suture was placed to ligate the left anterior descending artery (LAD) approximately 2 mm below the left atrial appendage. Criterion for occlusion success was an immediate color change in the left ventricular (LV) anterolateral wall (red turning to pale gray). In each animal group, two prepared collagen patches were implanted: 1- one patch was sutured (at two points) onto the ischemic/healthy myocardium, and 2- another patch was placed onto the subcutaneous muscle (no suture), after closing the intercostal space (Figure 1B). The incision was then closed and animals were kept on a heating pad until they were extubated and recovered.
Extraction of protein corona in vivo
Two hours after patch implantation into the various mice groups, the animals were scarified and the patches were extracted from heart and muscle tissues. The extracted collagen scaffolds were then briefly rinsed with saline (20 secs) to remove the blood and nonattached residues. To obtain the in vivo profile of (soft) protein corona complexes formed on the patches, patches were extensively rinsed with PBS (3 times, 5 mins each, on the shaker), and supernatant solutions were collected for further analysis. The patches were then immersed in 1ml of trypsin (0.05%) for 5 mins for removing the attached proteins with higher affinities (hard corona).
Sodium dodecyl sulfate poly-acrylamide gel electrophoresis (SDS-PAGE)
Supernatant solutions obtained from various rinsing steps described above were separately mixed with loading buffer [62.5 mM Tris-HCL pH 6.8, 2% (w/v) SDS, 10% glycerol, 0.04M DTT and 0.01% (w/v) bromophenol blue], followed by boiling for 5 minutes at 100°C. An equal sample volume was loaded in 12% gel polyacrylamide gel. Gel electrophoresis was performed at 120V, 400 mA for ~60 minutes each, until the proteins neared the end of the gel. The gels were stained with silver staining kit. Gels were then scanned using a calibrated densitometer scanner (Bio-Rad Molecular Imager Gel Doc XR+ Imaging System) and gel densitometry was performed using image J (1.410).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Proteins adsorbed on the collagen scaffolds were digested using Filter-Aided Sample Preparation (FASP) method.[45] Proteins reduction and alkylation were carried in presence of 8 M urea on filter by adding DTT (10 mM final concentration) and incubating at 32°C for 60 minutes using thermo-mixer set on 800 rpm. Iodoacetamide was added (to 30 mM final concentration) and proteins were alkylated for 20 minutes at room temperature, in the dark and on thermo-mixer set at 800 rpm. Mass spectrometry grade trypsin (Promega) was added (1:50 ratio) for overnight digestion at 32°C on thermo-mixer. After digestion and peptide elution, formic acid was added to the peptide solution (to 1%), followed by drying the sample to 150 μL and desalting by peptide microtrap (Michrom-Bruker, TR1/25109/02). Desalted peptides were resuspended in 50 μL of 5% ACN in 0.1% Formic Acid/Water; followed by on-line analysis of 20% of total digests by high-resolution, high-accuracy LC-MS/MS, consisting of a Michrom 1D HPLC, a 15-cm Michrom Magic C18 column, a captive spray Michrom source and LTQ-Orbitrap Velos Pro with ETD (Thermo Fisher Scientific). A 45-minute gradient of 10–30%B (0.1% formic acid, 100% acetonitrile) was used to separate the peptides. The LTQ-OrbiTrap Velos Pro was set to scan precursors in the OrbiTrap followed by data-dependent MS/MS of the top 10 precursors and a column heater set to 27°C during the LC-MS/MS runs.
Sorcerer Enterprise v.3.5 release (Sage-N Research Inc.) with SEQUEST algorithm was used as the search program for peptide/protein identification. SEQUEST parameters was set to search the target-decoy ipi.MOUSE.vs3.73 and ipi.BOVINE.vs3.72 fasta protein database; containing protein sequences using trypsin for enzyme with the allowance of up to 2 missed cleavages, Semi tryptic search and precursor mass tolerance of 50 ppm. Differential search included methionine oxidation (16 Da) and cysteines carboxyamidomethylation (57 Da). The search results were viewed, sorted, filtered, and statically analyzed by comprehensive proteomics data analysis software, Peptide/Protein prophet v.4.02 (ISB). In this study, the trans-proteomic pipeline (TPP) protein minimum probability was set to 0.95, to assure very low error (much less than FDR 1%) with reasonably good sensitivity. The differential spectral count analysis was done by QTools, an automated differential peptide/protein spectral count and Gene Ontology analysis tool.[46]
To determine the total number of the mass spectra for the all peptides attributed to a matched protein, a semi-quantitative assessment of the protein amount was conducted through application of the spectral counting (SpC) method by applying the following equation:
| Equation 1 |
Where NpSpCk is the normalised percentage of the spectral count for protein k, SpC is the spectral count identified, and Mw is the molecular weight (in kDa) of protein k.
