Abstract
Astrocytes use gliotransmitters to modulate neuronal function and plasticity. However, the role of small extracellular vesicles, called exosomes, in astrocyte-to-neuron signaling is mostly unknown. Exosomes originate in multivesicular bodies of parent cells and are secreted by fusion of the multivesicular body limiting membrane with the plasma membrane. Their molecular cargo, consisting of RNA species, proteins, and lipids, is in part cell type and cell state specific. Among the RNA species transported by exosomes, microRNAs (miRNAs) are able to modify gene expression in recipient cells. Several miRNAs present in astrocytes are regulated under pathological conditions, and this may have far-reaching consequences if they are loaded in exosomes. We propose that astrocyte-derived miRNA-loaded exosomes, such as miR-26a, are dysregulated in several central nervous system diseases; thus potentially controlling neuronal morphology and synaptic transmission through validated and predicted targets. Unraveling the contribution of this new signaling mechanism to the maintenance and plasticity of neuronal networks will impact our understanding on the physiology and pathophysiology of the central nervous system.
Keywords: microRNA, extracellular vesicles, astrocytes, neurons
Astrocytes in Neuronal Plasticity
Astrocytes, a major glial cell type in the central nervous system, have emerged as powerful regulators of brain function where they maintain ion, metabolic, and neurochemical homeostasis. Astrocytes form organized networks that communicate through gap junctions formed by hemichannel proteins called connexins, potentially creating a functional syncytium.1 The close structural and functional relationship between the presynaptic bouton and the postsynapse, both engulfed by astrocyte processes, has been named the tripartite synapse.2 Thus, by extending processes that wrap around brain capillaries and synapses, astrocytes control neurovascular coupling, as well as synaptic transmission.3,4 Increasing evidence has specified their important functions in the nervous system, regulating blood–brain barrier permeability, cellular responses to brain injury, formation and elimination of synapses, synaptic maintenance and plasticity, as well as axon myelination in the adult brain, among others.5–8 Table 1 shows a summary of the core functions of astrocytes in relation to neuronal function. Though most of this evidence has been obtained in vitro, encouraging additional evidence comes from in vivo models, as recently revealed by measures of astrocytic Ca2+ dynamics with high-resolution Ca2+ imaging in living mice.9 The nature of astrocyte-to-neuron interactions are mediated by direct cell-to-cell contact as well as by a complex array of astrocyte-derived molecules, ie, the astrocyte secretome, that varies according to neuronal activity and astrocyte activation and thus according to physiological or pathological conditions.3,10 It has been recently proposed and in part documented that the different cell types in the central nervous system could communicate by means of extracellular vesicles, among which small extracellular vesicles, also named exosomes, have received intense attention.11 However, the involvement of exosomes in astrocyte-to-neuron communication has not been explored in depth. Thus, in this brief commentary, we will discuss the new exciting possibilities related to the interaction of astrocyte-derived exosomes with neurons in the nervous system, focusing on the content of microRNAs (miRNAs) present in these vesicles.
Table 1.
Summary of neuron-related functions of astrocytes.
| NEURON RELATED FUNCTION | EXPERIMENTAL EVIDENCE | REFERENCES |
|---|---|---|
| Metabolic support | ||
| Lactate-shuttling | In vivo/in vitro | 12–15 |
| Excitatory/inhibitory control | ||
| Glutamate buffering | In vitro | 16,17 |
| Glutamate/glutamine cycle | In vitro/ex-vivo | 18,19 |
| GABAergic activity | In vitro/ex-vivo | 20–22 |
| D-serine release for LTP | In vitro | 23 |
| Myelination | In vivo/in vitro | 10 |
| Cognitive impact/function | In vivo | 24 |
| Trophic support with gliotransmitters | In vitro | 25,26 |
| Axon regeneration | In vivo | 27 |
| Neurovascular coupling | In vivo | 28,29 |
| Blood-brain barrier permeability | In vitro | 30 |
| Regulation of neural oscillations | In vivo | 31 |
Exosome Release by Astrocytes
Cells release a variety of extracellular vesicles that differ in their subcellular origin and, possibly, in their biology. Among them, exosomes are defined by their endosomal origin, storage in multivesicular bodies, and characteristic size of ~30–120 nm. By fusion of multivesicular bodies with the plasma membrane, exosomes are released by exocytosis from most eukaryotic cells.32 These vesicles carry a large diversity of molecules, such as RNA species, proteins, and lipids, that can modify the physiology of nearby or distant target cells.
The current knowledge on the mechanisms involved in the biogenesis and secretion of exosomes, as well as their possible functional interaction with target cells, has been described in several excellent recent reviews.32–35 Most of this knowledge relies on the isolation of exosomes from extracellular fluids by differential ultracentrifugation and further characterization by size and presence of protein markers.36 However, a major problem has more recently been identified: this isolated exosome-like small extracellular vesicle fraction contains a range of small vesicle subpopulations consisting of vesicles of larger size (>150 nm) or of nonendosomal origin, while in most exosome studies, their endosomal origin has not been demonstrated.37 Thus, the identification of specific protein markers and general protein markers among vesicle subpopulations as well as the implementation of additional purification steps leading to homogeneous preparations are under active research. In this commentary, we will focus on the available literature in the field that uses mostly small extracellular vesicle preparations obtained by ultracentrifugation that are enriched in exosomes, as revealed by the current available experimental tools.
Despite the large amount of evidence about exosomes derived from cells of the nervous system,38 specific research about astrocyte-derived exosomes is scarce. Some of the pioneers’ evidences for the presence of exosomes in astrocyte cultures came from Milligan’s and Agnati’s group in 2007 and 2010, respectively. They found the presence of classical exosome markers in a fraction obtained after the well-accepted differential centrifugation protocol.39,40 Three years later, it was shown that astrocyte-derived exosomes harbor synapsin-1, a glycoprotein that was released from these extracellular vesicles following an increase in extracellular potassium levels. Although they did not show a direct effect of these exosomes on neurons, it was suggested that released synapsin-1 could have a positive effect on neurite outgrowth and neuronal survival.41 Soon after, it was reported that astrocytes exposed to β-amyloid both in vitro and in vivo release exosomes that trigger apoptosis in other astrocytes, acting in an autocrine fashion.42 Using a different strategy, astrocytes may use extracellular vesicles, probably including exosomes, to transport the excitatory amino acid transporters EAAT-1 and EAAT-2, essential components in the maintenance of glutamate homeostasis. Interestingly, this extracellular vesicle fraction is able to actively take up aspartate, suggesting that in addition to their potential role in cell–cell communication, exosomes may participate in glutamate clearance.43 Interestingly, members of the heat shock protein 70 family as well as astrocyte-specific glycolitic enzyme aldolase C have also been found in this fraction, raising the question about the potential transfer of these proteins with functional consequences to neurons.39,44 Accordingly, it was recently shown that astrocytes release exosomes containing nonpathogenic prion protein that can be transferred to neurons, inducing protection after exposure to a series of stressors such as hypoxia, hypoglycemia, and ischemia.45 Thus, astrocyte-derived exosomes are capable of transferring proteins and lipids to target cells to modify their function.42 Strikingly, astrocyte-derived exosomes may have far-reaching consequences in vivo: they facilitate the outgrowth of metastatic cells in the brain by inhibiting the expression of the tumor suppressor phosphatase and tensin homolog (PTEN) through a functional transfer of miR-19a.46 Nevertheless, as far as we know, there is no functional evidence of exosome-mediated transfer of astrocytic miRNAs to neurons.
