Significance
Growth of mitochondria and their transmission during cytokinesis depends on protein import from the cytosol as well as on the faithful replication and segregation of their genomes. Here, we show that in the early diverging eukaryote Trypanosoma brucei, a single mitochondrial outer membrane protein controls both the assembly of the master protein translocase as well as the inheritance of the single-unit mitochondrial genome. Although the protein is unique to trypanosomatids, it shows functional similarity to an outer membrane protein of yeast. The concept that a single protein mediates the assembly of the major protein translocase and the transmission of mitochondrial DNA might therefore be conserved in other eukaryotes.
Keywords: mitochondria, protein import, mitochondrial genome, trypanosomes, evolution
Abstract
Trypanosomatids are one of the earliest diverging eukaryotes that have fully functional mitochondria. pATOM36 is a trypanosomatid-specific essential mitochondrial outer membrane protein that has been implicated in protein import. Changes in the mitochondrial proteome induced by ablation of pATOM36 and in vitro assays show that pATOM36 is required for the assembly of the archaic translocase of the outer membrane (ATOM), the functional analog of the TOM complex in other organisms. Reciprocal pull-down experiments and immunofluorescence analyses demonstrate that a fraction of pATOM36 interacts and colocalizes with TAC65, a previously uncharacterized essential component of the tripartite attachment complex (TAC). The TAC links the single-unit mitochondrial genome to the basal body of the flagellum and mediates the segregation of the replicated mitochondrial genomes. RNAi experiments show that pATOM36, in line with its dual localization, is not only essential for ATOM complex assembly but also for segregation of the replicated mitochondrial genomes. However, the two functions are distinct, as a truncated version of pATOM36 lacking the 75 C-terminal amino acids can rescue kinetoplast DNA missegregation but not the lack of ATOM complex assembly. Thus, pATOM36 has a dual function and integrates mitochondrial protein import with mitochondrial DNA inheritance.
Mitochondria are a hallmark of all eukaryotic cells and are involved in many important functions including oxidative phosphorylation (1). The evolutionary origin of mitochondria can be traced back to a single endosymbiotic event between a free-living bacterium and an archaeal host cell. This explains why all mitochondria have a genome, which however encodes a small number of genes only (2). As a consequence, more than 95% of the mitochondrial proteome derives from nuclear-encoded proteins, which are synthesized in the cytosol and subsequently imported into mitochondria. Although the number of mitochondrially encoded proteins is small, they are essential for organellar function. Mitochondrial protein import as well as mitochondrial DNA replication and segregation are therefore key processes required for growth, division, and faithful transmission of mitochondria to the daughter cell (3–7).
The parasitic protozoa Trypanosoma brucei represents one of the earliest diverging branches of the eukaryotic evolutionary tree and has a fully functional mitochondrion capable of oxidative phosphorylation (8, 9). T. brucei is only remotely related to yeast and mammals, in which mitochondrial biogenesis is best understood. This unique phylogenetic position explains why in trypanosomes basic biological processes including mitochondrial protein import and mitochondrial DNA inheritance function quite differently than in other eukaryotes (6, 10, 11).
The protein import complex in the outer membrane (OM) of the single trypanosomal mitochondrion has recently been characterized and termed archaic translocase of the OM (ATOM) complex (12). It consists of two core subunits, the β-barrel membrane protein ATOM40 (13) and the α-helically anchored ATOM14, which are remote orthologs of yeast Tom40 and Tom22, respectively, although ATOM40 also shows similarity to a bacterial β-barrel protein. The four remaining translocase subunits (ATOM11, ATOM12, ATOM46, and ATOM69) do not show similarity to TOM complex subunits of yeast or any other organism outside the Kinetoplastids. Two of them, ATOM46 and ATOM69, have large cytosolic domains and function as protein import receptors. Their function is analogous to Tom20 and Tom70 of yeast, even though they do not show sequence similarity with any of these proteins and thus arose by convergent evolution (12). In addition to the ATOM complex, an abundant integral mitochondrial OM protein, which is specific for Kinetoplastids, was shown to be involved in import of a subset of mitochondrial proteins. Because in pull-down experiments of the tagged protein a small fraction of ATOM40, the protein import pore of the ATOM complex, was detected, it was termed peripherally associated ATOM36 (pATOM36) (14). However, when tagged ATOM40 was used as bait, pATOM36 was not precipitated, indicating that it is not a subunit of the ATOM complex.
The mitochondrial genome of T. brucei, termed kinetoplast DNA (kDNA), is also unusual. It consists of a single-unit kDNA network—made of two genetic elements, the maxi- and the minicircles—that localizes to a distinct region within the mitochondrial matrix (15). The kDNA network is physically attached to the cytosolic basal body, the organizer of the eukaryotic flagellum, via a transmembrane structure termed tripartite attachment complex (TAC), the molecular composition of which is only poorly defined. Morphologically, the TAC is composed of (i) the unilateral filaments that attach the kDNA to the inner mitochondrial membrane, (ii) the differentiated mitochondrial membranes, and (iii) the exclusion zone filaments, which connect the OM to the basal body (16).
Replication of the kDNA network occurs at a defined stage of the cell cycle shortly before the onset of the nuclear S phase. After replication, the kDNA networks need to be positioned so that during mitochondrial division and cytokinesis each daughter cell receives a single organelle with a single kDNA network. This process depends on an intact TAC and is mediated by the movement of the basal bodies (15–17).
In this study, we show that the specific function of the previously identified protein import factor pATOM36 is to mediate assembly of a small subset of mitochondrial OM proteins, including ATOM complex subunits. Moreover, we demonstrate that pATOM36 interacts with TAC65, a previously uncharacterized component of the TAC, and that in line with this finding, a fraction of pATOM36 is stably associated with the TAC. Finally, we present evidence that ablation of pATOM36 not only affects assembly of the ATOM complex but also leads to a segregation defect of the kDNA, resulting in a rapid loss of mitochondrial DNA. Therefore, pATOM36 simultaneously mediates ATOM complex assembly and thus protein import, as well as mitochondrial DNA inheritance, as it is required for the formation of the TAC.
Results
Ablation of pATOM36 Primarily Affects OM Proteins.
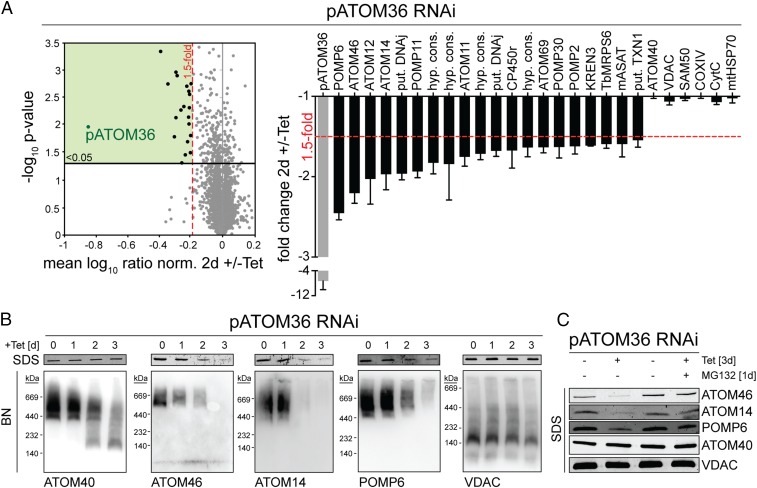
To more precisely study pATOM36 function, we analyzed the global changes its absence causes on the mitochondrial proteome. To that end, RNAi was combined with stable isotope labeling by amino acids in cell culture (SILAC) and high-resolution mass spectrometry (MS). Uninduced and induced pATOM36-RNAi cells were grown in medium containing different stable isotope-labeled variants of lysine and arginine. Subsequently, equal cell numbers of each population were mixed, and mitochondria-enriched fractions were prepared for quantitative MS analysis to compare the abundance of proteins in induced versus uninduced RNAi cells. To exclude indirect effects, we performed the SILAC analysis early after induction of RNAi, well before the onset of the growth phenotype. The results in Fig. 1A show that of 2,859 quantified proteins 21 were more than 1.5-fold down-regulated (P value < 0.05, n = 3). As expected, the most highly down-regulated protein is pATOM36 itself (sevenfold), the target of the RNAi. Interestingly, 17 out of the 21 proteins whose abundances were decreased are components of the previously characterized OM proteome (although two of them were only detected in one of the two OM preparations analyzed in ref. 18). Five of them, including the most strongly down-regulated ones, are subunits of the ATOM complex (12), indicating that pATOM36 plays an essential role in the biogenesis of this translocase.
