Abstract
Objective
Assess the suitability of high-resolution metabolomics (HRM) for measure of internal exposure and effect biomarkers from deployment related environmental hazards.
Methods
HRM provides extensive coverage of metabolism and data relevant to a broad spectrum of environmental exposures. This review briefly describes the analytic platform, workflow and recent applications of HRM as a prototype environmental exposure surveillance system.
Results
Building upon techniques available for contemporary occupational medicine and exposure sciences, HRM methods are able to integrate external exposures, internal body burden of environmental agents and relevant biological responses with health outcomes.
Conclusions
Systematic analysis of existing Department of Defense Serum Repository samples will provide a high-quality cross-sectional reference dataset for deployment-associated exposures while at the same time establishing a foundation for precision medicine.
Keywords: Mass spectrometry, biomonitoring, analytical chemistry, metabolomics, environmental exposure surveillance, exposome, Department of Defense serum repository
I. Critical need for deployment-associated exposure assessment
In 2011, the United States Institute of Medicine (IOM) recommended that the Department of Defense (DoD) collect individual breathing zone samples and conduct long-term studies of troop health outcomes to address concerns about perceived health risks from exposure during deployment (1–3). Realistically, there are inherent limits to exposure assessment in deployed settings. For example, the use of personal monitoring equipment limits mobility in active combat situations, logistics of sampler collection is challenging with large-scale troop movements, and assessment for biologically relevant dose requires additional molecular measurements. Furthermore, the post-exposure window of opportunity for measuring exposures or immediate consequences may range from hours to days for some agents. Therefore, valid and reliable measures are needed to characterize exposures that do not disrupt effective operation during deployment. Retrospective profiling of biological specimens collected pre- and post-deployment for biomarkers of exposure, effect and susceptibility provide a means of assessing the occurrence of chemical exposure related to poor health outcomes. Through the DoD Serum Repository (DoDSR), an extensive system exists for collection, cataloguing and storing of serum samples collected pre- and post-deployment from active duty armed forces personnel (4, 5). Incorporating chemical screening measures using serum samples collected under the current DoDSR framework could therefore be completed with minimum disruption to military operations.
To fully realize the potential benefits of environmental chemical surveillance and bioeffect monitoring in serum specimens collected from armed forces personnel, there is a need to identify biomarkers relevant to exposure during the deployment period. A number of methods are being evaluated to improve deployment-related exposure assessment using DoDSR samples (1, 6). For instance, biomarkers of combustion products, including dioxins, free and protein-bound polycyclic aromatic hydrocarbon (PAH) are being used to assess burn pit exposures (7); circulating micro-RNA (miRNA), which play an important role in gene expression (8), provide epigenetic measures of biological response to exposure (9); cytokines and cardiovascular health markers are being applied to assess pathophysiological changes during deployment, particularly those related to respiratory pathways. While providing a means to evaluate exposure and biological response to environmental hazards, the measurements discussed above are limited by the need to a priori select chemical targets. A more complete understanding of how environmental exposures contribute to disease susceptibility and progression is required to mitigate risk, develop effective treatment strategies and identify at risk populations. Currently, no unified method exists to characterize the cumulative contribution of environmental and chemical exposures in disease.
A variety of approaches using genomics, metabolomics, lipidomics, transcriptomics, and proteomics are being pursued to determine how external and internal exposures from the environment impact health (10, 11). The application of omic technologies in environmental health research has enabled a more comprehensive measure of the continuum of exposure and bioeffect occurring within human populations. Here, we focus on metabolomics as a general approach to provide a cost-effective means to measure body burden of known chemicals, detect unidentified exposures, and monitor a broad spectrum of metabolic perturbations that could predict potential adverse health outcomes. We first describe high-resolution metabolomics (HRM) using liquid chromatography (LC) coupled to high-resolution mass spectrometry (MS) as an analytical platform to simultaneously evaluate perturbations in metabolism and detect chemicals present at very low levels in biological samples (12). This methodology has been developed for precision medicine (13, 14), and is being refined for use to sequence exposures as part of a human exposome project (15). The exposome was introduced as a complement to the genome to account for environmental contributions to disease risk (16, 17), and is defined as the cumulative measure of environmental influences and biological responses throughout the lifespan, including environmental, dietary, microbiome, behavioral, therapeutic and endogenous processes (18). The second section of the review addresses the use of HRM for measurement of low-level chemical exposure biomarkers. The third section summarizes approaches to use HRM for deployment-associated exposure surveillance, where we address the challenges in using point measurements for reconstructing exposure history and the evolving concepts of exposure memory systems (19). Within the exposure memory framework, we discuss the need to pursue combined analyses of epigenetic changes and other biomarkers. Finally, we provide a brief perspective on opportunities and needs for development of HRM as an integral component of improved deployment exposure surveillance systems and the considerable societal benefit from having an in depth, cross-sectional reference database of high-quality metabolomics data in support of nation-wide precision medicine initiatives.
II. High-resolution metabolomics (HRM): advanced clinical chemistry
II. A Mass spectrometry for metabolic profiling
Mass spectrometry (MS) involves ionization of chemicals in the gas phase with subsequent detection of mass-to-charge ratio (m/z) and ion intensity. Because molecular mass is an absolute property of the chemical, the method is powerful for measurement of endogenous metabolites and environmental chemicals. Multiple types of mass spectrometers are available and have been recently reviewed (20–22). Nutrients and intermediate metabolites are often present in the micromolar (μM) to millimolar (mM) concentration range, and many analytic methods are available for targeted analysis (23–26). Using targeted approaches, analytes measured in biological samples are compared to a series of reference standards selected a priori and utilize optimized analytical workflows (including sample preparation and instrumental analysis) developed to maximize selectivity while providing sufficient sensitivity to detect metabolite levels observed over a prescribed concentration range (27–30). Such approaches are useful for routine evaluation of individual metabolic characteristics and changes in association with deployment, however, due to limited chemical coverage (50–100 analytes) and requirement that analytical targets are selected prior to analysis, there is the potential to not detect unidentified exposure markers and biosignatures occurring during deployment.