After the first evidences were provided by Valadi et al, the functional transfer of exosomal components such as proteins and nucleic acids to elicit responses on target cells has been reported in several studies47,48 and has been summarized recently.35 Next, we will focus here on the possible transfer of miRNAs from astrocytes to neurons by means of exosomes.
MiRNAs in Exosomes
Exosomes as well as other extracellular vesicles contain mRNAs and a plethora of noncoding RNAs, among which miRNAs stand out for their capacity of reprogramming protein expression in recipient cells. A long precursor miRNA molecule is transcribed in the nucleus, and after several sequential processing steps by RNase complexes, it is transported as pre-miRNA to the cytoplasm.49–51 Here, the pre-miRNA hairpin is cleaved by the RNase Dicer producing the ~22 nucleotides, double-stranded mature form of the miRNA.52 The mature form is loaded onto the RNA-induced silencing complex (RISC), where one of the strands (ie, the passenger strand) is degraded after recruiting argonaute proteins while the other, called the leading strand, is held inside the mature complex carrying argonaute 2 (Ago2).53 After this processing, a short sequence of 6–8 nucleotides or seed sequence of the miRNA is capable of targeting the 3′ UTR region of mRNAs to inhibit translation by two mechanisms: (1) the direct degradation of the mRNA or (2) translational repression by reducing the recognition capacity of ribosomes to mRNA.54 As each miRNA harbors target-recognition motifs in as many as hundreds of mRNAs, they are able to control complex biochemical processes in a coordinated manner and thus, disease and health states.
It is important to consider that genes coding for miRNAs tend to form clusters, defined as groups of genes that are separated by no more than 1 MB between them, in such a way that three to six miRNA genes are usually found in one cluster. All of these genes may be transcribed as polycistrons and, surprisingly, have preference for targeting functionally related genes.55,56 For example, one of the most studied mammalian clusters is miR-17-92, which is composed of six members: miR-17, miR-19b, miR-20a, miR-92, miR-18a, and miR-19a. All of them, except miR-18a, are demonstrated to downregulate PTEN, inducing axonal elongation.57 Interestingly, all of these miRNAs have been previously described in astrocytes, and miR-19a is contained in exosomes.46 In addition, miR-26a, although located outside this cluster, also favors neurite/axonal elongation by targeting PTEN as well as glycogen synthase kinase-3β (GSK-3β) (see below).58,59
The loading of miRNAs into exosomes leads to a specific enrichment of some miRNAs in them in comparison to their originating cells, mediated by sorting mechanisms that include: (1) the presence of specific miRNA sequences (including uridylated miRNAs and EXOmotifs), (2) miRNA binding to lipid raft-like regions in the cytoplasmatic face of the multivesicular body limiting membrane, and (3) interactions with proteins such as sumoylated heterogeneous nuclear ribonucleoproteins or Ago2.60,61 The participation of Ago2 in miRNA loading is further demonstrated because its KRAS–MEK–ERK pathway-dependent phosphorylation decreases the sorting of Ago2 and its associated miRNAs to exosomes.62,63 Although miRNAs are selectively retained or released by different cell types,48,64–66 the mechanisms underlying selective packaging of miRNAs into astrocyte exosomes needs to be further explored.
Exploring Astrocyte’s miRNA Content: Unraveling the Words to Understand Exosome-mediated Messages to Target Cells
Over the past few years several studies have provided a deeper insight into the miRNA species that are expressed or enriched in astrocytes, or even unique compared to other CNS cell types (eg, neurons, oligodendrocytes, and microglia).67–72 We will summarize some of the reported miRNAs and their functional relevance in different cellular contexts.
Some of the miRNAs found in astrocytes may be crucial to determine a glial fate (as opposed to a neuronal one) from progenitor cells.73 miR-146a, for example, may prevent astrocytes from expressing neuronal proteins,67 and provided that certain conditions are met, miR-34a can promote astrogliogenesis.74 MiR-9 and miR-124 provide another example of a miRNA in regulating cellular differentiation fate. If the miR-124 binding site in the 3′UTR of the histone methyltransferase enhancer of zeste homolog 2 (EZH2) is manipulated as to prevent the action of this miRNA, astrocytic differentiation is enhanced (and neuronal differentiation diminished).75 Consistent with these results, overexpression of miR-124a or miR-9 in neural precursor cells promotes neurogenesis and reduces gliogenesis.76 Studies performed in different brain regions of humans show that many miRNAs are common to all astrocytes, regardless of the brain region analyzed, whereas others are unique to certain areas (eg, miR-129 and miR-181a were not found in interlaminar astrocytes, but were detected in astrocytes from deep cortical layers), suggesting their involvement in the regulation of factors specific to those regions. In line with the previously mentioned results, a low expression of miRNAs involved with inflammatory responses was observed in fetal compared to adult white-matter astrocytes. A differential expression was also observed between adult interlaminar astrocytes compared to those found at the fetal germinal matrix, as the latter contains miRNAs involved in neurogenesis with antiapoptotic properties (eg, miR-210, miR-129, and miR-214).77
In various pathological contexts, astrocyte miRNAs have been shown to increase or decrease their levels, as summarized in Table 2. The highly expressed miR-29 family in astrocytes is transported with functional consequences in exosomes.82 When the expression of miR-29b increases by exposure to pathological conditions in the brain and in cultured astrocytes, its derived exosomes reduce neuronal viability by targeting the platelet-derived growth factor (PDGF-B).78 Surprisingly, transfecting a mimic of miR-29a into cultured astrocytes protects them from oxygen-glucose deprivation.79 As the targets of miR-29 are both pro- and antiapoptotic, this apparently controversial effect can be explained by different mRNAs being overexpressed and thus targeted under diverse physiological or pathological conditions.70
Table 2.