Fig. 1.
Ablation of pATOM36 primarily affects OM proteins. (A) Global changes in the proteome of procyclic pATOM36-RNAi cells were analyzed by SILAC-MS by comparing protein abundances in 2-d-induced versus uninduced cells. For proteins quantified in at least two of the three replicates, the mean log10 of normalized ratios (induced/uninduced) were plotted against the –log10 P value (two-sided t test). Proteins with a P value < 0.05 that were down-regulated more than 1.5-fold are depicted as black dots in the green square. (Right) Graph showing which proteins were down-regulated more than 1.5-fold. A selection of proteins that were not down-regulated is depicted on the right side of the graph. SDs are depicted. COXIV, cytochrome c oxidase subunit IV; CP450r, NADPH-cytochrome p450 reductase; CytC, cytochrome c; hyp. cons., hypothetical conserved; KREN3, RNA editing endoribonuclease; mASAT, mitochondrial aspartate aminotransferase; mtHSP70, mitochondrial heat shock protein 70; put., putative; Sam50, sorting and assembly machinery 50; TbMRPS6, mitochondrial ribosomal protein of small subunit 6; TXN1, tryparedoxin 1a. For a complete list of proteins, see Dataset S1. (B, Top) Immunoblots of whole-cell extracts analyzed by SDS/PAGE at the indicated time points after induction of pATOM36-RNAi. (B, Bottom) As in B, Top, but analysis was done with mitochondrial-enriched fractions by BN-PAGE. (C) Immunoblots of whole-cell extracts from the uninduced and 3-d-induced pATOM36-RNAi cell line separated by SDS/PAGE. The samples were either left untreated or incubated with 500 nM of the proteasome inhibitor MG132.
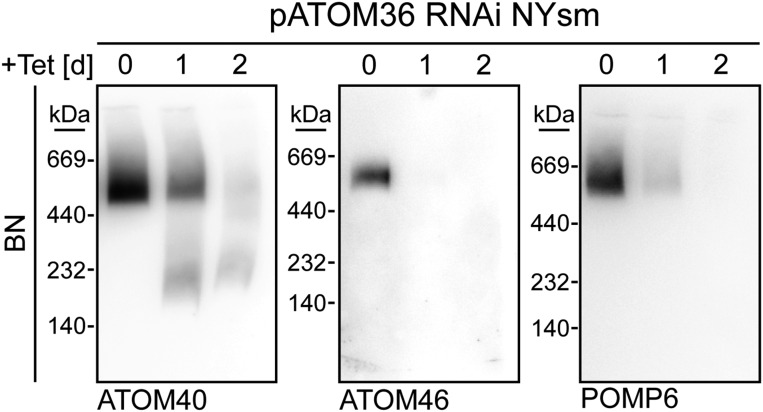
The SDS polyacrylamide gel electrophoresis (SDS/PAGE) immunoblot analysis in Fig. 1B, Top, shows that the steady state levels of ATOM14, ATOM46, and POMP6 in pATOM36-RNAi cells are affected not only in mitochondria-enriched fractions but also in whole cells. The levels of ATOM40 and voltage-dependent anion channel (VDAC), on the other hand, remained stable. Interestingly, the RNAi-induced decrease in the abundance of ATOM14, ATOM46, and POMP6 was considerably reduced in the presence of the proteasome inhibitor MG132, suggesting that it is caused by degradation of the proteins in the cytosol (Fig. 1C). The Blue Native (BN)-PAGE analysis of mitochondria-enriched fractions finally demonstrates that ablation of pATOM36, as a consequence of the decline of the various ATOM subunits, inhibits the assembly of the ATOM complex, whereas the complex formed by VDAC is only marginally decreased (Fig. 1B, Bottom). Essentially the same result was obtained in bloodstream form cells (Fig. S1).
Fig. S1.
Ablation of pATOM36 in bloodstream form (NYsm) cells inhibits ATOM complex assembly. Immunoblots of mitochondria-enriched fractions analyzed by BN-PAGE at the indicated time points after induction of pATOM36-RNAi in bloodstream form cells.
pATOM36 Is Required for Assembly but Not for Insertion of ATOM Subunits.
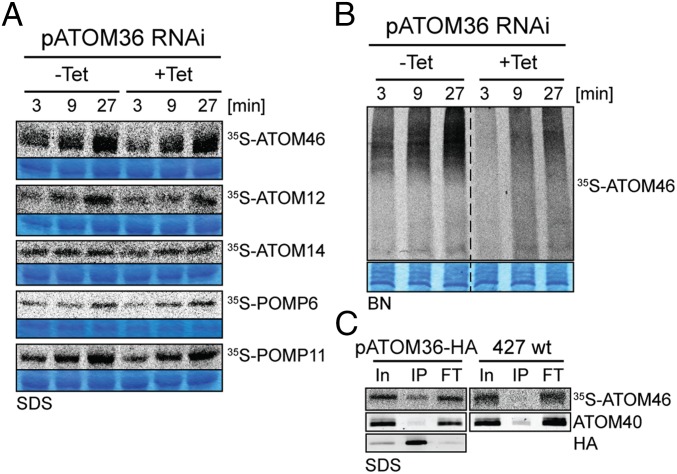
Our in vivo analysis of the pATOM36 RNAi cell line shows that the protein is essential for the biogenesis of the ATOM complex and of a number of OM proteins that are not subunits of the ATOM complex. To investigate at which stage pATOM36 may function, we performed membrane insertion assays using isolated mitochondria. To that end, putative pATOM36 substrate proteins, three ATOM subunits and two of the other OM proteins, were in vitro translated in the presence of [35S]methionine using reticulocyte lysate. The resulting radioactive proteins were then incubated with isolated mitochondria. As a means to measure insertion of the substrates into the mitochondrial OM, the reisolated organelles were extracted with sodium carbonate at pH 11.5. Such a treatment disrupts protein–protein interactions and extracts peripheral membrane proteins as well as soluble proteins. Integral membrane proteins, however, are retained in membrane sheets and thus are recovered in the pellet fraction (19). The SDS/PAGE analysis depicted in Fig. 2A shows a time-dependent increase of the radioactive signal in the carbonate insoluble pellet fractions for all tested substrates, indicating that they can be inserted into the OM of mitochondria isolated from a cell line containing wild-type levels of pATOM36. Interestingly, essentially the same result is obtained when mitochondria ablated for pATOM36 were used, demonstrating that the protein is not essential for OM insertion of the tested proteins when assayed in vitro.
Fig. 2.
pATOM36 is required for assembly but not for insertion of ATOM subunits. (A) In vitro insertion assay of the indicated 35S-Met–labeled OM proteins using mitochondria isolated from the uninduced (–Tet) and induced (+Tet, 2 d) procyclic pATOM36-RNAi cell line. Incubation time is indicated at the top. After alkaline carbonate extraction, the integral membrane proteins were separated by SDS/PAGE and analyzed by autoradiography. Sections of the Coomassie-stained gels serve as loading controls. (B) Assembly of 35S-Met–labeled ATOM46 into the ATOM complex is monitored by BN-PAGE followed by autoradiography. Incubation time is indicated at the top. A section of the Coomassie-stained gel serves as a loading control. (C) pATOM36 transiently interacts with ATOM46. 35S-labeled ATOM46 was incubated for 15 min with mitochondria isolated from wild-type cell (427 wt) and from a cell line that exclusively expresses C-terminally HA-tagged pATOM36. Subsequently mitochondria were solubilized and subjected to IP using anti-HA antiserum. Ten percent of the input (In), 100% of the eluate (IP), and 100% of the flow-through (FT) fractions were analyzed by SDS/PAGE. 35S-Met–ATOM46 and ATOM40, which serves as a control, were detected by autoradiography and immunoblots, respectively.
ATOM46, unlike the other ATOM subunits, can not only be inserted into the OM of isolated mitochondria but also becomes integrated into the ATOM complex. Thus, the in vitro assembly of ATOM46 was analyzed by BN-PAGE. The result in Fig. 2B shows that as expected, ATOM46 is integrated into the high-molecular weight ATOM complex in a time-dependent manner. However, complex formation is much reduced after ablation of pATOM36. This suggests that although not being required for membrane insertion, pATOM36 is essential for the integration of ATOM46 into the ATOM complex.
pATOM36 Transiently Interacts with ATOM46.