Scanning electron microscopy (SEM) of patch-corona
To investigate the ultrastructural changes in the collagen scaffolds as a result of interaction with the biologic milieu, patches pre- and post- incubation with biologic media were analyzed using scanning electron microscopy (Zeiss Sigma field emission microscope, FESEM, Carl Zeiss Microscopy, NY) operated at 1-2 kV, using InLens SE detection. Collagen scaffolds were fixed by immersion in 4% paraformaldehyde solution overnight and washed twice with 0.1 M sodium cacodylate buffer (pH 7.4) and then post-fixed in 1% aqueous osmium tetroxide (OsO4) for one hour. Samples were then washed twice in purified water and dehydrated in an increasing ethanol solution series (50, 70, 90, and 100%, 2 × 15 mins each). Finally, the specimens were critical-point dried (CPD) in liquid CO2, in a Tousimis 815B critical-point dryer (Tousimis, MD). CPD-dried samples were mounted on standard SEM stubs with adhesive copper tape and sputter-coated with 4 nm of Au/Pd in a Denton Desk II machine (Denton Vacuum, NJ).
Cell culture and metabolic activity assay
Human umbilical vein endothelial cells (HUVECs) were plated (at a density of 100,000 cells/ml) onto the collagen matrices obtained following 2-hr incubation with different biologic media and were cultured in CS-C medium (Sigma, MO, US) supplemented with 10% Fetal Bovine Serum (FBS, Sigma) in a humidified incubator at 37°C using 5% CO2. AlamarBlue™ (Invitrogen Inc., Carlsbad, CA) was used to measure the metabolic activity of HUVECs as an indicator of cell viability and growth. Following 24 hrs of culture, the medium was renewed and AlamarBlue™ reagent was added to each well at 10% of the culture volume. Samples were then incubated at 37°C for 4 hrs. Acellular scaffolds were used for background reference subtraction. The absorbance of 100 μL of medium was read at 550 and 595 nm using a microplate reader (Cytation 3 Cell Imaging Multi-Mode Reader, BioTek Instruments, Inc., VT, US) and the percentage of reduced AlamarBlue™ was calculated according to the manufacturer's instructions.
Luminex – eBiosciences/Affymetrix Magnetic bead Kit
This assay was performed in the Human Immune Monitoring Center at Stanford University. Patch without cells was used as background signal. Values smaller than background were considered as negative and omitted. Moreover, mouse cytokines that were cross-reactive with human panel were disregarded. Median florescence intensities of 63 human cytokines were probed in the supernatant culture media of the HUVEC cells after incubation for 24 hrs. Human 63-plex kits were purchased from eBiosciences/Affymetrix and used according to the manufacturer's recommendations with modifications as described below. Briefly, beads were added to a 96-well plate and washed in a Biotek ELx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and incubated at room temperature for 1 hr followed by overnight incubation at 4°C with shaking. Cold and Room temperature incubation were performed on an orbital shaker at 500-600 rpm. Following the overnight incubation, plates were washed in a Biotek ELx405 washer and then biotinylated detection antibody added for 75 mins at room temperature with shaking. Plate was washed and streptavidin-PE was added. Following incubation for 30 mins at room temperature, wash was performed as above and reading buffer was added to the wells. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument with a lower bound of 50 beads per sample per cytokine. Custom assay Control beads by Radix Biosolutions were added to all wells.
Statistical analysis
The number of samples (n) used in each experiment is noted in the text. Dependent variables are expressed as means ± SEM. The differences in the means were tested using ANOVA and Student T-test to check for statistical significance (P < 0.05).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants HL65484 and HL086879 (to PRL) and NIH R01 HL095571 (to JCW). VS was an Oak Foundation postdoctoral fellow at Stanford. DB was supported by NIG grant HL061535. KW was a SBMRI California Institute for Regenerative Medicine postdoctoral fellow (TG2-01162).