Regulation of miRNAs in astrocytes.
| CONDITION | miRNA IN ASTROCYTES | EFFECT | SOURCE | REF |
|---|---|---|---|---|
| Spinal cord injury | 21 | ⬆ around lesion | In situ (mouse) | 81 |
| 145 | ⬇ around lesion | In situ (rat) | 88 | |
| Glioneuronal lesions (from epileptic patients) | 146a | ⬆ | In situ (tissue from epileptic patients) | 82 |
| Forebrain ischemia | 29a | ⬆ dentate gyrus | Hippocampus | 79 |
| ⬇ CA1 | ||||
| Multiple sclerosis lesions patients | 155, 160 | ⬆ | Laser capture microdissection from white matter | 83 |
| Oxygen Glucose Deprivation | 21, 29b, 30b, 107, 137, 210 | ⬆ | Cultured rat astrocytes | 84 |
| 7 | ⬇ | Cultured mice astrocytes | 89 | |
| Transfection with human heme oxygenase 1 | 140, 17, 16 | ⬆ | Cultured rat astrocytes | 68 |
| 297, 206, 187, 181a, 138, 29c | ⬇ | |||
| Ammonia | 26a, 30b, 30e, 125, 135, 145, 425… (43 in total) | ⬆ | Cultured rat astrocytes | 69 |
| Aβ 42 | 146a | ⬆ | Cultured human astrocytes | 85 |
| LPS | 145 | ⬇ | Cultured rat astrocytes | 88 |
| LPS and IFN-γ | 146a, 155 | ⬆ | Cultured mice astrocytes | 73 |
| 149, 455, 351, 298 | ⬇ | |||
| 146a, 155 | ⬆ | Cultured marmoset astrocytes | ||
| 149, 455, 125b | ⬇ | |||
| IL-10 or IL-4 | 145 | ⬆ | Cultured rat astrocytes | 88 |
| IL-β | 146a | ⬆ | Cultured human astrocytes | 82,85 |
| Inflammatory cytokines (e.g. IL-β, IFN-γ, TNF-α, etc) | 23a, 146a, 155 | ⬆ | Cultured human astrocytes | 83 |
| IL-6 | 125b | ⬆ | Cultured human astrocytes | 86 |
| IL-1/IFN-γ | 155, 483-3p, 147, 27a, 147b, solexa-578-1915, 23a, 155, 29b-1, 33b, 146a | ⬆ | Cultured human astrocytes | 87 |
| 296-3p, 767-3p | ⬇ |
Another frequently addressed neuropathological phenomenon is astrogliosis. For example, after spinal cord injury, the activation of astrocytes involving the overexpression of astrocyte-specific genes such as the glial fibrillary acid protein favors the repair and limits the inflammation of the damaged zone.80 The concomitant increase of miR-21, however, attenuates the beneficial astrocytic response to injury and limits axon numbers in the scar zone. Although the targets of miR-21 explaining this effect are not clear, inhibiting miR-21 facilitates recovery and increases axon density.81 The expression of miR-146 is upregulated in tissue obtained from patients with intractable epilepsy, especially in areas of noticeable gliosis. This miRNA regulates inflammation: in cultured astrocytes, its upregulation reduces the interleukin-1β (IL-1β) mediated increase of interleukin-6 (IL-6) and cyclooxygenase 2 (COX-2), two molecules associated with inflammatory gliosis.82 In tissue obtained from multiple sclerosis patients, most of the miRNAs induced by the lesions are present in astrocytes, with a subset of these being upregulated by cytokines (eg, miR-23a, miR-146a, and miR-155). Some of these miRNAs (miR-155, miR-326, and miR-34a) regulate the expression of CD47, leading to the release of macrophages and promoting myelin phagocytosis.83 A differential expression of astrocytic miRNAs has been observed after oxygen-glucose deprivation, and this expression profile changed with time after deprivation, as was the case with miR-29b and miR-21, upregulated after 12 hours, or of miR-30b and miR-107, upregulated after 6 and 8 hours after oxygen-glucose deprivation, respectively.84 The levels of miRNAs in astrocytes also change after exposure to various stressors that increase oxidative stress such as Aβ-42,85 ammonia,69 and transfection with human heme oxygenase 1, an enzyme induced by oxidative stress that is overexpressed in a number of CNS disorders.68 In line with these observations, exposing astrocytes to cytokines and pro-inflammatory molecules (eg, interleukins, tumor necrosis factors, and interferon gamma) has a profound impact on the expression of miRNAs.73,82,83,85–88
Though still speculative, the prospect that some of the abovementioned miRNAs may be found in exosomes circulating in body fluids and thus amenable to be used as biomarkers, or the possibility of engineering exosomes with sequences that either mimic or antagonize these miRNAs to target astrocytes under pathological conditions remains an exciting topic of exploration.
The miRNA Content of Astrocyte-derived Exosomes: Focusing on miR-26a
An extensive RNA sequencing profile of astrocyte-derived exosomes under different physiological or pathological conditions is a must to predict the main functional pathways that exosomal miRNAs modify on target cells. On the next part of this commentary, we will focus on the possible transfer of miR-26a within exosomes based on the following arguments: (1) it is highly expressed in astrocytes when compared with neurons,72 (2) we have found miR-26a to be present in astrocyte-derived exosomes (unpublished data), (3) this miRNA is dysregulated in several CNS diseases (see below), and (4) miR-26a can be sorted to exosomes and transported by these vesicles in the plasma, serum, whole blood, urine, or secreted in vitro by human umbilical vein endothelial cells.90–94
Altered levels of miR-26 have been reported for some of the most prevalent disorders of the CNS. In human postmortem tissue, this miRNA can be upregulated or down-regulated, depending on the brain region, at different stages of Alzheimer’s disease (AD)95–99 as well as in biofluids (eg, blood, serum, and cerebrospinal fluid) obtained from AD patients.100,101 Increased levels of miR-26b have been observed in the substantia nigra of patients with Parkinson’s disease (PD),102,103 as well as in the blood of PD patients who underwent treatment compared with those who did not.104 Levels of miR-26a are enhanced in the relapsing phase of patients with relapsing-remitting multiple sclerosis compared to the remitting phase and to healthy controls.105 Patients diagnosed with major depression showed lower blood levels of miR-26a/b,101 while treatment with the antidepressant escitalopram increased its blood levels.106 Patients diagnosed with schizophrenia showed lower blood levels of miR-26a/b,101 while the precursor and mature miR-26b was upregulated in postmortem tissue from the superior temporal gyrus and the dorsolateral prefrontal cortex107 and downregulated in postmortem tissue from the prefrontal cortex.108