To demonstrate a direct interaction of pATOM36 with its substrate proteins, we performed in vitro import assays using radioactive substrate proteins, as described above. These experiments were done using mitochondria either isolated from wild-type cells or from a cell line in which both alleles of pATOM36 had been tagged with the HA-epitope. After a short coincubation of organelles with their substrates, mitochondria from both cell lines were reisolated, solubilized by digitonin, and subjected to immunoprecipitation (IP) using anti-HA antibodies. Fig. 2C , Bottom Left shows that HA-tagged pATOM36 is efficiently immunoprecipitated in the cell line that expresses the tagged protein. Moreover, a small fraction of radiolabeled ATOM46 is specifically coimmunoprecipitated in the cell line expressing HA-tagged pATOM36, whereas the large majority of ATOM40, which is not a substrate for pATOM36, is not recovered in the eluate (Fig. 2C). These results are consistent with the idea that during the import and assembly reaction, there is a transient interaction between pATOM36 and ATOM46.
pATOM36 Interacts with a Previously Uncharacterized TAC Component.
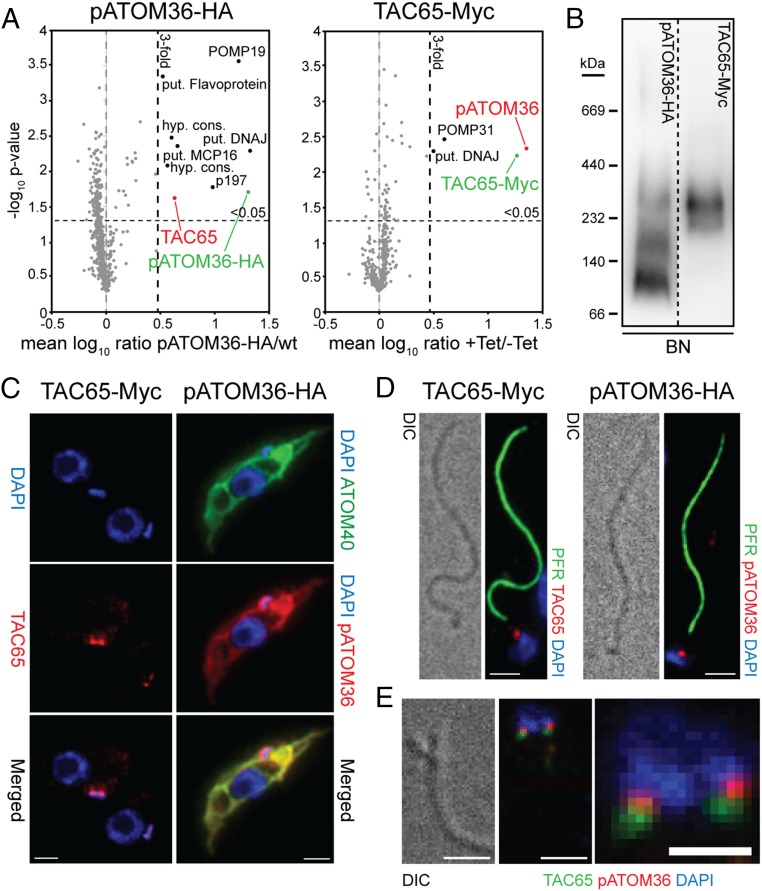
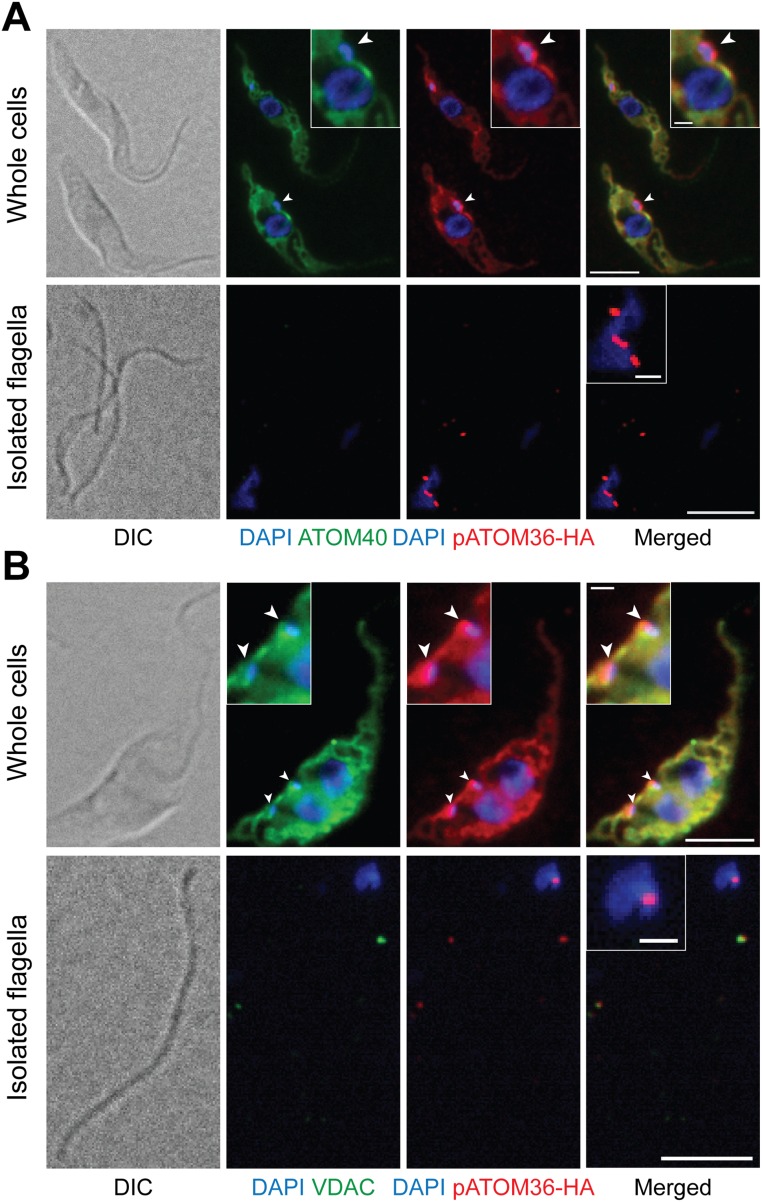
We have previously shown by BN-PAGE that pATOM36 is present in protein complexes ranging in molecular mass from 100 to 300 kDa, with the most abundant fraction being present in the 100 kDa band (14). To analyze the composition of these complexes, we performed SILAC-based affinity purification MS experiments. T. brucei wild-type cells and a cell line exclusively expressing HA-tagged pATOM36 were grown in the presence of either heavy or light isotope-labeled variants of lysine and arginine. Equal cell numbers of both populations were mixed and mitochondria-enriched fractions thereof subjected to IP using anti-HA antibodies. The resulting eluates were subsequently analyzed by quantitative MS. Fig. 3A, Left shows that nine proteins were threefold or more enriched (P value < 0.05, n = 3). Among these, we selected a 65-kDa protein which we termed TAC65 and produced a cell line allowing inducible expression of a Myc-tagged version of the protein to perform a second set of SILAC-IPs. In these reciprocal co-IPs, the most highly enriched protein was pATOM36, indicating that TAC65 and pATOM36 are present in the same complex (Fig. 3A, Right). This was further confirmed by BN-PAGE analysis, which shows that the protein complex containing Myc-tagged TAC65 comigrates with the 300-kDa complex that contains pATOM36 (Fig. 3B). Immunofluorescence (IF) analysis indicates that tagged TAC65 is localized to a dot-like structure between the kDNA and the basal body of the flagellum (Fig. 3C, Left). The fact that the TAC resists extraction by nonionic detergents allows us to isolate flagella that are still attached to the kDNA. IF analysis of such a fraction reveals the same staining pattern that is observed in intact cells (Fig. 3D, Left). This suggests that TAC65 is a stable component of the TAC that links the kDNA to the flagellum, which is why it was termed TAC65.
Fig. 3.
pATOM36 interacts with a previously uncharacterized TAC component. (A) Reciprocal SILAC-IPs of C-terminally HA-tagged pATOM36 and C-terminally Myc-tagged TAC65 from digitonin-solubilized mitochondrial extracts. Mean log10 ratios (induced/uninduced) of proteins detected by quantitative MS in three independent experiments are plotted against the corresponding –log10 P values (one-sided t test). Horizontal dashed lines indicate a P value of 0.05, whereas the vertical black dashed lines mark a threefold-enrichment. The bait proteins are indicated in green. For a complete list of proteins, see Datasets S2 and S3. (B) BN-PAGE of mitochondria-enriched fractions of the procyclic cells expressing either pATOM36-HA or TAC65-Myc. Immunoblots were probed with anti-HA or anti-Myc antibodies, respectively. (C) IF analysis of procyclic T. brucei cells expressing either TAC65-Myc or pATOM36-HA, respectively. DNA is stained with DAPI (blue). ATOM40 serves as a mitochondrial marker (green). HA- or Myc-tagged proteins are shown in red. (Scale bar, 2 µm.) (D) Isolated flagella from procyclic cells expressing TAC65-Myc or pATOM36-HA, respectively. DNA is stained with DAPI (blue). The anti-PFR antiserum stains the paraflagellar rod (green). HA- and Myc-tagged proteins are shown in red. Differential interference contrast (DIC). (Scale bar, 2 µm.) (E) Costaining of TAC65-Myc and pATOM36-HA in isolated flagella. TAC65-Myc is shown in green, pATOM36-HA is shown in red, and the DNA is stained with DAPI (blue). [Scale bar, (Left) 2 µm and (Right) 1 µm.]