REFERENCES AND NOTES
- 1.Monopoli MP, Åberg C, Salvati A, Dawson KA. Nat. Nanotechnol. 2012;7:779. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 2.Monopoli MP, Bombelli FB, Dawson KA. Nat. Nanotechnol. 2011;6:11. doi: 10.1038/nnano.2011.267. [DOI] [PubMed] [Google Scholar]
- 3.Lynch I, Salvati A, Dawson KA. Nat. Nanotechnol. 2009;4:546. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 4.Laurent S, Burtea C, Thirifays C, Rezaee F, Mahmoudi M, Colloid Interface Sci J. 2013;392:431. doi: 10.1016/j.jcis.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Mahmoudi M, Saeedi-Eslami SN, Shokrgozar MA, Azadmanesh K, Hassanlou M, Kalhor HR, Burtea C, Rothen-Rutishauser B, Laurent S, Sheibani S, Vali H. Nanoscale. 2012;4:5461. doi: 10.1039/c2nr31185b. [DOI] [PubMed] [Google Scholar]
- 6.Kelly PM, Åberg C, Polo E, O’Connell A, Cookman J, Fallon J, Krpetić Ž, A K. Dawson, Nat. Nanotechnol. 2015 doi: 10.1038/nnano.2015.47. In Press, doi:10.1038/NNANO.2015.47. [DOI] [PubMed] [Google Scholar]
- 7.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J. Am. Chem. Soc. 2010;132:5761. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 8.Mahmoudi M, Shokrgozar MA, Sardari S, Moghadam MK, Vali H, Laurent S, Stroeve P. Nanoscale. 2011;3:1127. doi: 10.1039/c0nr00733a. [DOI] [PubMed] [Google Scholar]
- 9.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF. Nat. Nanotechnol. 2011;6:39. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 10.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Chem. Rev. 2011;111:5610. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 11.Mahmoudi M, Abdelmonem AM, Behzadi S, Clement JH, Dutz S, Ejtehadi MR, Hartmann R, Kantner K, Linne U, Maffre P, Metzler S, Moghadam M, Pfeiffer C, Rezaie M, Ruiz-Lozano P, Serpooshan V, Shokrgozar M, Nienhaus GU, Parak WJ. ACS Nano. 2013 doi: 10.1021/nn305337c. [DOI] [PubMed] [Google Scholar]
- 12.Mahmoudi M, Lohse SE, Murphy CJ, Fathizadeh A, Montazeri A, Suslick KS. Nano Lett. 2014;14:6. doi: 10.1021/nl403419e. [DOI] [PubMed] [Google Scholar]
- 13.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Nat. Nanotechnol. 2013;8:772. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 14.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA. J. Am. Chem. Soc. 2011;133:2525. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 15.Biological Interactions on Materials Surfaces - Understanding and Controlling Protein, Cell, and [Google Scholar]
- 16.Serpooshan V, Julien M, Nguyen O, Wang H, Li A, Muja N, Henderson JE, Nazhat SN. Acta Biomater. 2010;6:3978. doi: 10.1016/j.actbio.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Breuls RG, Jiya TU, Smit TH. Open Orthop J. 2008;2:103. doi: 10.2174/1874325000802010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 19.Murphy CM, O'Brien FJ, Little DG, Schindeler A. Eur. Cell. Mater. 2013;26:120. doi: 10.22203/ecm.v026a09. [DOI] [PubMed] [Google Scholar]
- 20.Venugopal J, Low S, Choon AT, Ramakrishna S. J. Biomed. Mater. Res. B Appl. Biomater. 2008;84:34. doi: 10.1002/jbm.b.30841. [DOI] [PubMed] [Google Scholar]
- 21.Spector M. Tissue Eng. 2002;8:351. doi: 10.1089/107632702760184628. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Liu Y, Jiang Z, Chen W, Yu Y, Hu Q. Exp. Cell Res. 2014;323:346. doi: 10.1016/j.yexcr.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Monopoli MP, Åberg C, Salvati A, A K. Dawson, Nat. Nanotechnol. 2012;7:779. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 24.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J. Am. Chem. Soc. 2010;132:5761. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 25.Ghavami M, Saffar S, Emamy BA, Peirovi A, Shokrgozar MA, Serpooshan V, Mahmoudi M. RSC Adv. 2012;3:1119. [Google Scholar]