During neurogenesis, an increase in the level of some miR-26 family members has been reported,109–111 in contrast to pre-miR-26,72 which remains at constant levels.110 In zebrafish, miR-26b targets the mRNA of the C-terminal domain of the small phosphatases (ctdsp2) gene. As the sequence for miR-26b is localized in one of the introns of ctdsp2, this is a case where a gene is transcribed simultaneously with the miRNA that regulates its expression. For neuronal differentiation to occur, miR-26b has to inhibit ctdsp2 translation, therefore allowing for genes containing the repressor element 1 (RE-1; otherwise inhibited by Ctspd2) sequence to be expressed. MiR-26a is also able to repress the expression of ctdsp2 and shows a upregulation similar to miR-26b during progression of neurogenesis.110,112
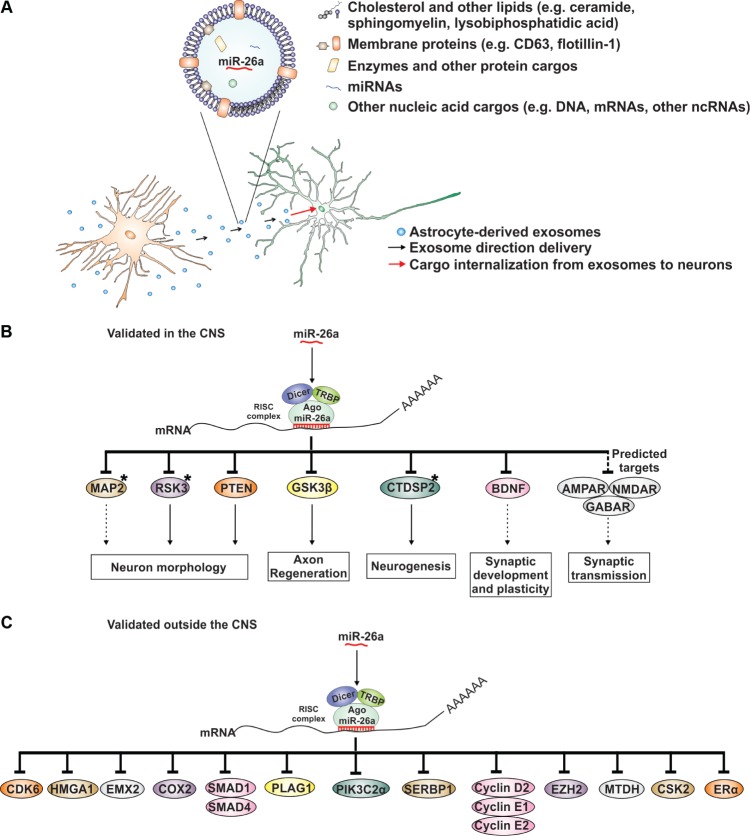
The predicted mRNA targets of a miRNA are commonly validated by tools such as quantitative real-time polymerase chain reaction (qRT-PCR), Western blots, or luciferase reporter assays. The first two do not exclude that the decrease of a given mRNA or protein is secondarily associated with the downregulation of the miRNA target sequence. However, the luciferase reporter assay provides direct evidence of mRNA targets, because the expression of the luciferase reporter 3′-UTR construct will decrease in the presence of a regulatory miRNA, which is able to interact with the introduced 3′-UTR sequence.113 Figure 1 contains the proposal of the present commentary, in which astrocyte-derived exosomes carrying miR-26a will influence neuronal function and brain physiology through validated and predicted targets.
Figure 1.
Exosomes are released by astrocytes and their cargo internalized by neurons, resulting in the regulation of neuronal function according to the cargo identity. (A) A simplified diagram of an exosome and its principal components. miR-26a is highlighted as an example of an miRNA that is highly expressed in astrocytes and transported by exosomes. (B) Targets of miR-26a in the central nervous system. miR-26a is incorporated into the RNA-induced silencing complex, where it can recognize a mRNA sequence complementary to its seed region, leading to RNA silencing. Those mRNA targets of miR-26a that have not been validated by luciferase assays are marked with an asterisk (*). Included in the diagram are targets found in silico that need further validation. Solid arrows show the reported impact on neuronal physiology. Dashed arrows show a possible impact on neuronal physiology that needs to be corroborated by experimental evidence after modulation of miR-26a levels. Protein names: MAP2, microtubule associated protein 2; RSK3, ribosomal protein S6 kinase; PTEN, tumor suppressor phosphatase and tensin homolog; GSK-3β, glycogen synthase kinase-3β; CTDSP2, C-terminal domain of small phosphatases 2; BDNF, brain-derived neurotrophic factor; NMDAR, N-methyl-d-aspartate receptor subunits; AMPAR, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunits; GABA, (ionotropic) gamma-aminobutyric acid receptor subunits. (C) MiR-26a targets outside the central nervous system. Their possible regulation in the brain needs to be explored experimentally.
Abbreviations: CDK6, cyclin-dependent kinase 6; HMGA1, high mobility group AT-hook 1; EMX2, empty spiracles homeobox 2; COX-2, cyclooxygenase 2; SMAD1, mothers against decapentaplegic homolog 1 and 4; PLAG1, pleiomorphic adenoma gene 1; PIK3C2α, phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide; SERBP1, plasminogen activator inhibitor 1 RNA-binding protein; EZH2, enhancer of zeste homolog 2; MTDH, metadherin; CSK2, cyclin-dependent kinases regulatory subunit 2; Erα, estrogen receptor alpha.
One of the validated targets of miR-26 members in neurons and other cell types under physiological/pathological conditions is GSK-3β.59,114–116 GSK-3β is a cytosolic protein that takes part in the canonical Wnt pathway, and it is capable of forming a complex with β-catenin and two other proteins: axin and adenomatous polyposis coli.117 In the absence of Wnt signaling, β-catenin is phosphorylated by GSK-3β and degraded.118 However, under Wnt signaling, GSK-3β is inhibited, allowing the stabilization of β-catenin and its translocation into the nucleus to activate the expression of Wnt target genes.119,120
Another target of miR-26 detected in neurons is the phosphatase PTEN, a negative regulator of the phosphoinositide 3-kinase (PI3K)/AKT pathway.58 PI3K/AKT inhibits the previously mentioned target of miR-26a GSK-3β, by phosphorylation of a serine residue.121 Inhibition of PTEN increases neurite outgrowth both in vitro and in vivo,122–125 and these changes resemble those observed under miR-26a regulation, with the concomitant decrease in PTEN levels and increase in AKT protein levels.58,126
Other targets validated by luciferase reporter assays include Wnt5a,127,128 a ligand of the noncanonical, β-catenin independent pathway,129 and brain-derived neurotrophic factor (BDNF),130 a growth factor that controls a wide range of mechanisms in the CNS, including plasticity and synaptic maturation.131
Potential and Demonstrated miR-26a Functional Consequences on Neurons
In hippocampal neurons, the exogenous addition of miR-26a mimics induces an increment in axonal length;111 consistently, electroporation of an inhibitor of miR-26a prevents axonal regeneration in vivo after sciatic nerve crush.59 Neurite outgrowth is also enhanced by the addition of miR-26a in dorsal root ganglia and in primary cortical neurons.58,126 In mature primary hippocampal neurons (ie, 14DIV), transfection with a miR-26a-expressing plasmid or a miR-26a sponge did not affect spine area or density, but miR-26a did inhibit the spine enlargement induced after 90 minutes of chemical long term potentiation (LTP). Reduction in the levels of miR-26a is necessary for maintenance of LTP but not for its induction, and the effects of miR-26a on both, spine remodeling and LTP, was mediated by the miR-26a targeting of ribosomal S6 kinase 3 (RSK3).132 This evidence suggests that this miRNA could have a differential effect in two different neuronal compartments: the dendritic and the axonal arborization.