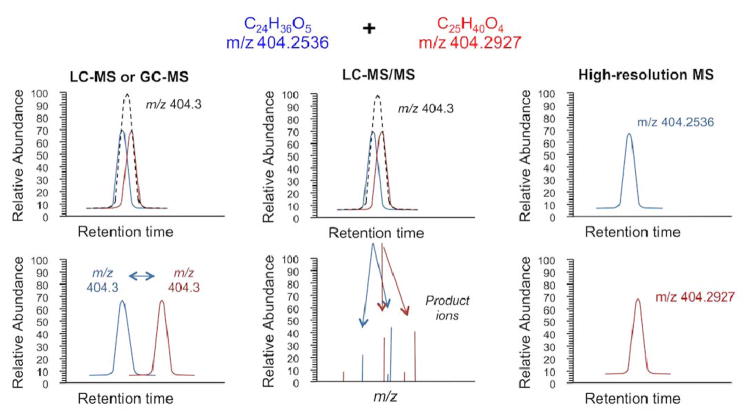
Central metabolic pathways include approximately 2000 known metabolites, and improved coverage can be obtained using high performance liquid chromatography (HPLC) coupled to high-resolution MS for HRM analysis. The benefits of high-resolution MS instruments for screening are presented in Figure 1. Low resolution mass spectrometers, often utilized for targeted analysis, are capable of unit m/z accuracy (1 atomic mass unit (AMU)), which is not sufficient to distinguish compounds with very similar molecular mass, requiring chromatographic separation prior to detection (i.e., gas- or liquid chromatography (GC; LC). Tandem MS (MS/MS or MS2) involves combined use of mass spectrometry components to obtain m/z measurements on an ion and then subsequent measurement of m/z for product ions generated following ion dissociation, enabling quantification of specific chemicals based upon product ions even when the precursor ion is not separated from chemicals with very similar mass (31, 32). High-resolution accurate MS (e.g. Fourier Transform Ion Cyclotron Resonance (FTICR) or Orbitrap based mass spectrometers) resolve ions and measure m/z with 0.0002–0.005 AMU accuracy (where mass accuracy is commonly referred to as parts-per-million, defined as: relative m/z error × 106). The high level of mass resolution achieved simplifies analyte separation requirements and provides improved capability to measure low abundance chemicals in complex biological samples. Hence, high-resolution MS provides for identification and quantification of a broad spectrum of m/z features, facilitating the discovery of metabolic alterations since multiple metabolites in the same pathway can be measured simultaneously and tested for enrichment (33, 34) while not requiring selection of specific analytical targets prior to analysis.
Figure 1.
High-resolution MS supports untargeted measurement of metabolic chemicals by reducing requirements for chemical separation and sample preparation while providing improved capability to measure low abundance chemicals in biological samples (14). Used with copyright permission from Annual Review of Nutrition.
A broader view of metabolism includes more than a million biochemicals derived from the diet, microbiome and environment, as well as a large number of lipids, peptides, glycans, peptidolipids, glycolipids, and peptidoglycans (14, 35). These chemicals have the potential to act as specific markers of exposure, toxicity and/or disease. Systematic knowledge of these biomarkers is not available, but information is rapidly accumulating and may provide useful early disease or disease-risk indicators. Many of the m/z features detected by HRM platforms are currently unidentified and include uncharacterized complex carbohydrates, environmental chemicals and their metabolites, complex lipids and amino acid metabolites derived from covalently modified proteins (36, 37).
II. B Quantification
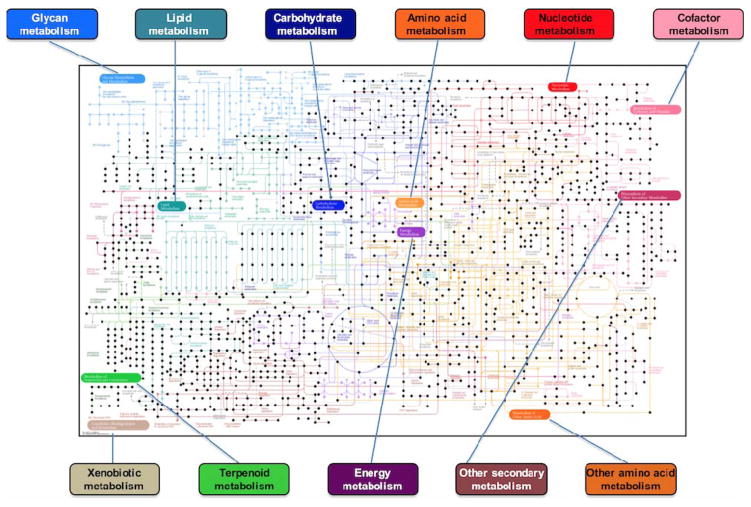
High resolution MS offers an advantage for quantification of many metabolites since mass resolution is sufficient to separately quantify co-eluting ions when the accurate mass differs by >10 ppm (13). Over 90% of metabolites in the Kyoto Encyclopedia of Genes and Genomes (KEGG) have accurate mass m/z values that differ by >10 ppm (12). HRM profiling of blood plasma samples obtained from healthy individuals has indicated measure of metabolites from >80% of the pathways present in the KEGG database (Figure 2), enabling quantification with >85% accuracy based upon ion intensity in terms of the integral of precursor ions (MS1) (15, 38). Since HRM profiling is typically applied in an untargeted manner, analytical standards for absolute quantification are often not included within the analytical methodology, however, a number of strategies for quantification have been developed allowing determination of absolute concentrations. For example, different options are available using internal standardization with stable isotope dilution (39), surrogate standardization (40) or by external standardization with a method of additions (38). Systematic comparison of these methods shows that reference standardization, a procedure using a quantitatively calibrated pooled reference material, such as National Institute of Standards and Technology SRM1950, can support quantification of thousands of chemicals in a single analysis (15). Using reference standardization, quantification post-data acquisition is possible by referencing the pooled sample analyzed within each batch of samples. Known concentrations within the reference standard can be used to determine a chemical response factor and calculate analyte concentrations based on single-point calibration. Typically, intensity of the most reliable ion corresponding to a given chemical species is used for quantification, since combining multiple ions would result in error propagation. The benefit of reference standardization is that targeted quantification is only required in the reference sample, chemicals selected for quantification do not need to be selected a priori and population-wide estimates of plasma chemical concentrations can be determined without having to re-analyze samples using a targeted approach. Such results support development of cumulative databases to evaluate time- and intensity-dependent changes in exposure and related metabolic perturbations.
Figure 2.
HRM profiling of plasma obtained from healthy individuals has indicated that metabolic intermediates from >80% of pathways present in the KEGG database can be detected (represented by black dots) using standardized sample preparation methods and dual-column chromatography.
II. C Applications for precision medicine
In addition to reproducibility and accurate quantification, other practical issues for routine use of HRM in precision medicine include cost and throughput. Assuming single instrument operation at 24 h/day for 250 days/year (which allows 25% time for holidays, vacations, servicing and repair) and a 3-year instrument lease, cost for triplicate analysis using dual chromatography and (41) is approximately $125/sample and would enable analysis of 5500 samples per instrument-year. Due to improved mass resolution and increased scan speeds available with next-generation instruments, cost per sample can be reduced to $50/sample and instrument-year throughput doubled by decreasing analytical run time to 5 minutes. Although detection of chemicals arising from exposure to environmental chemicals would be minimized, further cost reduction is also possible through focused analysis of high abundance metabolites (e.g., amino acids, lipids, vitamins/co-factors, fatty acids) that can be used for advanced clinical chemistry purposes. For example, routine analysis by HRM reliably detects approximately 1000 common, endogenous metabolites with coefficient of variation (CV) <10% (14, 15, 42–44). By limiting m/z detection to high abundance, endogenous metabolites, single replicate analysis on one column is sufficient, reducing analysis cost to approximately $15 per sample. The resulting throughput for single replicate analyses per instrument is 6 samples per hour, with analysis capacity of 150 samples/day (37,500 samples-per instrument year) possible. Thus, sufficient metabolic coverage for precision medicine purposes and detection of exposure related bioeffect could be obtained with sufficient data quality for lower costs using available instruments and appropriate automation.