The interaction of TAC65 with pATOM36 was surprising, as the latter plays an essential role in the biogenesis of the ATOM complex, which does not colocalize with the TAC but is distributed over the whole mitochondrial OM. Thus, we analyzed the intramitochondrial localization of an HA-tagged version of pATOM36 using IF. The results show the expected staining of the entire mitochondrial network (Fig. 3C, Right). However, in addition, a strong dot-like staining was found that colocalizes with TAC65 and thus the TAC in general. Interestingly, the same staining was also detected in isolated flagella (Fig. 3D, Right). IF analysis of other abundant mitochondrial OM proteins, such as the import channel ATOM40 and the metabolite transporter VDAC, shows that although these proteins stain the entire mitochondrion, they are excluded from the TAC (Fig. S2). Consequently, ATOM40 and VDAC, in contrast to pATOM36, could not be detected in isolated flagella. Double staining of flagella isolated from cells simultaneously expressing both Myc-tagged TAC65 and HA-tagged pATOM36 shows a distinct and in part overlapping localization, with pATOM36 being more proximal to the kDNA (Fig. 3E). This indicates that, in regard to the tripartite structure of the TAC (16), pATOM36 is a component of the “differentiated membranes,” whereas TAC65 localizes to the “exclusion zone filaments.” TAC65 is therefore not a mitochondrial protein and does not need to be imported into the organelle.
Fig. S2.
IF analysis of procyclic T. brucei cells expressing pATOM36-HA. (A) Whole cells (Top) and isolated flagella (Bottom) were stained for HA-tagged pATOM36 (red) and ATOM40 (green), respectively. DNA is stained with DAPI (blue). [Scale bar, 5 µm and (Insets) 1 µm.] (B) As in A, but staining was done for HA-tagged pATOM36 (red) and VDAC (green), respectively.
In summary, we show that pATOM36 is dually localized: It is present in the OM of the entire mitochondrial network, but it is also a stable component of the TAC, where it is present in the same protein complex as TAC65, which exclusively localizes to the TAC.
Ablation of TAC65 and pATOM36 Causes Missegregation of kDNA.
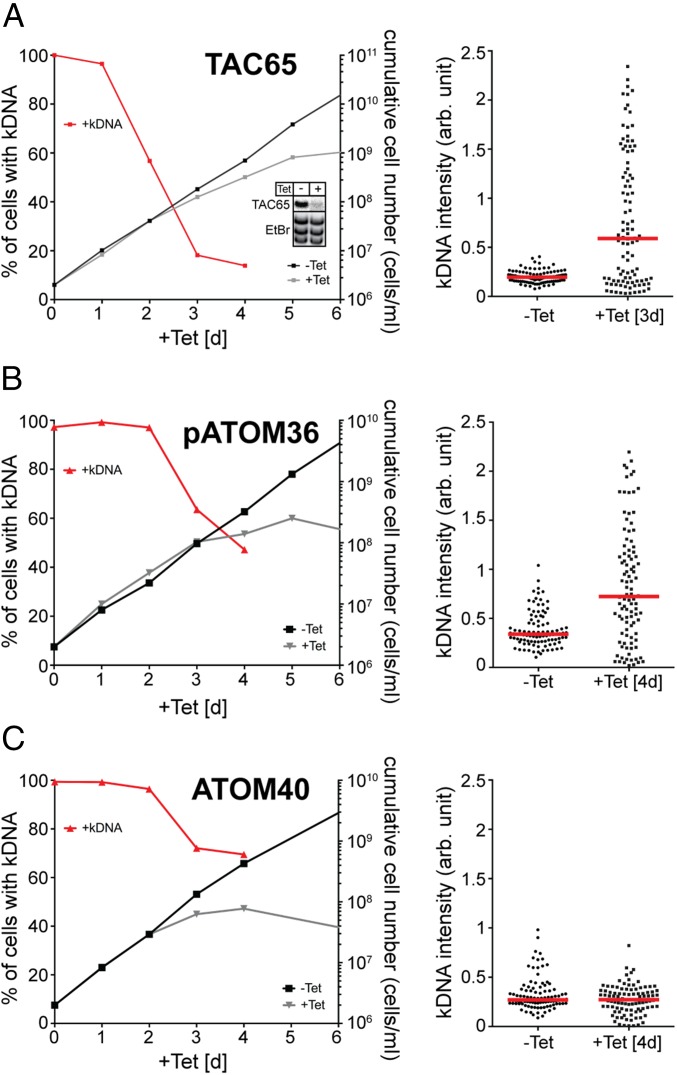
Ablation of TAC65 affects fitness of procyclic (Fig. 4A) and bloodstream forms of T. brucei, but the ATOM complex remains intact in induced cells (Fig. S3). When the protein is ablated in a transgenic bloodstream form cell line that due to a nuclear mutation can grow in the absence of kDNA (20), no inhibition of growth is observed (Fig. S4). This indicates that the essential function of TAC65 must be directly or indirectly linked to mitochondrial gene expression. In line with this, analysis of DAPI-stained cells in the procyclic form revealed a rapid loss of kDNA after induction of TAC65 RNAi. Moreover, microscopic analysis of DAPI-stained samples shows that although most cells have lost their kDNAs, there is a fraction of cells that contain enlarged kDNA networks and a few cells that contain smaller kDNA networks (Fig. 4A). This indicates that ablation of TAC65 does not affect kDNA replication but rather the segregation of the replicated kDNA networks.
Fig. 4.
Ablation of TAC65 and pATOM36 causes missegregation of kDNA. (Left) Graphs showing growth curves and loss of kDNA of the indicated knockdown cell lines. Red lines depict percentage of cells still having the kDNA. (Right) Fluorescent intensities of kDNA networks were measured in the indicated knockdown cell lines. Red lines mark the median. Time of induction is indicated. (A) TAC65-RNAi cell line. (Inset) Northern blot showing ablation of TAC65 mRNA. EtBr-stained rRNAs serve as a loading control. (B) pATOM36-RNAi cell line targeting the ORF. (C) ATOM40-RNAi cell line. Northern blots confirming the ablation of ATOM40 and pATOM36 in the corresponding RNAi cell lines have been published previously (13, 14).
Fig. S3.
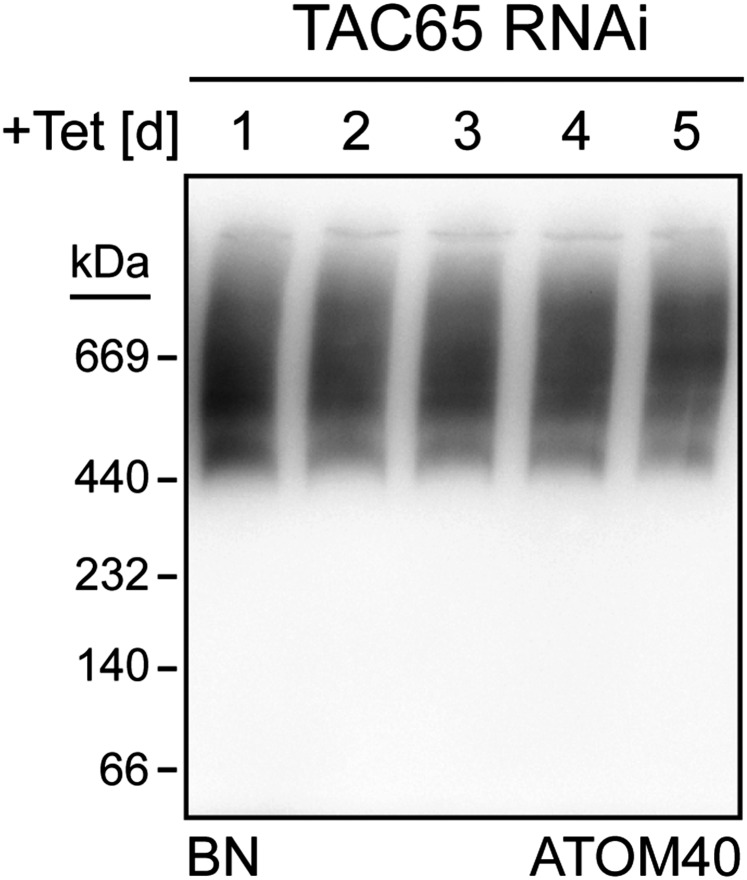
Ablation of TAC65 does not affect assembly of the ATOM complex. Immunoblots of mitochondria-enriched fractions analyzed by BN-PAGE at the indicated time points after induction in the procyclic TAC65-RNAi cell line.