Outside the CNS, other targets of miR-26a have been validated: plasminogen activator inhibitor 1 RNA-binding protein (serbp1),133 phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (PIK3C2α),134 and mothers against decapentaplegic homolog 1 and 4 (Smad1 and Smad4).135–137 Cyclooxgenase-2 (COX-2),138 metadherin (MTDH),139,140 cyclin-dependent kinases regulatory subunit 2 (CSK2),141 enhancer of zeste homolog 2 (EZH2),139,142,143 estrogen receptor alpha (ERα),144 pleiomorphic adenoma gene 1 (PLAG1),145 empty spiracles homeobox 2 (emx2),146 high mobility group AT-hook 1 (HMGA1),147 cyclin-dependent kinase 6 (CDK6), cyclin E1,148 and cyclins D2 and E2.149 Further experiments will be needed to study if miR-26a is also regulating some of these targets in the CNS. Last, it is worth mentioning that in silico studies of predicted targets for miR-26a include mRNAs that express subunits for receptors involved in synaptic transmission, such as GABAR, AMPAR, and NMDAR. These need to be validated in the CNS and the impact of this modulation remains to be assessed.
Conclusion
The role of exosomes derived from astrocytes on neuronal physiology promises to be an exciting area of study. Such a mechanism of communication suggests a new level of complexity in the processing of information in the CNS. MiRNAs stand out as key regulators of cellular processes, due to their capacity to inhibit hundreds of different mRNA targets. We have focused on miR-26a in this review, as it targets mRNAs that may impact neuronal function and morphology, and its dysregulation has been implicated in many neurological disorders, such as depression and AD. As such, it may be an interesting therapeutic target and biomarker for pathologies of the CNS. This and other miRNAs packaged in exosomes of astrocytic origin may open the doors to a better understanding of how astrocytes impact on neuronal functions, and it may also provide us with new tools to compensate for cellular malfunctions under pathological conditions.
Acknowledgments
We are grateful to Soledad Sandoval and Teresa Gomez for their technical and administrative support.
Footnotes
ACADEMIC EDITOR: Lora Talley Watts, Editor in Chief
PEER REVIEW: Two peer reviewers contributed to the peer review report. Reviewers’ reports totaled 698 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by Fondecyt Program 1140108. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: JPR, CL, and AL. Developed the structure and arguments of the paper: CL, AL, and UW. Produced figures and tables: JPR, AF, CL, AL, and UW. Agreed with manuscript proposal and conclusions: CL, JPR, AL, AF, and UW. Wrote the final version of the manuscript: UW. Made critical revisions: AL. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Giaume C, Koulakoff A, Roux L, Holcman D, Rouach N. Astroglial networks: a step further in neuroglial and gliovascular interactions. Nat Rev Neurosci. 2010;11(2):87–99. doi: 10.1038/nrn2757. [DOI] [PubMed] [Google Scholar]
- 2.Araque A, Parpura V, Sanzgiri RP, Haydon PG. Tripartite synapses: Glia, the unacknowledged partner. Trends Neurosci. 1999;22(5):208–215. doi: 10.1016/s0166-2236(98)01349-6. [DOI] [PubMed] [Google Scholar]
- 3.Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci. 2013;14(5):311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koehler RC, Roman RJ, Harder DR. Astrocytes and the regulation of cerebral blood flow. Trends Neurosci. 2009;32(3):160–169. doi: 10.1016/j.tins.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Rinholm JE, Bergersen LH. Neuroscience: the wrap that feeds neurons. Nature. 2012;487(7408):435–436. doi: 10.1038/487435a. [DOI] [PubMed] [Google Scholar]
- 6.Allen NJ. Role of glia in developmental synapse formation. Curr Opin Neurobiol. 2013;23(6):1027–1033. doi: 10.1016/j.conb.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Chung W-S, Clarke LE, Wang GX, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504(7480):394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakely PK, Hussain S, Carlin LE, Irani DN. Astrocyte matricellular proteins that control excitatory synaptogenesis are regulated by inflammatory cytokines and correlate with paralysis severity during experimental autoimmune encephalomyelitis. Front Neurosci. 2015 Oct;9:1–11. doi: 10.3389/fnins.2015.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanemaru K, Sekiya H, Xu M, et al. In vivo visualization of subtle, transient, and local activity of astrocytes using an ultrasensitive Ca2+ indicator. Cell Rep. 2014;8(1):311–318. doi: 10.1016/j.celrep.2014.05.056. [DOI] [PubMed] [Google Scholar]
- 10.Kıray H, Lindsay SL, Hosseinzadeh S, Barnett SC. The multifaceted role of astrocytes in regulating myelination. Exp Neurol. 2016:1–9. doi: 10.1016/j.expneurol.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budnik V, Ruiz-Cañada C, Wendler F. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci. 2016;17(3):160–172. doi: 10.1038/nrn.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotelo-Hitschfeld T, Niemeyer MI, Machler P, et al. Channel-mediated lactate release by K+-stimulated astrocytes. J Neurosci. 2015;35(10):4168–4178. doi: 10.1523/JNEUROSCI.5036-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91(22):10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mächler P, Wyss MT, Elsayed M, et al. In vivo evidence for a lactate gradient from astrocytes to neurons. Cell Metab. 2016;23(1):94–102. doi: 10.1016/j.cmet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Barros LF. Metabolic signaling by lactate in the brain. Trends Neurosci. 2013;36:396–404. doi: 10.1016/j.tins.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Bergles DE, Diamond JS, Jahr CE. Clearance of glutamate inside the synapse and beyond. Curr Opin Neurobiol. 1999;9(3):293–298. doi: 10.1016/s0959-4388(99)80043-9. [DOI] [PubMed] [Google Scholar]
- 17.Murphy-Royal C, Dupuis JP, Varela JA, et al. Surface diffusion of astrocytic glutamate transporters shapes synaptic transmission. Nat Neurosci. 2015;18(2):219–226. doi: 10.1038/nn.3901. [DOI] [PubMed] [Google Scholar]
- 18.Westergaard N, Sonnewald U, Schousboe A. Metabolic trafficking between neurons and astrocytes: the glutamate/glutamine cycle revisited. Dev Neurosci. 1995;17:203–211. doi: 10.1159/000111288. [DOI] [PubMed] [Google Scholar]
- 19.Tani H, Dulla CG, Farzampour Z, Taylor-Weiner A, Huguenard JR, Reimer RJ. A local glutamate-glutamine cycle sustains synaptic excitatory transmitter release. Neuron. 2014;81(4):888–900. doi: 10.1016/j.neuron.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benedetti B, Matyash V, Kettenmann H. Astrocytes control GABAergic inhibition of neurons in the mouse barrel cortex. J Physiol. 2011;589(pt 5):1159–1172. doi: 10.1113/jphysiol.2010.203224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christian CA, Huguenard JR. Astrocytes potentiate GABAergic transmission in the thalamic reticular nucleus via endozepine signaling. Proc Natl Acad Sci U S A. 2013;110(50):20278–20283. doi: 10.1073/pnas.1318031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Schwab C, Mcgeer PL. Astrocytes are GABAergic cells that modulate microglial activity. Glia. 2011;59(1):152–165. doi: 10.1002/glia.21087. [DOI] [PubMed] [Google Scholar]