The most important implication of these considerations is the potential feasibility of applying HRM to biological specimens collected for the DoDSR to obtain a comprehensive understanding of normal variations in metabolism and environmental exposures in young healthy adults from across the US. For instance, by selecting a random set of one million samples from the repository, one would be able to evaluate population differences due to geography, occupation, health habits, age, gender and disease, as well as trends over time. Using the cost structure discussed above, it would be possible to analyze one million samples (5 minute runtime with triplicate analyses) in five years using 20 instruments at a cost of $10 million/year; focused analysis of high-abundance metabolites could be completed using five instruments at a cost of $3 million/year. The data obtained from such an analyses would provide a resource for DoD researchers to evaluate possible health concerns and provide a large reference database of normal values for future precision medicine initiatives involving diet and metabolism.
III. HRM for environmental chemical analysis
III. A Detection of low abundance chemicals
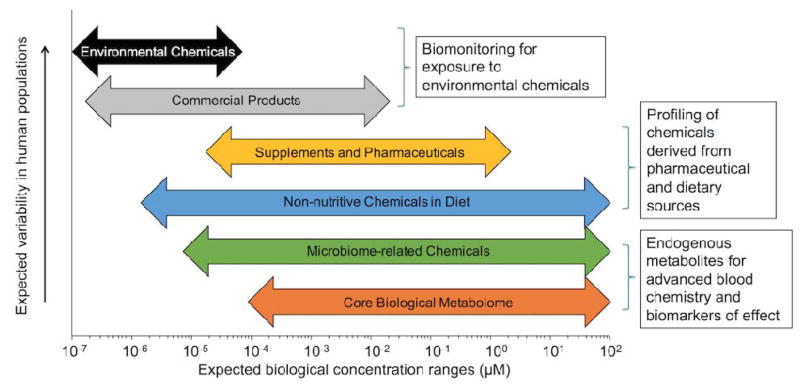
By increasing the number of replicate injections to three or more, the number of ions that can be reliably quantified in a biological sample is increased and noise reduction is improved. Improved sensitivity is important for measurement of environmental chemicals in biological samples, which are often present at four or five-orders-of-magnitude lower abundance than endogenous metabolites (Figure 3). A chemical present in only one sample out of 100 can be reliably measured if it is present in three replicate analyses of a sample (15, 43, 45), even though it is absent from the preceding 99 samples. With further relaxation of precision requirements, such as increasing CV thresholds from 50% to 100% and allowable number of samples with non-detected intensity values (which is justified by the usefulness of information on low abundance and low frequency exposures), >20,000 ions can be measured routinely on a single column; use of dual chromatography increases this to >30,000 ions (46). For high-abundance species, multiple adducts and isotopic forms will be detected based upon the ionization scheme used and molecule functional groups. The act of pairing different m/z features corresponding to the same chemical is referred to as deconvolution, and can be used as additional criteria for confirming identification. For example, the presence of an M+2 isotope corresponding to 37Cl confirms the presence of at least one chlorine functional group, which can qualify whether the identification is correct based upon chemical formula. Numerous strategies exist for deconvoluting high-resolution mass spectra, with rule-based methods relying on ion correlation, retention time matching, known adduct spacing, mass defect pairing and comparison of expected isotopic distributions to detected isotopes resulting in the lowest number of false groupings. Due to the low concentration of many chemicals present in biological samples (23), multiple features from highly abundant metabolic species represents a small portion of detected features and most low intensity ions appear to be exogenous chemicals detected as a single form. Thus, approximately >20,000 chemicals are uniquely detected using a dual column configuration. Although half of these ions do not match known chemicals in metabolomics databases, most are reproducible when analyzed on different LC-MS systems, indicating actual chemical signals and not instrumental noise or sample preparation artifacts. Therefore, HRM platforms, in addition to providing measure of endogenous metabolites, have multiple applications for profiling low abundance chemicals present in biological samples such as plasma (15, 47), urine (48, 49) or tissue biopsies (50, 51).
Figure 3.
While a core metabolome consisting of metabolites required for life will be detected universally, environmental, drug and dietary chemicals in the metabolic profile are expected to vary greatly in concentration and presence. Adapted from (45).
III. B Measurement of exposure and linking to body burden
Incorporating exposure information into population research has traditionally relied on monitoring approaches with varying levels of uncertainty. For example, heuristic models calibrated to chemical monitoring surveys have been employed to prioritize toxicity screening (52), geospatial models have been used to predict respiratory exposures (53), recall surveys have been used to estimate dietary exposures (54), ambient exposure measurements can be useful for estimating exposure to large groups, passive silicone and breathing zone samplers have been developed to estimate exposure over short and long-term periods (55, 56). While these are useful measures for evaluating the occurrence of environmental exposure, they represent generalized estimates and cannot be used to assess internal exposure and biological relevance. Furthermore, their implementation in active duty situations would negatively impact the efficacy of combat operations and daily functioning of a fighting force.
Measurement of exposure biomarkers within biological samples obtained from humans can reduce uncertainty in exposure assessment, however interpretation within an environmental health context can be challenging. Models used for risk assessment (57) as well as toxicokinetic studies (58, 59) provide frameworks for analysis, but more detailed studies are needed to interpret the meaning of spot measurements of environmental chemicals in single samples. Without knowledge of intensity, duration or route of exposure, inferences concerning abundance of chemicals in human samples are problematic. Similarly, for unidentified chemicals without time course data, the biologic half-life, the apparent volume of distribution and other toxicokinetic parameters cannot be estimated. Thus, one cannot readily evaluate the likelihood that a measurement represents an acute or long-term exposure from a single point measurement. On the other hand, routine, periodic measurement can provide the ability to detect regional differences and time-dependent differences in specific chemicals. Ongoing measurements, even for a small fraction of individuals, can therefore approximate a real-time surveillance tool to detect new or unexpected exposures. Importantly, the power of such a tool increases with time as the surveillance database expands with different exposure scenarios.