Fig. S4.
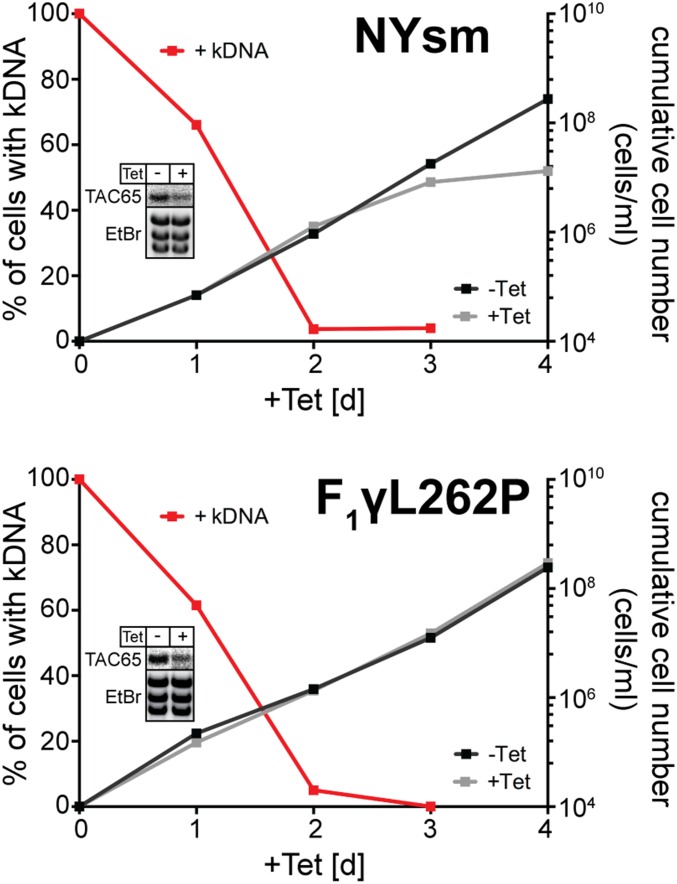
TAC65 is essential in bloodstream form cells (NYsm) but dispensable in an engineered bloodstream from a cell line that can grow in the absence of the kDNA (F1γL262P). Graphs show growth curves and loss of kDNA of the indicated bloodstream form cell lines. Red lines depict percentage of cells still having the kDNA. (Insets) Northern blots demonstrating ablation of endogenous TAC65 mRNA. The corresponding EtBr-stained rRNAs serve as loading controls.
To investigate whether the presence or absence of the kDNA during TAC65 ablation correlates with the presence or absence of TAC65, we produced an TAC65-RNAi cell line that simultaneously expressed c-terminally Myc-tagged TAC65. The results show that during early times of TAC65 depletion, as expected for a TAC subunit, the tagged TAC65 was exclusively found in association with the enlarged kDNA. Moreover, basal bodies that were not in contact with the kDNA always lacked TAC65 (Fig. S5).
Fig. S5.
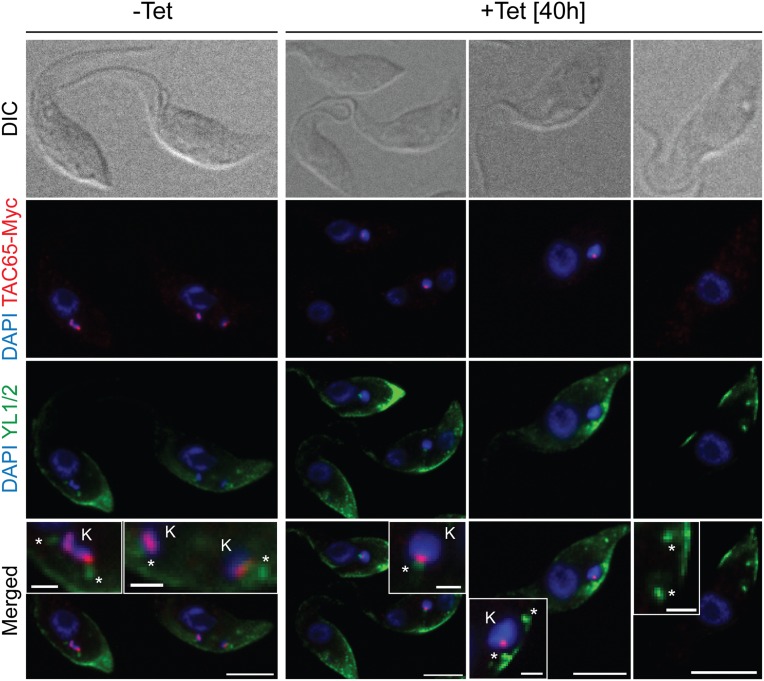
IF analysis of a procyclic T. brucei TAC65-RNAi cell line in which one allele of TAC65 is tagged with the Myc epitope. Whole cells of uninduced and 40 h-induced cells were stained for Myc-tagged TAC65 using anti Myc antibodies (red) and for the basal bodies using the monoclonal anti–α-tubulin antibody YL1/2 (green). DNA is stained with DAPI (blue). In the merged pictures, the positions of the kDNA (K) and the basal bodies (*) are indicated. [Scale bar, 5 µm and (Insets) 1 µm.]
Thus, in the absence of TAC65, a new complete TAC cannot be formed and therefore the connection between the basal body and the kDNA is disrupted. Subsequent cell division results in two daughter cells: one in which the mitochondrion contains an overreplicated kDNA network and another one that lacks kDNA altogether.
These features are hallmarks of TAC components (21, 22). If pATOM36 indeed is essential for formation of the TAC, its ablation should cause similar phenotypes. Fig. 4B shows that this is the case. Ablation of pATOM36 causes a rapid loss of the kDNA, although to a lesser extent than observed in the absence of TAC65. Moreover, in the remaining 45% of pATOM36-ablated cells that still have kDNA, the networks are generally much larger than in uninduced cells. This indicates that, as in the induced TAC65-RNAi cell line, kDNA replication continues in the absence of proper segregation of the replicated networks.
pATOM36 also functions in the biogenesis of select OM proteins, including most ATOM complex subunits. To demonstrate that the observed kDNA missegregation is linked to the TAC function of pATOM36, we analyzed the fate of the kDNA in a cell line ablated for ATOM40, the protein translocation channel of the ATOM complex. Should the kDNA loss caused by the lack of pATOM36 be due to an indirect effect of the impairment of protein import, we would expect it to be even more pronounced in the induced ATOM40 RNAi cell line. However, Fig. 4C shows that although ablation of ATOM40 causes loss of kDNA, the extent is much smaller than seen in the absence of pATOM36. Moreover, unlike in the TAC65 and the pATOM36 RNAi cell lines, no overreplication of the kDNA is observed, suggesting that the loss of kDNA in the absence of ATOM40 is due to the lack of replication and not due to missegregation.