- 23.Henneberger C, Papouin T, Oliet SHR, Rusakov DA. Long-term potentiation depends on release of D-serine from astrocytes. Nature. 2010;463(7278):232–236. doi: 10.1038/nature08673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lima A, Sardinha VM, Oliveira AF, et al. Astrocyte pathology in the prefrontal cortex impairs the cognitive function of rats. Mol Psychiatry. 2014;19(7):834–841. doi: 10.1038/mp.2013.182. [DOI] [PubMed] [Google Scholar]
- 25.Ricci G, Volpi L, Pasquali L, Petrozzi L, Siciliano G. Astrocyte-neuron interactions in neurological disorders. J Biol Phys. 2009;35(4):317–336. doi: 10.1007/s10867-009-9157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takemoto T, Ishihara Y, Ishida A, Yamazaki T. Neuroprotection elicited by nerve growth factor and brain-derived neurotrophic factor released from astrocytes in response to methylmercury. Environ Toxicol Pharmacol. 2015;40(1):199–205. doi: 10.1016/j.etap.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Berg A, Zelano J, Pekna M, Wilhelmsson U, Pekny M, Cullheim S. Axonal regeneration after sciatic nerve lesion is delayed but complete in GFAP- and vimentin-deficient mice. PLoS One. 2013;8(11):e79395. doi: 10.1371/journal.pone.0079395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otsu Y, Couchman K, Lyons DG, et al. Calcium dynamics in astrocyte processes during neurovascular coupling. Nat Neurosci. 2015;18(2):210–218. doi: 10.1038/nn.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowianski P, Lietzau G, Steliga A, Waskow M, Morys J. The astrocytic contribution to neurovascular coupling—still more questions than answers? Neurosci Res. 2013;75(3):171–183. doi: 10.1016/j.neures.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Janzer RC, Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature. 1987;325(6101):253–257. doi: 10.1038/325253a0. [DOI] [PubMed] [Google Scholar]
- 31.Poskanzer KE, Yuste R. Astrocytes regulate cortical state switching in vivo. Proc Natl Acad Sci U S A. 2016;113(19):E2675–E2684. doi: 10.1073/pnas.1520759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 33.McKelvey KJ, Powell KL, Ashton AW, Morris JM, McCracken SA. Exosomes: mechanisms of uptake. J Circ Biomarkers. 2015;4(7):1. doi: 10.5772/61186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77(1):13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 35.Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 36.Thery C, Clayton A, Amigorena S, Raposo G. Isolation and characterization of exosomes from cell culture supernatants. Curr Protoc Cell Biol. 2006:1–29. doi: 10.1002/0471143030.cb0322s30. Chapter 3. [DOI] [PubMed] [Google Scholar]
- 37.Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frühbeis C, Fröhlich D, Krämer-Albers EM. Emerging roles of exosomes in neuronglia communication. Front Physiol. 2012;3:1–7. doi: 10.3389/fphys.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor AR, Robinson MB, Gifondorwa DJ, Tytell M, Milligan CE. Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev Neurobiol. 2007;67(13):1815–1829. doi: 10.1002/dneu.20559. [DOI] [PubMed] [Google Scholar]
- 40.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117(1):1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Cesca F, Loers G, et al. Synapsin I is an oligomannose-carrying glycoprotein, acts as an oligomannose-binding lectin, and promotes neurite outgrowth and neuronal survival when released via glia-derived exosomes. J Neurosci. 2011;31(20):7275–7290. doi: 10.1523/JNEUROSCI.6476-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Dinkins M, He Q, et al. Astrocytes secrete exosomes enriched with proapoptotic ceramide and Prostate Apoptosis Response 4 (PAR-4): potential mechanism of apoptosis induction in Alzheimer Disease (AD) J Biol Chem. 2012;287(25):21384–21395. doi: 10.1074/jbc.M112.340513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gosselin R-D, Meylan P, Decosterd I. Extracellular microvesicles from astrocytes contain functional glutamate transporters: regulation by protein kinase C and cell activation. Front Cell Neurosci. 2013 Dec;7:251. doi: 10.3389/fncel.2013.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandoval M, Luarte A, Herrera-Molina R, et al. The glycolytic enzyme aldolase C is up-regulated in rat forebrain microsomes and in the cerebrospinal fluid after repetitive fluoxetine treatment. Brain Res. 2013;1520:1–14. doi: 10.1016/j.brainres.2013.04.049. [DOI] [PubMed] [Google Scholar]
- 45.Guitart K, Loers G, Buck F, Bork U, Schachner M, Kleene R. Improvement of neuronal cell survival by astrocyte-derived exosomes under hypoxic and ischemic conditions depends on prion protein. Glia. 2016;64(6):896–910. doi: 10.1002/glia.22963. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Zhang S, Yao J, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Nedawi K. The yin-yang of microvesicles (exosomes) in cancer biology. Front Oncol. 2014;4(12):172. doi: 10.3389/fonc.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 49.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 51.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 52.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293(5538):2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130(2):299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10(12):842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 55.Altuvia Y, Landgraf P, Lithwick G, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions—beyond repression of gene expression. Nat Rev Genet. 2014;15(9):599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Ueno Y, Liu XS, et al. The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci. 2013;33(16):6885–6894. doi: 10.1523/JNEUROSCI.5180-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Sun H. MiR-26a promotes neurite outgrowth by repressing PTEN expression. Mol Med Rep. 2013;8(2):676–680. doi: 10.3892/mmr.2013.1534. [DOI] [PubMed] [Google Scholar]
- 59.Jiang J-J, Liu C-M, Zhang B-Y, et al. MicroRNA-26a supports mammalian axon regeneration in vivo by suppressing GSK3β expression. Cell Death Dis. 2015;6(8):e1865. doi: 10.1038/cddis.2015.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janas T, Janas MM, Sapoń K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015;589(13):1391–1398. doi: 10.1016/j.febslet.2015.04.036. [DOI] [PubMed] [Google Scholar]
- 62.Cha DJ, Franklin JL, Dou Y, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKenzie AJ, Hoshino D, Hong NH, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15(5):978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pigati L, Yaddanapudi SCS, Iyengar R, et al. Selective release of MicroRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5(10):e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13(1):357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palma J, Yaddanapudi SC, Pigati L, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40(18):9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jovičić A, Roshan R, Moisoi N, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. Ann Intern Med. 2013;158(6):5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin S-HH, Song W, Cressatti M, Zukor H, Wang E, Schipper HM. Heme oxygenase-1 modulates microRNA expression in cultured astroglia: implications for chronic brain disorders. Glia. 2015;63(7):1270–1284. doi: 10.1002/glia.22823. [DOI] [PubMed] [Google Scholar]