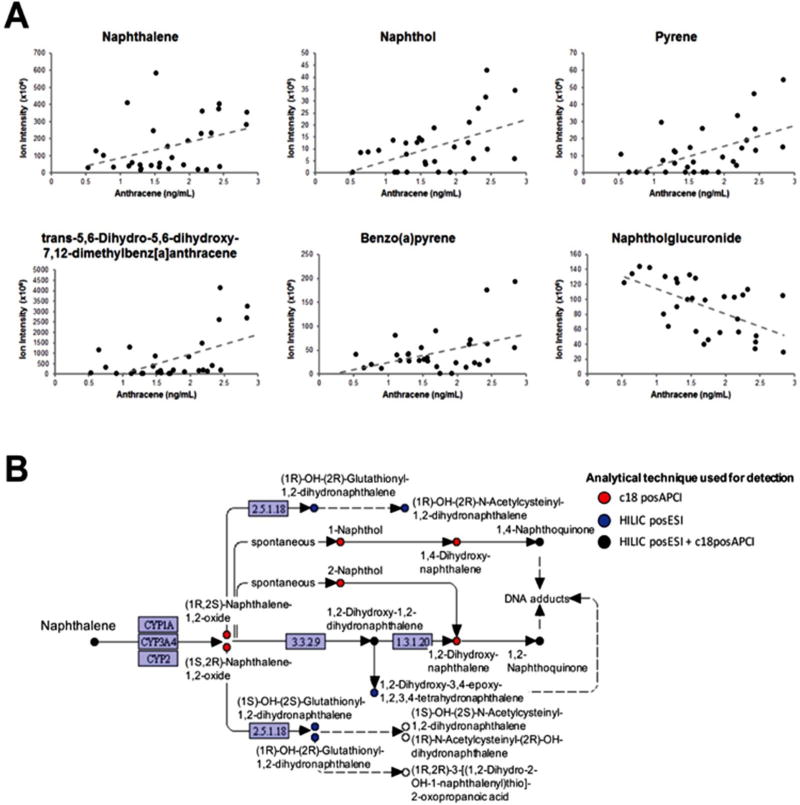
As discussed above, there is a need to measure exposure biomarkers arising from a range of environmental chemicals. Targeted biomonitoring utilizes measurement in biological specimens to estimate body burden of specific chemicals, providing information on internal dose and prevalence in a population. While biomonitoring has proven invaluable in population surveys, chemical coverage is often limited. For example, the National Health and Nutrition Examination Survey (NHANES) applied targeted biomonitoring to measure 212 chemicals in a cross-section of the US population. However, over 100,000 chemicals are registered for commercial use with the Environmental Protection Agency, with recent estimates from the Toxic Substance Control Act suggesting 70,000 are commonly used. A recent survey by Dionisio, Frame (60) identified approximately 20,000 chemicals used directly in consumer products. The ability to provide chemical monitoring of this magnitude far exceeds the capability of targeted biomonitoring platforms and requires advanced chemical profiling techniques to identify and monitor occurrence of environmental exposures. The ability of HRM to advance measurement of exposure biomarkers in human samples is highlighted in Figure 4A. In this analysis, quantified PAH levels of anthracene measured using GC-MS in 30 non-identified serum samples (6, 61) were tested for association with m/z ions detected by HRM matching known adduct masses of PAH metabolites from the KEGG database. Twenty-four metabolites detected using HILIC chromatography with positive electrospray ionization and 18 using reverse-phase chromatography with positive atmospheric chemical ionization (46) were found to be significantly associated with serum PAH levels, supporting the use of HRM for a more comprehensive measure of exposure biomarkers than available from targeted biomonitoring alone. Interestingly, anthracene levels were found to be predominantly associated with naphthalene metabolites (Figure 4B), suggesting possible co-exposures or synergistic metabolism. Successful applications of HRM in environmental chemical surveillance (15, 62–65) further support the use of this approach in exposome and environmental health research.
Figure 4.
Cross-platform comparison of targeted polycyclic aromatic hydrocarbon (PAH) measurements using gas chromatography MS and suspect screening for PAH metabolites by HRM (A). Combining liquid chromatography and ionizations schemes maximized coverage of PAH metabolism (B).
III. C Characterizing internal dose response
Considerable data are available to show that environmental chemicals are present in all human blood and urine samples (15, 23, 62, 63, 66, 67). As indicated above, detection of chemicals within the body does not provide information concerning the source of exposure, and relatively little information is available concerning the distribution chemicals within human tissues and rates of elimination. Furthermore, analytical methods are very sensitive so that detection alone does not provide information concerning risk from exposure and biological relevance.
In addition to measurement of chemical surveillance biomarkers, HRM profiling can be used to elucidate biological response to chemical exposures. Association of exposure biomarkers with HRM measured metabolic alterations provides insight into toxicant targets, biomarkers of effect and chemical biological fate, such as metabolism, distribution and excretion within human populations. Recent examples using human populations exposed to high levels of environmental chemicals highlight the ability of HRM in identifying biological response. In the study by Jeanneret, Boccard (68), metabolic profiling of dioxin poisoning was completed to identify the metabolic changes related to extreme cases of exposure. The resulting series of metabolites were then tested as discriminatory biomarkers in 11 workers previously exposed (in the late 1960’s) to dioxin residues in a pesticide production plant and matched, healthy controls. Twenty-four endogenous metabolites associated with dioxin exposure and related to steroid metabolism were found to distinguish exposed workers from controls, and were consistent with the known aryl hydrocarbon receptor (AhR) binding of dioxins. Importantly, the data indicated biological response occurring due to a recent, acute dioxin exposure was still detected in individuals exposed 40 years prior. Thus, in addition to biomonitoring, measurement of metabolic derangement by HRM can be used to identify biosignatures indicating a history of exposure. Additional studies applying metabolic profiling of human exposure to cadmium (69, 70), pesticides (66), PAH exposure (71), welding fumes (72) and arsenic (73) support the use of retrospective chemical measurement for effect markers and evaluation of whether exposure has occurred at a biologically relevant dose.