- 26.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. J. Am. Chem. Soc. 2011;133:2525. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 27.Carr MW, Roth SJ, Luther E, Rose SS, Springer TA. Proc. Natl. Acad. Sci. 1994;91:3652. doi: 10.1073/pnas.91.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binder BR, Christ G, Gruber F, Grubic N, Hufnagl P, Krebs M, Mihaly J, Prager GW. News Physiol. Sci. Int. J. Physiol. Prod. Jointly Int. Union Physiol. Sci. Am. Physiol. Soc. 2002;17:56. doi: 10.1152/nips.01369.2001. [DOI] [PubMed] [Google Scholar]
- 29.Panahloo A, Mohamed-Ali V, Gray RP, Humphries SE, Yudkin JS. Atherosclerosis. 2003;168:297. doi: 10.1016/s0021-9150(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 30.de la Cuesta F, Alvarez-Llamas G, Gil-Dones F, Darde VM, Calvo E, López JA, Vivanco F, G M. Barderas, Methods Mol. Biol. Clifton NJ. 2013;1005:237. doi: 10.1007/978-1-62703-386-2_19. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Ren Y, Sorokin V, Poh KK, Ho HH, Lee CN, de Kleijn D, Lim SK, Tam JP, Sze SK, Proteomics J. 2014;98:138. doi: 10.1016/j.jprot.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 32.Shimano M, Ouchi N, Walsh K. Circulation. 2012;126:e327. doi: 10.1161/CIRCULATIONAHA.112.150656. [DOI] [PubMed] [Google Scholar]
- 33.Vivanco F, Martín-Ventura JL, Duran MC, Barderas MG, Blanco-Colio L, Dardé VM, Mas S, Meilhac O, Michel JB, Tuñón J, Egido J. J. Proteome Res. 2005;4:1181. doi: 10.1021/pr0500197. [DOI] [PubMed] [Google Scholar]
- 34.Duffy PA, Betton G, Horner S, Horner J, Cotton P, McMahon N, Lawrence C, Prior H, Armstrong D, Philp K, Roberts RA. Eur. J. Cancer Suppl. 2007;5:143. [Google Scholar]
- 35.Tardif J-C, Heinonen T, Orloff D, Libby P. Circulation. 2006;113:2936. doi: 10.1161/CIRCULATIONAHA.105.598987. [DOI] [PubMed] [Google Scholar]
- 36.Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, Ponimaskin E, Schmiedl A, Yin X, Mayr M, Halder R, Fischer A, Engelhardt S, Wei Y, Schober A, Fiedler J, Thum T. J. Clin. Invest. 2014;124:2136. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serpooshan V, Quinn TM, Muja N, Nazhat SN. Acta Biomater. 2012 doi: 10.1016/j.actbio.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 38.Serpooshan V, Zhao M, Metzler SA, Wei K, Shah PB, Wang A, Mahmoudi M, Malkovskiy AV, Rajadas J, Butte MJ, Bernstein D, Ruiz-Lozano P. Biomaterials. 2013;34:9048. doi: 10.1016/j.biomaterials.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serpooshan V, Muja N, Marelli B, Nazhat SN. J Biomed Mater Res A. 2011;96:609. doi: 10.1002/jbm.a.33008. [DOI] [PubMed] [Google Scholar]
- 40.Artym VV, Matsumoto K. Curr. Protoc. Cell Biol. Editor. Board Juan Bonifacino Al. 2010 CHAPTER, Unit. [Google Scholar]
- 41.Abou Neel EA, Cheema U, Knowles JC, Brown RA, Nazhat SN. Soft Matter. 2006;2:986. doi: 10.1039/b609784g. [DOI] [PubMed] [Google Scholar]
- 42.Brown RA, Wiseman M, Chuo CB, Cheema U, Nazhat SN. Adv. Funct. Mater. 2005;15:1762. [Google Scholar]
- 43.Serpooshan V, Quinn TM, Muja N, Nazhat SN. Soft Matter. 2011;7:2918. [Google Scholar]
- 44.Serpooshan V, Zhao M, Metzler SA, Wei K, Shah PB, Wang A, Mahmoudi M, Malkovskiy AV, Rajadas J, Butte MJ, Bernstein D, Ruiz-Lozano P. Bioengineered. 2014;5:193. doi: 10.4161/bioe.27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Nat. Methods. 2009;6:359. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 46.Brill LM, Motamedchaboki K, Wu S, Wolf DA. Methods San Diego Calif. 2009;48:311. doi: 10.1016/j.ymeth.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.