- 69.Oenarto J, Karababa A, Castoldi M, Bidmon HJ, Görg B, Häussinger D. Ammonia-induced miRNA expression changes in cultured rat astrocytes. Nat Publ Gr. 2016:1–12. doi: 10.1038/srep18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ouyang YB, Xu L, Yue S, Liu S, Giffard RG. Neuroprotection by astrocytes in brain ischemia: importance of microRNAs. Neurosci Lett. 2014;565:53–58. doi: 10.1016/j.neulet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butovsky O, Jedrychowski MP, Moore CS, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17(1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21(6):1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 73.Mor E, Cabilly Y, Goldshmit Y, et al. Species-specific microRNA roles elucidated following astrocyte activation. Nucleic Acids Res. 2011;39(9):3710–3723. doi: 10.1093/nar/gkq1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aranha MM, Santos DM, Solá S, Steer CJ, Rodrigues CMP. miR-34a regulates mouse neural stem cell differentiation. PLoS One. 2011;6(8):e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neo WH, Yap K, Lee SH, et al. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J Biol Chem. 2014;289(30):20788–20801. doi: 10.1074/jbc.M113.525493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krichevsky AM, Sonntag K-C, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24(4):857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rao VTS, Ludwin SK, Fuh S, et al. MicroRNA expression patterns in human astrocytes in relation to anatomical location and age. J Neuropathol Exp Neurol. 2016:1–11. doi: 10.1093/jnen/nlv016. [DOI] [PubMed] [Google Scholar]
- 78.Hu G, Yao H, Chaudhuri AD, et al. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis. 2012;3(8):e381. doi: 10.1038/cddis.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ouyang YB, Xu L, Lu Y, et al. Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia. 2013;61(11):1784–1794. doi: 10.1002/glia.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119(1):7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhalala OG, Pan L, Sahni V, et al. microRNA-21 regulates astrocytic response following spinal cord injury. J Neurosci. 2012;32(50):17935–17947. doi: 10.1523/JNEUROSCI.3860-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iyer A, Zurolo E, Prabowo A, et al. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS One. 2012;7(9):17–19. doi: 10.1371/journal.pone.0044789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Junker A, Krumbholz M, Eisele S, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132(12):3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 84.Ziu M, Fletcher L, Rana S, Jimenez DF, Digicaylioglu M. Temporal differences in microRNA expression patterns in astrocytes and neurons after ischemic injury. PLoS One. 2011;6(2):e14724. doi: 10.1371/journal.pone.0014724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem. 2010;285(50):38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476(1):18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- 87.Tarassishin L, Loudig O, Bauman A, Shafit-Zagardo B, Suh HS, Lee SC. Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155. Glia. 2011;59(12):1911–1922. doi: 10.1002/glia.21233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang CY, Yang SH, Tzeng SF. MicroRNA-145 as one negative regulator of astrogliosis. Glia. 2015;63(2):194–205. doi: 10.1002/glia.22743. [DOI] [PubMed] [Google Scholar]
- 89.Dong Y, Chen Z, Zhao Z, et al. Potential role of microRNA-7 in the anti-neuroinflammation effects of nicorandil in astrocytes induced by oxygen-glucose deprivation. J Neuroinflammation. 2016;13(1):60. doi: 10.1186/s12974-016-0527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, Zhang L, Liu F, Xiang G, Jiang D, Pu X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis Markers. 2015;2015:893594. doi: 10.1155/2015/893594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin X, He Y, Hou X, Zhang Z, Wang R, Wu Q. Endothelial cells can regulate smooth muscle cells in contractile phenotype through the miR-206/ARF6&NCX1/exosome axis. PLoS One. 2016;11(3):e0152959. doi: 10.1371/journal.pone.0152959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu S-C, Yang JC-S, Rau C-S, et al. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS One. 2013;8(10):e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ichii O, Otsuka-Kanazawa S, Horino T, et al. Decreased miR-26a expression correlates with the progression of podocyte injury in autoimmune glomerulonephritis. PLoS One. 2014;9(10):1–12. doi: 10.1371/journal.pone.0110383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3:1–14. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013;33(37):14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang W-X, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121(2):193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 98.Prendecki M, Dorszewska J. The role of MicroRNA in the pathogenesis and diagnosis of neurodegenerative diseases. Austin Alzheimers Parkinsons Dis. 2014;1(3):1–10. [Google Scholar]
- 99.Satoh J. Molecular network of microRNA targets in Alzheimer’s disease brains. Exp Neurol. 2012;235(2):436–446. doi: 10.1016/j.expneurol.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 100.Kumar S, Reddy PH. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim Biophys Acta. 2016;1862(9):1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leidinger P, Backes C, Deutscher S, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14(7):R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ. Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci. 2013;7:178. doi: 10.3389/fncel.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alvarez-Erviti L, Seow Y, Schapira AHV, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell Death Dis. 2013;4(3):e545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Margis R, Margis R, Rieder CRM. Identification of blood microRNAs associated to Parkinson’s disease. J Biotechnol. 2011;152(3):96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 105.Honardoost MA, Kiani-Esfahani A, Ghaedi K, Etemadifar M, Salehi M. MiR-326 and miR-26a, two potential markers for diagnosis of relapse and remission phases in patient with relapsing-remitting multiple sclerosis. Gene. 2014;544(2):128–133. doi: 10.1016/j.gene.2014.04.069. [DOI] [PubMed] [Google Scholar]
- 106.Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol. 2013;23(7):602–611. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 107.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15(12):1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perkins DO, Jeffries CD, Jarskog LF, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Busskamp V, Lewis NE, Guye P, et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol. 2014;10:760. doi: 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26(1):25–30. doi: 10.1101/gad.177774.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Spronsen M, van Battum EY, Kuijpers M, et al. Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS One. 2013;8(10):e74907. doi: 10.1371/journal.pone.0074907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han J, Denli AM, Gage FH. The enemy within: intronic miR-26b represses its host gene, ctdsp2, to regulate neurogenesis. Genes Dev. 2012;26(1):6–10. doi: 10.1101/gad.184416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuhn DE, Martin MM, Feldman DS, Terry AV, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44(1):47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suh JH, Choi E, Cha M-JJ, et al. Up-regulation of miR-26a promotes apoptosis of hypoxic rat neonatal cardiomyocytes by repressing GSK-3β protein expression. Biochem Biophys Res Commun. 2012;423(2):404–410. doi: 10.1016/j.bbrc.2012.05.138. [DOI] [PubMed] [Google Scholar]