HRM profiling of biological samples can also be applied in an environment wide association study framework to systematically examine chemical associations with disease, which can then be utilized to identify susceptibility to poor health outcomes. In a HRM study of Parkinson’s Disease (PD), groups of subjects (80 from 146 PD and 20 from controls) were matched by age, gender, smoking status and pesticide exposure to identify metabolic alterations associated with slow and rapid disease progression (38). Significant differences between 80 patients and 20 controls included matches to a polybrominated diphenyl ether (PBDE), tetrabromobisphenol A, octachlorostyrene and pentachloroethane. The chemical feature corresponding to PBDE had a mean intensity 50% above controls, while the match to 2-amino-1,2-bis(p-chlorophenyl)ethanol, was more than 50% higher in individuals with rapid disease progression. Although the population is too small to make firm conclusions and the identification of the chemicals were not confirmed by comparison to authentic reference standards, the presence of disease-exposure associations provides potential risk factors for further study
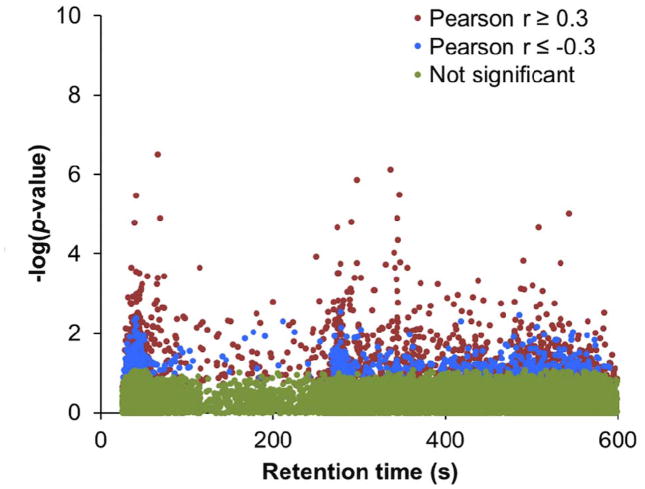
Important to the development of HRM in routine screening for biomarkers of exposure and effect has been the creation of software tools enabling a systems biology approach to understanding exposure-associated metabolic perturbations. For example, MetabNet (74), a software routine written in R, allows rapid analysis of metabolites and metabolic pathways correlating with individual chemicals in cross-sectional analyses. The analyses possible by MetabNet enable testing for correlations with environmental chemicals directly within the same metabolomics analyses, thereby avoiding the need for phenotypic biomarkers, which are often not available. For instance, HRM profiling was recently used for targeted measurement of the flame retardant triethylphosophate and untargeted metabolomics of plasma obtained from 150 healthy adults (15). By testing all metabolites for correlations with triethylphosphate, one obtains a targeted metabolome wide association study (MWAS), shown in Figure 5 as a Manhattan plot of −log p-value for the correlation as a function of retention time obtained using reverse phase LC. By evaluating feature association significance as a function of retention time, one can see that many of the ions correlated with the flame retardant exhibited retention times consistent with lipid species (retention time > 300 seconds), which was verified by database matches to phospholipids and sterols. Metabolites identified through MWAS can then be utilized for metabolic pathway enrichment analysis, providing a starting point to begin studies of metabolic perturbations linked to body measures of environmental chemicals.
Figure 5.
A targeted metabolome wide association study (MWAS) was completed by testing each metabolite for significant associations with triethylphosphate levels in plasma. Significant metabolites can be tested for pathway enrichment, providing a starting point in identifying perturbations linked to chemical exposure.
III. D Exposure memory
The need to include environment in understanding human disease led Christopher Wild to introduce the concept of the exposome in 2005 (17), which he defined as “encompassing life-course environmental exposures (including lifestyle factors), from the pre-natal period onwards.” The exposome is envisioned as a complement to the genome, where a life course of exposure and interaction with the genome defines risk for disease development. Unlike the genome, exposures are transient and change on both short and long-term time scales, making quantitative assessment challenging. A more tractable definition of the exposome was proposed by Miller and Jones (18): “The cumulative measure of environmental influences and associated biological responses throughout the lifespan, including exposures from the environment, diet, behavior, and endogenous processes.” Exposures in this framework are not only limited to external chemical exposures, but also include processes internal to the body (host factors), wider socioeconomic influences and maladaptations to external influences, with the interaction of these factors linking environment to health and disease (16, 75).
Accumulating research indicates that multiple exposure memory systems exist to allow adaptation to environmental challenges over a lifetime (19). A consequence is that response can result in decreased flexibility and adaptability to subsequent challenges. Many of the body’s response to environmental insults are short-term and reversible; some, such as scarring, provide a long-term and sometimes permanent change in structure and function. For example, inhalation injury to the lungs can result in permanent change in structure and function, which may not be evident except in response to challenge. Computational methods are available to use the regularity/irregularity present in metabolomics data to measure health (76) and, in principle; these methods could be used with HRM to study metabolic resilience following deployment. Such methods could be applied to screen service personnel for exposure histories and identify individuals who would benefit from nutrition or medical intervention.
III. E Addressing the dark matter of the exposome
A critical aspect of post-deployment surveillance lies in addressing unknown exposures. While contemporary MS methods are powerful, characterization of unidentified chemicals in human samples has seriously lagged behind the progress in understanding human genetics. At least half of the ions detected by HRM are uncharacterized; if one pushes data extraction to the limits of contemporary methods, the fraction of uncharacterized ions is approximately 80% (13). This represents the dark matter of the exposome, which includes detected but uncharacterized analytes and a spectrum of unknown unknowns, for which little effort has been made to distinguish. Resources to address this daunting challenge are currently unavailable, as emphasis has traditionally been placed on analysis of a relatively small number of recognized hazardous chemicals. As a consequence, risks associated with uncharacterized chemicals are largely unknown.
HRM methods provide a useful approach to this challenging problem. Already, data are available within the Emory University Clinical Biomarkers Laboratory for more than 100,000 ions obtained from approximately 20,000 biological samples. All samples have been analyzed with rigorous standard operating procedures so that the accurate mass, retention time and ion intensity enable collation into a reference database. This type of database will provide a resource for comparison to post-deployment measurement for the detection of new ions, which could represent unidentified or unknown exposures and capabilities are now available to establish a rigorous analytical framework to regularly monitor exposures. With appropriate commitment and resources, an analytical resource can be established to support routine exposure surveillance in armed forces personnel.
IV. HRM in deployment exposure surveillance
IV. A Retrospective analysis using the DoDSR
For effective real-time analyses, databases representing normal exposures and ranges of unexpected exposures will be needed to provide reference values. Previous studies have established the integrity of samples stored within the DoDSR (61, 77) and therefore highlight the opportunity for further advancement of the repository as an environmental health resource. The recent development of reference standardization (15) to obtain estimates of absolute concentrations of endogenous metabolites, health indicators and environmental chemicals, establishes an affordable approach. Additionally, the concepts are developed to use metabolic correlations for retrospective chemical identification (74), where correlations of a metabolite are obtained in reference populations so that the correlation structure can be used to identify chemicals using data from previously analyzed samples. Due to the use of rigorous sample collection, preparation and analysis procedures, the data from these analyses can be stored in a cumulative database for future analysis and data mining. As new hazards and exposures are recognized, the data will be available for retrospective analysis of individual exposures, their trends and associated health outcomes.
IV. B Prospective use for exposure surveillance
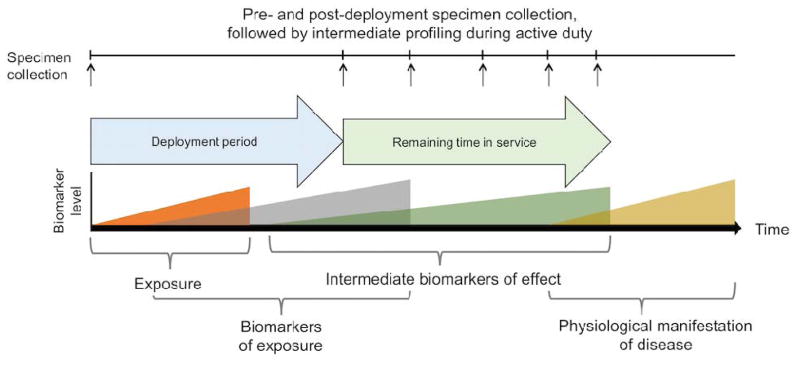
HRM is sufficiently developed to allow implementation on a test basis for ongoing deployment surveillance. For instance, metabolic indicators of nutrition (vitamins, amino acids), liver function (bilirubin), renal function (creatinine, uric acid) and other health phenotypes can be readily measured using routine analysis. At the same time, a range of PAH metabolites, persistent halogenated chemicals, insecticides, herbicides, flame retardants and other chemicals are detected (12, 15, 41, 51, 63, 78, 79). Thus, implementation of routine HRM for exposure surveillance would provide a means to address uncertainties concerning exposure. An example framework for specimen collection and implementation of HRM profiling that is consistent with current DoDSR protocols is provided in Figure 6. In this framework, chemical profiling of samples obtained pre- and post-deployment and over the course of service will enable evaluation for biomarkers of exposure, effect and poor health outcome. Routine detection of biomarkers will enable identification of individuals at risk for environment-associated disease, enabling intervention and preventive measures. Ultimately, such an analytical structure could facilitate improved management of risk associated with work in adverse environments.