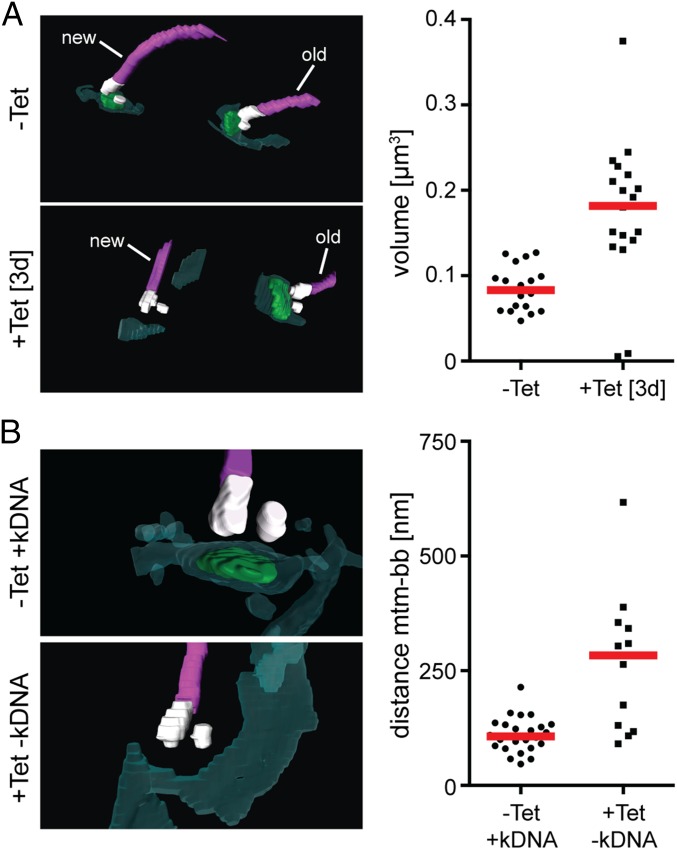
To analyze the kDNA missegregation in pATOM36 ablated cells in more detail, we used serial block-face scanning electron microscopy (SBF-SEM), which allows us to analyze the 3D arrangement of flagella, basal bodies, kDNAs, and the mitochondrial membranes at high resolution as well as precise measurement and quantification of volumes and distances (23). The results showed that the volume of the kDNA in induced cells was approximately twice as large as in uninduced cells (Fig. 5A). Moreover, in all dividing cells, there was an unequal distribution of mitochondrial genomes: With a single exception, the large kDNA remained attached to the old flagellum, whereas there was no or only minimal amounts of kDNA associated with the new flagellum (Fig. 5A). This lack of attachment of basal bodies to kDNA was further supported by the fact that the median of the distances between the basal body and the OM was increased in cells where no kDNA was associated with the basal body. This suggests that without pATOM36 the link between the OM and the basal body is not formed or becomes disrupted (Fig. 5B).
Fig. 5.
pATOM36 is necessary for kDNA segregation and connects the outer mitochondrial membrane to the basal body. (A) pATOM36 ablation leads to accumulation of unsegregated kDNAs that are connected to the old flagellum. kDNA/basal body regions of uninduced and induced pATOM36-RNAi cells were visualized by SBF-SEM. (Top Left) Uninduced cell in which a single kDNA network each is connected to the old and the new flagellum. (Bottom Left) Induced cell (3 d) in which a large kDNA network is connected only to the old flagellum, indicating a kDNA segregation defect. (Right) Graph depicts volume measurements of kDNAs from uninduced (n = 19) and induced (3 d, n = 19) pATOM36-RNAi cells. Red line, median. (B) Ablation of pATOM36 prevents the formation of the connection between the outer mitochondrial membrane and the basal body. kDNA/basal body regions of uninduced and induced pATOM36-RNAi cells were visualized by SBF-SEM and used to measure distances between the basal body and the mitochondrial membranes. (Top Left) Uninduced cell in which a single kDNA network is connected to a single flagellum. (Bottom Left) kDNA/basal body region of an induced cell (3 d) that lacks kDNA. (Right) Graph depicts the distances between the mitochondrial membrane (mtm) and the basal body (bb) from uninduced (–Tet, 22 bb analyzed) and from induced pATOM36-RNAi cells (+Tet, 12 bb analyzed). For induced cells, only basal bodies lacking an attached kDNA were included in the analysis. Dark green, Mitochondrion; light green, kDNA; purple, flagella; red line, median; white, basal and probasal body.
pATOM36 Has Distinct Functions in the OM and the TAC.
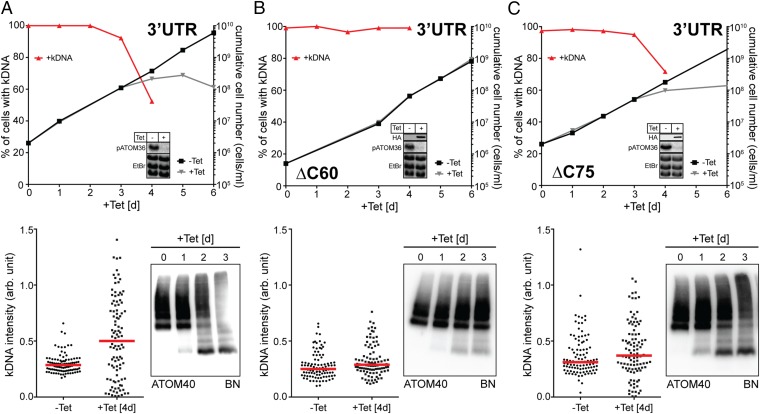
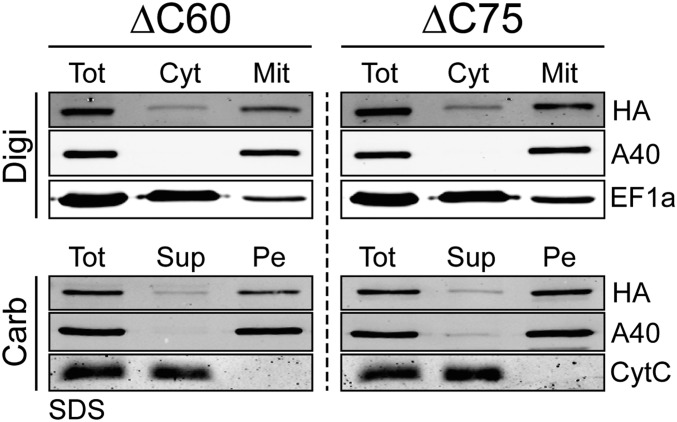
pATOM36 is involved in OM protein biogenesis and TAC maintenance. To uncouple the two functions, we produced a cell line in which the RNAi was targeting the 3′-flanking region of the pATOM36 transcript (Fig. 6A). This cell line allows us to test whether variants of pATOM36 are able to complement the observed growth, ATOM complex assembly, and kDNA segregation phenotypes. Two truncated versions of pATOM36 lacking either 60 (ΔC60) or 75 (ΔC75) amino acids at their C termini were tested. Both variants were shown to be integral mitochondrial membrane proteins (Fig. S6). If cells ablated for the endogenous pATOM36 were complemented with the ΔC60 variant of the protein, both growth and assembly of the ATOM complex reverted to the wild-type situation. Moreover, no kDNA loss or missegregation was observed. Thus, the C-terminal 60 amino acids of pATOM36 are dispensable for both OM protein biogenesis and TAC maintenance (Fig. 6B). If the same parental cell line (3′UTR) was complemented with the ΔC75 variant of pATOM36, a very different result is obtained. Growth was still impaired, and BN-PAGE indicates that ATOM assembly is also strongly inhibited in these cells. However, kDNA loss is reduced, and most importantly, no missegregation of the kDNA is observed anymore. In fact, the phenotype in these cells represents a phenocopy of the one observed if the dedicated protein import factor ATOM40 is ablated (Fig. 6C). We therefore conclude that the C-terminal protein segment, between amino acids 60 and 75, of pATOM36 is essential for the biogenesis of OM proteins but dispensable for TAC maintenance. In summary, these results show that the dual localization of pATOM36 reflects the fact that pATOM36 has distinct functions in OM protein biogenesis and TAC maintenance.
Fig. 6.
pATOM36 has distinct functions in the OM and the TAC. (A) pATOM36-3′UTR-RNAi cell line. (B) pATOM36-3′UTR-RNAi cell line complemented with an ectopically HA-tagged version of pATOM36 lacking the C-terminal 60 amino acids (ΔC60). (C) Same as in B, but complementation was done with a pATOM36 version lacking the last 75 amino acids (ΔC75). (Upper) Graphs showing growth curves and loss of kDNA of the indicated knockdown and complemented 3′UTR-RNAi cell lines. Red lines depict percentage of cells still having the kDNA. (Insets) Northern blots demonstrating ablation of endogenous pATOM36 mRNA. The corresponding EtBr-stained rRNAs serve as loading controls. For the complemented cell lines, immunoblots showing the expression of the inducible HA-tagged versions of pATOM36 are also shown. (Bottom Left) Graph of the fluorescent intensities of kDNA networks in the indicated knockdown cell lines. Time of induction is indicated. Red lines mark the median. (Bottom Right) BN-PAGE (BN) immunoblots of mitochondrial-enriched fractions at the indicated time points after induction of pATOM36-RNAi stained with the anti-ATOM40 antiserum.
Fig. S6.