- 115.Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates MicroRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3β. J Biol Chem. 2010;285(38):29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun J, Yan P, Chen Y, et al. MicroRNA-26b inhibits cell proliferation and cytokine secretion in human RASF cells via the Wnt/GSK-3β/β-catenin pathway. Diagn Pathol. 2015;10(1):72. doi: 10.1186/s13000-015-0309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Valvezan AJ, Zhang F, Diehl JA, Klein PS. Adenomatous polyposis coli (APC) regulates multiple signaling pathways by enhancing glycogen synthase kinase-3 (GSK-3) activity. J Biol Chem. 2012;287(6):3823–3832. doi: 10.1074/jbc.M111.323337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβ-TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13(3):270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fagotto F. Looking beyond the Wnt pathway for the deep nature of β-catenin. EMBO Rep. 2013;14(5):422–433. doi: 10.1038/embor.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59(1):3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 121.Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11(8):539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ohtake Y, Park D, Abdul-Muneer PM, et al. The effect of systemic PTEN antagonist peptides on axon growth and functional recovery after spinal cord injury. Biomaterials. 2014;35(16):4610–4626. doi: 10.1016/j.biomaterials.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mao L, Jia J, Zhou X, et al. Delayed administration of a PTEN inhibitor BPV improves functional recovery after experimental stroke. Neuroscience. 2013;231:272–281. doi: 10.1016/j.neuroscience.2012.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lewandowski G, Steward O. AAVshRNA-mediated suppression of PTEN in adult rats in combination with salmon fibrin administration enables regenerative growth of corticospinal axons and enhances recovery of voluntary motor function after cervical spinal cord injury. J Neurosci. 2014;34(30):9951–9962. doi: 10.1523/JNEUROSCI.1996-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30(27):9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cui C, Xu G, Qiu J, Fan X. Up-regulation of miR-26a promotes neurite outgrowth and ameliorates apoptosis by inhibiting PTEN in bupivacaine injured mouse dorsal root ganglia. Cell Biol Int. 2015;39(8):933–942. doi: 10.1002/cbin.10461. [DOI] [PubMed] [Google Scholar]
- 127.Zhao S, Ye X, Xiao L, et al. MiR-26a inhibits prostate cancer progression by repression of Wnt5a. Tumour Biol. 2014;35(10):9725–9733. doi: 10.1007/s13277-014-2206-4. [DOI] [PubMed] [Google Scholar]
- 128.Li S, Hu C, Li J, et al. Effect of miR-26a-5p on the Wnt/Ca2+ pathway and osteogenic differentiation of mouse adipose-derived mesenchymal stem cells. Calcif Tissue Int. 2016;99(2):174–186. doi: 10.1007/s00223-016-0137-3. [DOI] [PubMed] [Google Scholar]
- 129.Kikuchi A, Yamamoto H, Sato A, Matsumoto S. Wnt5a: its signalling, functions and implication in diseases. Acta Physiol. 2012;204(1):17–33. doi: 10.1111/j.1748-1716.2011.02294.x. [DOI] [PubMed] [Google Scholar]
- 130.Caputo V, Sinibaldi L, Fiorentino A, et al. Brain derived neurotrophic factor (BDNF) expression is regulated by microRNAs miR-26a and miR-26b allele-specific binding. PLoS One. 2011;6(12):e28656. doi: 10.1371/journal.pone.0028656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sakata K. Brain-derived neurotrophic factor and major depression. Neurobiol Depress. 2012;64(2):391–417. [Google Scholar]
- 132.Gu Q-H, Yu D, Hu Z, et al. miR-26a and miR-384-5p are required for LTP maintenance and spine enlargement. Nat Commun. 2015;6:6789. doi: 10.1038/ncomms7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 134.Chai ZT, Kong J, Zhu XD, et al. MicroRNA-26a inhibits angiogenesis by down-regulating VEGFA through the PIK3C2α/Akt/HIF-1α pathway in hepatocellular carcinoma. PLoS One. 2013;8(10):1–12. doi: 10.1371/journal.pone.0077957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Trompeter HI, Dreesen J, Hermann E, et al. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics. 2013;14:111. doi: 10.1186/1471-2164-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23(2):287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 137.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kwon Y, Kim Y, Eom S, et al. MicroRNA-26a/-26b-COX-2-MIP-2 loop regulates allergic inflammation and allergic inflammation-promoted enhanced tumorigenic and metastatic potential of cancer cells. J Biol Chem. 2015;290(22):14245–14266. doi: 10.1074/jbc.M115.645580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang B, Liu X, He J, et al. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011;32(1):2–9. doi: 10.1093/carcin/bgq209. [DOI] [PubMed] [Google Scholar]
- 140.Liu P, Tang H, Chen B, et al. MiR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett. 2015;357(1):384–392. doi: 10.1016/j.canlet.2014.11.050. [DOI] [PubMed] [Google Scholar]
- 141.Lv M, Zhang X, Li M, et al. miR-26a and its target CKS2 modulate cell growth and tumorigenesis of papillary thyroid carcinoma. PLoS One. 2013;8(7):1–11. doi: 10.1371/journal.pone.0067591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Sander S, Bullinger L, Klapproth K, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112(10):4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 143.Song QIC, Shi ZB, Zhang YT, et al. Downregulation of microRNA-26a is associated with metastatic potential and the poor prognosis of osteosarcoma patients. Oncol Rep. 2014;31(3):1263–1270. doi: 10.3892/or.2014.2989. [DOI] [PubMed] [Google Scholar]
- 144.Shen W, Song M, Liu J, et al. MiR-26a promotes ovarian cancer proliferation and tumorigenesis. PLoS One. 2014;9(1):e86871. doi: 10.1371/journal.pone.0086871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Volinia S, Calin GA, Liu C-G, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yin C, Zhang J, Shi Z, Sun W, Zhang H, Fu Y. Identification and expression of the target gene emx2 of miR-26a and miR-26b in Paralichthys olivaceus. Gene. 2015;570(2):205–212. doi: 10.1016/j.gene.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 147.Zhao XX, Yuan QZ, Mu DP, et al. MicroRNA-26a inhibits proliferation by targeting high mobility group AT-hook 1 in breast cancer. Int J Clin Exp Pathol. 2015;8(1):368–373. [PMC free article] [PubMed] [Google Scholar]
- 148.Zhu Y, Lu Y, Zhang Q, et al. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40(10):4615–4625. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]