Figure 6.
HRM profiling of specimens collected from active duty individuals could readily be integrated into DoDSR protocols. In this example, HRM can evaluate the presence of exposure and effect biomarkers, identifying individuals at risk for exposure related diseases. Adapted from (45).
IV. C Integration with targeted phenotypic and other omics platforms
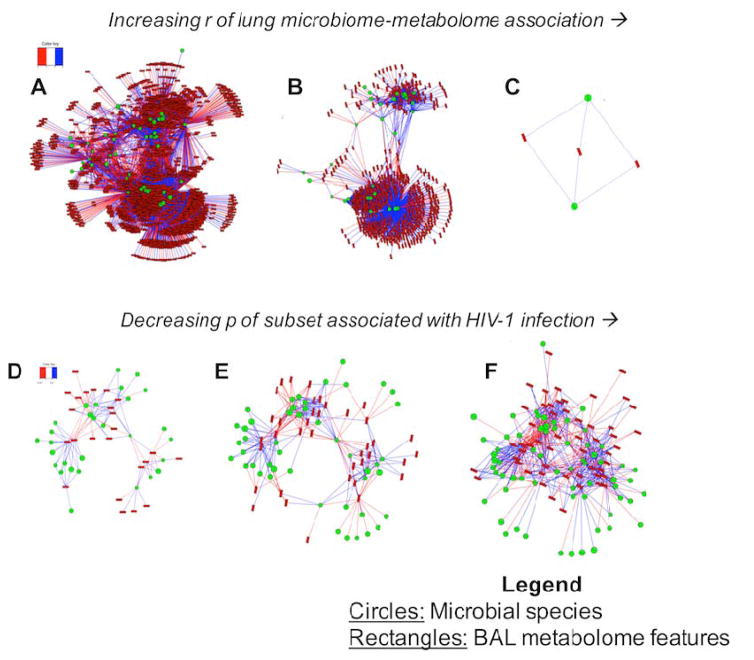
Many mechanistic studies show that the combination of HRM with other phenotypic platforms (e.g. genomics, proteomics, transcriptomics) provides a powerful approach to understand mechanisms of toxicity and disease (75, 80–82). Although deployment surveillance does not necessarily require mechanistic understanding, the integrative approaches may be suitable for improved risk assessment. For instance, transcriptome-metabolome wide association study (TMWAS) showed interaction hubs reflecting toxic reactions as well as early stress response and adaptive responses (83). Gene expression and metabolism in peripheral white blood cells may be sufficiently persistent to monitor adverse exposures for several weeks (84). Similarly, circulating miRNA in combination with metabolomics could provide a useful means to evaluate prior adverse exposures. The utility of integrative approaches was recently shown by the interaction of the lung microbiome with the metabolome measured in lung bronchoalveolar lavage fluid from healthy controls and HIV-1 infected individuals (85). In this study, increasing association stringency for microbiome-metabolome interaction using all individuals simplified a large number of significant associations to the top two genera of bacteria and top 3 metabolites, with the associations centered on bacteria causing opportunistic infection (Figure 7A–C). Microbiome-metabolome associations were then evaluated based upon significance of association with HIV-1 status (Figure 7D–F). By decreasing the p-value threshold, clear visualization of the genera-metabolite hubs was obtained, which include the top 3 groups of bacteria specifically causing opportunistic infections in individuals with HIV-1. Thus, there are considerable opportunities to utilize HRM in combination with other powerful contemporary methods to enhance detection, understanding and management of post-deployment health risks.
Figure 7.
Microbiome-MWAS of bronchoalveolar lavage fluid in HIV-1: A to C represent increasing stringency to identify network associations with greatest r meeting significance criteria. D to F contains the network subset associated with HIV-1, in order of increasing leniency. From (86).
V. HRM: summary and perspective
High-resolution metabolomics (HRM) provides an advanced clinical chemistry platform for precision medicine that could be of considerable utility for exposure surveillance of armed forces personnel. HRM not only provides extensive coverage of metabolism but also detects a broad spectrum of exposure biomarkers, including both known and currently unidentified chemicals. Building upon contemporary occupational medicine and exposure science, HRM can be integrated into environmental health research through connecting external exposures and health outcomes to internal body burden of environmental agents and respective biological responses. Analytic platforms, workflow and available applications establish the suitability of HRM for development into an environmental exposure surveillance system. Systematic analysis of existing DoDSR samples using HRM would provide a high-quality cross-sectional reference dataset for deployment-associated exposures, while development of real-time analytical capabilities using HRM would provide a demonstration project for use in precision medicine. Furthermore, use of HRM is expected to improve the ability of healthcare practitioners to include exposure-related measurements in management and treatment of disease in active duty and retired armed forces personnel.
Acknowledgments
The authors gratefully acknowledge the technical expertise of ViLinh Tran for mass spectrometry analyses. This public health surveillance project was supported with funding from the Department of Defense (306889-1.00-64239) and the National Institute of Health (P30 ES019776).