C-terminally truncated HA-tagged versions of pATOM36 cofractionate with crude mitochondria and are integral membrane proteins. (Top) Immunoblots of total (Tot), digitonin-extracted cytosolic (Cyt), and crude mitochondrial (Mit) fractions from cell lines expressing C-terminally truncated HA-tagged versions (ΔC60, ΔC75) of pATOM36 probed with anti-HA antibodies (HA). ATOM40 and elongation factor 1a (EF-1a) serve as mitochondrial and cytosolic markers, respectively. (Bottom) Immunoblots of an alkaline carbonate extraction from mitochondria-enriched fractions of the indicated transgenic cell lines. Pe, pellet; Sup, supernatant; Tot, total. The integral membrane protein ATOM40 and the peripheral membrane protein CytC serve as markers.
Discussion
pATOM36 is an integral trypanosomatid-specific mitochondrial OM protein that is not only present on the entire surface of the mitochondrion but also enriched in the specialized membrane region found in the TAC, from which other OM proteins are excluded. The dual localization of pATOM36 reflects the two functions it has in biogenesis of OM proteins and mitochondrial DNA inheritance.
It has previously been reported that ablation of pATOM36 selectively affects import of a subpopulation of matrix proteins (14). In the present study, we show that pATOM36 is essential for the biogenesis of a subset of OM proteins, including most ATOM subunits. A likely explanation for the previously reported effects on matrix proteins is the fact that in induced pATOM36-RNAi cells, the receptors ATOM46 and ATOM69 are affected to different extents (Fig. 1A).
Interestingly, the lack of pATOM36 does not interfere with membrane insertion of the tested substrates when assayed in vitro. Rather, as demonstrated for ATOM46, it may promote the assembly of ATOM subunits into the high-molecular weight ATOM complex. pATOM36 was shown to transiently bind to ATOM46, but it is not a stoichiometric component of the ATOM complex. This suggests that it may stabilize newly inserted ATOM subunits before they are integrated into the ATOM complex. The reduction of the steady-state levels of the ATOM subunits in the induced pATOM36 RNAi cell line is caused by the activity of the cytosolic proteasome, suggesting that, if ATOM subunits are not stabilized by pATOM36, they are extracted from the OM and directed to the proteasome. Recently, an AAA-ATPase termed Msp1 that is able to extract mislocalized proteins from the OM has been described in yeast and humans (24, 25). Interestingly, the mitochondrial OM proteome of T. brucei contains an ortholog of this protein (18).
In regard to its role in OM protein biogenesis, pATOM36 shows a number of similarities to the fungi-specific mitochondrial OM proteins Mim1 and Mim2, which are essential for assembly of the yeast TOM complex, the functional homolog of the trypanosomal ATOM complex (26–30). However, whereas both pATOM36 and Mim1/Mim2 are required for the assembly of their respective OM protein translocases, it was shown for Mim1 that for some substrates it also mediates membrane insertion (31, 32). Structurally, there are no similarities between Mim1/Mim2 and pATOM36, and their molecular masses are also very different (Mim1, 13 kDa; Mim2, 11 kDa; and pATOM36, 36 kDa). The yeast proteins have a single membrane-spanning domain each, whereas for pATOM36 at least two such domains are predicted. The only structural features pATOM36 shares with Mim1 are two GxxxG(A) motifs known to mediate dimerization of membrane helices (32). However, whereas in Mim1 the two motifs are found in the single membrane-spanning domain, we find a single motif each in the two predicted transmembrane domains of pATOM36 (Fig. S7). Finally, it is worth mentioning that the absence of pATOM36 or Mim1/Mim2 in their respective systems causes a drastic alteration of mitochondrial morphology (28, 30). In summary, pATOM36 and Mim1/Mim2 are key factors required for the biogenesis of the ATOM and the TOM complex, respectively. However, in both cases, the molecular mechanism by which they exert their function remains presently unclear.
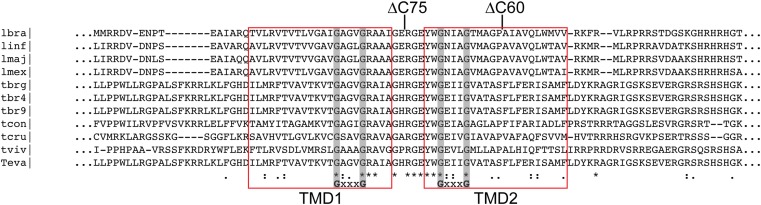
Fig. S7.
pATOM36 has two predicted transmembrane domains that contain conserved GxxxG motifs. Shown is the protein sequence alignment of pATOM36 orthologs. Lbra, Leishmania braziliensis; linf, Leishmania infantum; lmaj, Leishmania major; lmex, Leishmania mexicana; tbrg, T. brucei gambiense; tbr4, T. brucei brucei 427; tbr9, T. brucei brucei 927; tcon, Trypanosoma congolense; tcru, Trypanosoma cruzi; tviv, Trypanosoma vivax; teva, Trypanosoma evansi. Predicted transmembrane domains (phobius.sbc.su.se) are highlighted in red rectangles (TMD1, TMD2). Conserved residues are marked with asterisks, and GxxxG motifs are highlighted with gray bars. Sites of deletion for truncated versions of pATOM36 are indicated (ΔC60, ΔC75).
pATOM36 is not only implicated in the biogenesis of the ATOM complex and other OM proteins, but it is also required to build the TAC, and thus, unlike Mim1/Mim2 in yeast, it is required for the segregation of the replicated mitochondrial genome of T. brucei. Complementation experiments have shown that the C-terminal 60 amino acids of pATOM36 are dispensable for either of its two functions. In contrast, removal of the C-terminal 75 amino acids, which include one of the two GxxxG(A) motifs (Fig. S7), selectively abolishes the function of pATOM36 in ATOM complex assembly, whereas its function in the segregation of the replicated kDNA networks was not affected. These results suggest that the two functions of pATOM36 are distinct, indicating that the protein might be an obligate structural component of TAC rather than being involved in its assembly. Alternatively pATOM36 might function as an assembly factor for both the ATOM complex as well as for the TAC. However, should this be the case, the molecular mechanisms by which the protein assists the assembly are likely different for the two structures as they require in part different domains of pATOM36.
The dual function of pATOM36 is reminiscent of the fungi-specific Mdm10 (33), which is a subunit of both the ER mitochondria encounter structure (ERMES) (34) and the sorting and assembly machinery (SAM) (35). ERMES tethers mitochondria with the ER and has multiple functions, one of which is marking mitochondrial division sites. Because ERMES associates with a fraction of actively replicating nucleoids, it is also implicated in mitochondrial genome inheritance (36). The fraction of Mdm10 that is present in the SAM mediates the association of TOM40 with the receptor TOM22 and small TOM proteins (37). Thus, as pATOM36 in trypanosomes, Mdm10 in yeast integrates mitochondrial protein import with mitochondrial genome inheritance. However, Mdm10 is a β-barrel membrane protein, whereas pATOM36 is not.
Trypanosomatids and yeast, two essentially unrelated groups of eukaryotes, both have factors that coregulate the biogenesis of the master protein translocase in the OM, and thus ultimately mitochondrial protein import, with mitochondrial genome inheritance. Although the two factors in the different systems are structurally unrelated, the concept that a single protein participates in both processes is conserved. Thus, it is tempting to speculate that coregulation of protein import and mitochondrial DNA inheritance might be of physiological importance in mitochondria of a wide variety of different eukaryotes.
Materials and Methods
Transgenic Cell Lines.
Transgenic procyclic cell lines are based on T. brucei 427 or 29–13 (38) and were grown at 27 °C in SDM-79 supplemented with 10% (vol/vol) FCS. Transgenic bloodstream form cell lines are based on the New York single marker strain or a derivative thereof termed F1γL262P (20). Bloodstream form cells were cultured at 37 °C in HMI-9 containing 10% (vol/vol) FCS. Both pATOM36 alleles were tagged in situ at the C terminus with a triple HA-epitope in T. brucei 427 (14). The resulting cell line was used to in situ tag a single TAC65 (Tb927.5.830) allele at the C terminus with a triple Myc-tag (39). For inducible expression of tagged TAC65, the full-length ORF was cloned into a modified pLew100 expression vector (40) containing a puromycine resistance gene in which a cassette had been inserted allowing C-terminal triple Myc-tagging (40). pATOM36 or the truncated versions thereof, termed ΔC60 and ΔC75, in which the C-terminal 60 or 75 amino acids were deleted, were inserted into the modified pLew100 vector and expressed with a C-terminal triple HA-tag. RNAi was done using pLew100-derived stemloop vectors containing the blasticidine resistance gene (40). The RNAi cell lines targeting the pATOM36 and ATOM40 ORFs have been described before (13, 14). The pATOM36 3′UTR-RNAi cell line was targeting nucleotides 88–371 after the stop codon, whereas the TAC65-RNAi cell line was directed against nucleotides 150–648 of the ORF.