Footnotes
Conflicts of interest: None to declare
References
- 1.Lindler LE. Enhancing the Department of Defense’s Capability to Identify Environmental Exposures Into the 21st Century. Mil Med. 2015;180(10 Suppl):5–9. doi: 10.7205/MILMED-D-14-00723. [DOI] [PubMed] [Google Scholar]
- 2.Tollerud D, et al. Long-Term Health Consequences of Exposure to Burn Pits in Iraq and Afghanistan. Washington DC: 2011. [DOI] [PubMed] [Google Scholar]
- 3.Baird CP. Review of the Institute of Medicine report: long-term health consequences of exposure to burn pits in Iraq and Afghanistan. US Army Med Dep J. 2012:43–7. [PubMed] [Google Scholar]
- 4.Rubertone MV, Brundage JF. The Defense Medical Surveillance System and the Department of Defense serum repository: glimpses of the future of public health surveillance. Am J Public Health. 2002;92(12):1900–4. doi: 10.2105/ajph.92.12.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perdue CL, Eick-Cost AA, Rubertone MV. A Brief Description of the Operation of the DoD Serum Repository. Mil Med. 2015;180(10 Suppl):10–2. doi: 10.7205/MILMED-D-14-00739. [DOI] [PubMed] [Google Scholar]
- 6.Mallon TM, et al. Introduction to Department of Defense research on burn pits, biomarkers and health outcomes related to deployment in Iraq and Afghanistan. Journal of Occupational and Environmental Medicine. 2016 doi: 10.1097/JOM.0000000000000775. Submitted; DoD Biomarkers Supplement. [DOI] [PubMed] [Google Scholar]
- 7.Xia X, et al. Polycyclic Aromatic Hydrocarbons and Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans in Microliter Samples of Human Serum as Exposure Indicators. Journal of Occupational and Environmental Medicine. 2016 doi: 10.1097/JOM.0000000000000743. Submitted; DoD Biomarkers Supplement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425(19):3582–600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect. 2015;123(5):399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DelRaso NJ, et al. Air Force Research Laboratory Integrated Omics Research. Mil Med. 2015;180(10 Suppl):67–75. doi: 10.7205/MILMED-D-15-00051. [DOI] [PubMed] [Google Scholar]
- 11.Bradburne C, et al. Overview of ‘Omics Technologies for Military Occupational Health Surveillance and Medicine. Mil Med. 2015;180(10 Suppl):34–48. doi: 10.7205/MILMED-D-15-00050. [DOI] [PubMed] [Google Scholar]
- 12.Park YH, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295(1–3):47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JM, et al. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135(11):2864–70. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Go YM, et al. Reference Standardization for Mass Spectrometry and High-Resolution Metabolomics Applications to Exposome Research. Toxicol Sci. 2015 doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330(6003):460–1. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1847–50. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 18.Miller GW, Jones DP. The nature of nurture: refining the definition of the exposome. Toxicol Sci. 2014;137(1):1–2. doi: 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones DP. Redox theory of aging. Redox biology. 2015;5:71–79. doi: 10.1016/j.redox.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schymanski EL, et al. Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Anal Bioanal Chem. 2015;407(21):6237–55. doi: 10.1007/s00216-015-8681-7. [DOI] [PubMed] [Google Scholar]
- 21.Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:579–99. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- 22.Makarov A, et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78(7):2113–20. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 23.Rappaport SM, et al. The blood exposome and its role in discovering causes of disease. Environ Health Perspect. 2014;122(8):769–74. doi: 10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krumsiek J, et al. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012;8(10):e1003005. doi: 10.1371/journal.pgen.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menni C, et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int J Epidemiol. 2013;42(4):1111–9. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scalbert A, et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr. 2014;99(6):1286–308. doi: 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- 27.Roberts LD, et al. Targeted metabolomics. Curr Protoc Mol Biol. 2012;Chapter 30(Unit 30 2):1–24. doi: 10.1002/0471142727.mb3002s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barr DB, Needham LL. Analytical methods for biological monitoring of exposure to pesticides: a review. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2002;778(1–2):5–29. doi: 10.1016/s1570-0232(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 29.Sandau CD, et al. Comprehensive solid-phase extraction method for persistent organic pollutants. Validation and application to the analysis of persistent chlorinated pesticides. Anal Chem. 2003;75(1):71–7. doi: 10.1021/ac026121u. [DOI] [PubMed] [Google Scholar]
- 30.Sirimanne SR, et al. Quantification of polycyclic aromatic hydrocarbons and polychlorinated dibenzo-p-dioxins in human serum by combined micelle-mediated extraction (cloud-point extraction) and HPLC. Anal Chem. 1996;68(9):1556–60. doi: 10.1021/ac951028+. [DOI] [PubMed] [Google Scholar]
- 31.Vogeser M, Seger C. Pitfalls associated with the use of liquid chromatography-tandem mass spectrometry in the clinical laboratory. Clin Chem. 2010;56(8):1234–44. doi: 10.1373/clinchem.2009.138602. [DOI] [PubMed] [Google Scholar]
- 32.Yan Z, et al. Isobaric metabolite interferences and the requirement for close examination of raw data in addition to stringent chromatographic separations in liquid chromatography/tandem mass spectrometric analysis of drugs in biological matrix. Rapid Commun Mass Spectrom. 2008;22(13):2021–8. doi: 10.1002/rcm.3577. [DOI] [PubMed] [Google Scholar]
- 33.Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38(Web Server issue):W71–7. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart DS, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowen BP, Northen TR. Dealing with the unknown: metabolomics and metabolite atlases. J Am Soc Mass Spectrom. 2010;21(9):1471–6. doi: 10.1016/j.jasms.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 37.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roede JR, et al. Serum metabolomics of slow vs. rapid motor progression Parkinson’s disease: a pilot study. PLoS One. 2013;8(10):e77629. doi: 10.1371/journal.pone.0077629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JM, et al. A rapid LC-FTMS method for the analysis of cysteine, cystine and cysteine/cystine steady-state redox potential in human plasma. Clin Chim Acta. 2008;396(1–2):43–8. doi: 10.1016/j.cca.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greizerstein HB, et al. Standardization of a method for the routine analysis of polychlorinated biphenyl congeners and selected pesticides in human serum and milk. J Anal Toxicol. 1997;21(7):558–66. doi: 10.1093/jat/21.7.558. [DOI] [PubMed] [Google Scholar]
- 41.Soltow Q, et al. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013 doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uppal K, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DP. Sequencing the exposome: A call to action. Toxicology Reports. 2016;3:29–45. doi: 10.1016/j.toxrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Go YM, et al. Metabolome-wide association study of phenylalanine in plasma of common marmosets. Amino Acids. 2014 doi: 10.1007/s00726-014-1893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker DI, Nicholson JK, et al., editors. Metabolic Phenotyping in Personalized and Public Healthcare. Elsevier; 2016. Population Screening for Biological and Environmental Properties of the Human Metabolic Phenotype: Implications for Personalized Medicine. in press. [Google Scholar]
- 46.Liu KH, et al. High resolution metabolomics assessment of military personnel. Journal of Occupational and Environmental Medicine. 2016 doi: 10.1097/JOM.0000000000000773. Submitted; DoD Biomarkers Supplement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frediani JK, et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS One. 2014;9(10):e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edmands WM, et al. Polyphenol metabolome in human urine and its association with intake of polyphenol-rich foods across European countries. Am J Clin Nutr. 2015;102(4):905–13. doi: 10.3945/ajcn.114.101881. [DOI] [PubMed] [Google Scholar]
- 49.Khamis MM, Adamko DJ, El-Aneed A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom Rev. 2015 doi: 10.1002/mas.21455. [DOI] [PubMed] [Google Scholar]
- 50.Dunn W, et al. The metabolome of human placental tissue: investigation of first trimester tissue and changes related to preeclampsia in late pregnancy. Metabolomics. 2012;8(4):579–597. [Google Scholar]
- 51.Go YM, et al. Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods Mol Biol. 2014;1198:43–73. doi: 10.1007/978-1-4939-1258-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wambaugh JF, et al. High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ Sci Technol. 2014;48(21):12760–7. doi: 10.1021/es503583j. [DOI] [PubMed] [Google Scholar]
- 53.Lane KJ, et al. Effect of time-activity adjustment on exposure assessment for traffic-related ultrafine particles. J Expo Sci Environ Epidemiol. 2015;25(5):506–16. doi: 10.1038/jes.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chadeau-Hyam M, et al. Meeting-in-the-middle using metabolic profiling - a strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers. 2011;16(1):83–8. doi: 10.3109/1354750X.2010.533285. [DOI] [PubMed] [Google Scholar]
- 55.Lan Q, et al. Occupational exposure to trichloroethylene is associated with a decline in lymphocyte subsets and soluble CD27 and CD30 markers. Carcinogenesis. 2010;31(9):1592–6. doi: 10.1093/carcin/bgq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connell SG, Kincl LD, Anderson KA. Silicone wristbands as personal passive samplers. Environ Sci Technol. 2014;48(6):3327–35. doi: 10.1021/es405022f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patton AP, et al. Transferability and generalizability of regression models of ultrafine particles in urban neighborhoods in the Boston area. Environ Sci Technol. 2015;49(10):6051–60. doi: 10.1021/es5061676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiu WA, et al. Toxicokinetics of inhaled trichloroethylene and tetrachloroethylene in humans at 1 ppm: empirical results and comparisons with previous studies. Toxicol Sci. 2007;95(1):23–36. doi: 10.1093/toxsci/kfl129. [DOI] [PubMed] [Google Scholar]
- 59.Sobus JR, et al. Biomarker variance component estimation for exposure surrogate selection and toxicokinetic inference. Toxicol Lett. 2010;199(3):247–53. doi: 10.1016/j.toxlet.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dionisio KL, et al. Exploring consumer exposure pathways and patterns of use for chemicals in the environment. Toxicology Reports. 2015;2:228–237. doi: 10.1016/j.toxrep.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker DI, et al. Pilot metabolome-wide association study of benzo(a)pyrene in serum from military personnel. Journal of Occupational and Environmental Medicine. 2016 doi: 10.1097/JOM.0000000000000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roca M, et al. Comprehensive analytical strategy for biomonitoring of pesticides in urine by liquid chromatography-orbitrap high resolution masss pectrometry. J Chromatogr A. 2014;1374:66–76. doi: 10.1016/j.chroma.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Jamin EL, et al. Untargeted profiling of pesticide metabolites by LC-HRMS: an exposomics tool for human exposure evaluation. Anal Bioanal Chem. 2014;406(4):1149–61. doi: 10.1007/s00216-013-7136-2. [DOI] [PubMed] [Google Scholar]
- 64.Bletsou AA, et al. Targeted and non-targeted liquid chromatography-mass spectrometric workflows for identification of transformation products of emerging pollutants in the aquatic environment. TrAC Trends in Analytical Chemistry. 2015;66:32–44. [Google Scholar]
- 65.Schymanski EL, et al. Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ Sci Technol. 2014;48(3):1811–8. doi: 10.1021/es4044374. [DOI] [PubMed] [Google Scholar]
- 66.Bonvallot N, et al. Metabolomics tools for describing complex pesticide exposure in pregnant women in Brittany (France) PLoS One. 2013;8(5):e64433. doi: 10.1371/journal.pone.0064433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wishart D, et al. T3DB: the toxic exposome database. Nucleic Acids Res. 2015;43(Database issue):D928–34. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeanneret F, et al. Human urinary biomarkers of dioxin exposure: analysis by metabolomics and biologically driven data dimensionality reduction. Toxicol Lett. 2014;230(2):234–43. doi: 10.1016/j.toxlet.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 69.Ellis JK, et al. Metabolic profiling detects early effects of environmental and lifestyle exposure to cadmium in a human population. BMC Med. 2012;10:61. doi: 10.1186/1741-7015-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao Y, et al. Identifying early urinary metabolic changes with long-term environmental exposure to cadmium by mass-spectrometry-based metabolomics. Environ Sci Technol. 2014;48(11):6409–18. doi: 10.1021/es500750w. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, et al. Human metabolic responses to chronic environmental polycyclic aromatic hydrocarbon exposure by a metabolomic approach. J Proteome Res. 2015;14(6):2583–93. doi: 10.1021/acs.jproteome.5b00134. [DOI] [PubMed] [Google Scholar]
- 72.Wei Y, et al. Global metabolomic profiling reveals an association of metal fume exposure and plasma unsaturated fatty acids. PLoS One. 2013;8(10):e77413. doi: 10.1371/journal.pone.0077413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, et al. Urinary metabolomics revealed arsenic internal dose-related metabolic alterations: a proof-of-concept study in a Chinese male cohort. Environ Sci Technol. 2014;48(20):12265–74. doi: 10.1021/es503659w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uppal K, et al. MetabNet: An R Package for Metabolic Association Analysis of High-Resolution Metabolomics Data. Front Bioeng Biotechnol. 2015;3:87. doi: 10.3389/fbioe.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012;41(1):24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 76.Park Y, et al. Multifractal analysis for nutritional assessment. PLoS One. 2013;8(8):e69000. doi: 10.1371/journal.pone.0069000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perdue CL, et al. Description and utilization of the United States department of defense serum repository: a review of published studies, 1985–2012. PLoS One. 2015;10(2):e0114857. doi: 10.1371/journal.pone.0114857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Osborn MP, et al. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS One. 2013;8(8):e72737. doi: 10.1371/journal.pone.0072737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roede JR, et al. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS One. 2012;7(3):e33020. doi: 10.1371/journal.pone.0033020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gomez-Cabrero D, et al. Data integration in the era of omics: current and future challenges. BMC Syst Biol. 2014;8(Suppl 2):I1. doi: 10.1186/1752-0509-8-S2-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vineis P, et al. Advancing the application of omics-based biomarkers in environmental epidemiology. Environ Mol Mutagen. 2013;54(7):461–7. doi: 10.1002/em.21764. [DOI] [PubMed] [Google Scholar]
- 82.Wild CP, Scalbert A, Herceg Z. Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ Mol Mutagen. 2013;54(7):480–99. doi: 10.1002/em.21777. [DOI] [PubMed] [Google Scholar]
- 83.Roede JR, et al. Transcriptome–metabolome wide association study (TMWAS) of maneb and paraquat neurotoxicity reveals network level interactions in toxicologic mechanism. Toxicology Reports. 2014;1(0):435–444. doi: 10.1016/j.toxrep.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Leeuwen DM, et al. Transcriptome analysis in peripheral blood of humans exposed to environmental carcinogens: a promising new biomarker in environmental health studies. Environ Health Perspect. 2008;116(11):1519–25. doi: 10.1289/ehp.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cribbs SK, et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4(1):3. doi: 10.1186/s40168-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]