Antibodies.
Polyclonal rabbit VDAC [Western blot (WB) 1:1,000], ATOM40 (WB 1:10,000, IF 1:1,000), cytochrome c (WB 1:1,000), ATOM46 (WB 1:50), ATOM14 (WB 1:500) antisera, and the polyclonal rat PFR2 (IF 1:500) antiserum have previously been described (12, 41). Commercially available monoclonal antibodies used in this study were as follows: mouse Myc (Invitrogen, 132500; WB 1:2,000, IF 1:50), mouse HA (Enzo Life Sciences AG, CO-MMS-101 R-1000; WB 1:5,000, IF 1:1,000), and mouse EF1a (Merck Millipore, product no. 05–235; WB 1:10,000). A polyclonal rabbit Myc antiserum was obtained from Bethyl Laboratories, Inc. (A190-105 A; IF 1:200). Mouse anti-POMP6 (TbLOK1) (WB 1:1,000) was a generous gift from Robert E. Jensen, Johns Hopkins University, School of Medicine, Baltimore (42).
Secondary antibodies for WB analysis were IRDye 680LT goat anti-mouse, IRDye 800CW goat anti-rabbit (LI-COR Biosciences; 1:20,000), and HRP-coupled goat anti-mouse and anti-rabbit (Sigma-Aldrich; 1:5,000). Secondary antibodies for the IF analysis were goat anti-mouse Alexa Fluor 633, goat anti-mouse Alexa Fluor 596, goat anti-rat Alexa Fluor 488, goat anti-rabbit Alexa Fluor 488 (all from ThermoFisher Scientific; IF 1:1,000), and goat anti-rabbit FITC-conjugated (Sigma, product no. F0382; IF 1:100).
SILAC Proteomics and IPs.
For the SILAC RNAi experiment, the pATOM36-RNAi cell line was grown in SDM-80 (43) containing 5.55 mM glucose as well as arginine (1.1 mM) and lysine (0.4 mM) either in their light (unlabeled), medium–heavy (13C614N4-arginine; 4H2-lysine), or heavy (13C615N4-l-arginine; 13C615N2-lysine) versions (Cambridge Isotope Laboratories). All cultures were grown in the presence of 10–15% (vol/vol) dialyzed FCS (BioConcept) for ∼10 doubling times to ascertain complete labeling of proteins with medium–heavy or heavy amino acids. Samples were collected from uninduced cultures and from cultures that had been induced with tetracycline for 1 d and 2 d each. Before the preparation of the digitonin-extracted crude mitochondrial fractions, uninduced cells and cells induced for 1 and 2 d, respectively, were mixed in a 1:1:1 ratio. The mitochondria-enriched fractions were subsequently analyzed by liquid chromatography (LC)–MS. In the study presented here, only the comparison between uninduced cells and cells induced for 2 d is shown.
For SILAC-IP experiments, T. brucei 427 wild-type cells and cell lines allowing exclusive expression of in situ HA-tagged pATOM36 or inducible expression of Myc-tagged TAC65 were used. Expression of the tagged TAC65 was induced for 1 d. Cells were grown as described above in either light or heavy arginine- and lysine-containing SDM-80 medium. Digitonin-extracted crude mitochondrial fractions were solubilized in 20 mM Tris·HCl, pH 7.4, 0.1 mM EDTA, 100 mM NaCl, 10% (vol/vol) glycerol containing 1% (wt/vol) digitonin, and 1× Protease Inhibitor mix (EDTA-free, Roche) for 15 min at 4 °C. The extract was centrifuged (20,000 × g, 15 min, 4 °C), and the resulting supernatant was incubated with anti-HA affinity matrix (Roche) or anti-Myc affinity matrix (EZview red, Sigma) that had been equilibrated in the same buffer as above except that it contained 0.25% (wt/vol) of digitonin. After 2 h of incubation at 4 °C, the supernatant was removed and the resin was washed three times with 0.5 mL of the same buffer. Bound proteins were eluted by boiling the resin for 5 min in 60 mM Tris·HCl, pH 6.8, containing 0.1% SDS and analyzed by LC–MS.
SILAC RNAi and IP experiments were performed in three biological replicates including a label-switch each.
Quantitative MS.
Mitochondria-enriched fractions derived from differentially SILAC-labeled pATOM36-RNAi cells were resuspended in 8 M urea dissolved in 50 mM NH4HCO3. Proteins (30 µg per replicate) were separated by SDS/PAGE using a 4–12% NuPAGE BisTris gradient gel (Life Technologies) and visualized by colloidal Coomassie Brilliant Blue. Gel lanes were cut into 12 equal slices, and proteins were processed for LC–MS analysis including reduction of disulfide bonds, alkylation of free thiol groups, and tryptic digestion, as described previously (44). Proteins coimmunoprecipitated with HA-tagged pATOM36 and Myc-tagged TAC65 were acetone precipitated, reduced, alkylated, and tryptically digested in solution as described before (45).
LC–MS analyses were performed on an LTQ-Orbitrap XL or Orbitrap Elite instrument (Thermo Scientific) directly coupled to an UltiMate 3000 RSLCnano HPLC system (Thermo Scientific). For LC-based peptide separation, a binary solvent system consisting of 4% (vol/vol) DMSO/0.1% formic acid (solvent A) and 48% (vol/vol) methanol/30% (vol/vol) acetonitrile/4% (vol/vol) DMSO/0.1% formic acid (solvent B) was used; the flow rate was 250 nL/min. Peptides of pATOM36-RNAi cells (analyzed on the Orbitrap Elite) were separated applying a linear 50-min gradient ranging from 1% to 65% solvent B. For the separation of peptides derived from SILAC-IPs (LTQ-Orbitrap XL), a 130-min gradient of 1–70% solvent B was used. The MS instruments were operated with the following parameters: acquisition of MS survey scans (m/z 370–1,700) in the orbitrap at a resolution of 60,000 (LTQ-Orbitrap XL) or 120,000 (Orbitrap Elite) at m/z 400, fragmentation of the 5 (LTQ-Orbitrap XL) or 15 (Orbitrap Elite) most intense multiply charged precursor ions by collision-induced dissociation in the linear ion trap, and dynamic exclusion time of 45 s.
For protein identification and SILAC-based relative quantification, the software MaxQuant (version 1.4.1.2) (46) and its integrated search algorithm Andromeda (47) were used. MS/MS data were correlated with all entries in the TriTryp database (version 7.0 for SILAC-IP data; version 8.1 for pATOM36 RNAi data) using MaxQuant default parameters including Arg10 and Lys8 as heavy labels and, for the analysis of the pATOM36 RNAi data, additionally Arg6/Lys4 as medium–heavy labels. Protein identification was based on at least one unique peptide and a false discovery rate of 0.01 on peptide and protein level. For the calculation of protein abundance ratios, only unique peptides were taken into account, and the minimum number of ratio counts was one. Abundance ratios were log10-transformed, mean log10 ratios were determined, and P values were calculated for all proteins quantified in at least two replicates using a two-sided (pATOM36 RNAi data) or one-sided Student’s t test (SILAC IP data). Proteins with a P value < 0.05, a sequence coverage of ≥4%, and a fold change of ≤–1.5 were considered down-regulated in pATOM36 RNAi experiments; proteins with a fold change of ≥3 (P < 0.05, sequence coverage ≥ 4%) were considered specifically enriched in SILAC-IP experiments.
In Vitro Insertion of Protein into the Mitochondrial OM.
We synthesized 35S-Met–labeled ATOM46, ATOM12, ATOM14, POMP6, and POMP11 using the TNT T7 Quick for PCR (Promega) in vitro translation kit according to the instruction manual. For the coupled transcription and translation, gel-eluted PCR fragments consisting of the T7 RNA polymerase promoter fused to the complete ORF of the corresponding substrate proteins were used. We resuspended 100 μg each of isotonically isolated mitochondria (48) from uninduced and induced (2 d) pATOM36-RNAi cell lines in 50 μL of import buffer (20 mM Hepes–KOH, pH 7.4, 0.6 M sorbitol, 25 mM KCl, 10 mM MgCl2, 1 mM EDTA, 2 mM KH2PO4, 5 mg/mL fatty acid-free BSA) containing 4 mM ATP, 0.5 µg creatine kinase, and 20 mM phosphocreatine and incubated it with 6–10 μL of the corresponding 35S-Met–labeled substrates for the indicated times. The reaction was stopped by the addition of 4 μM valinomycin and 100 μM carbonyl cyanide 3-chlorophenylhydrazone, and mitochondria were reisolated by centrifugation. To monitor membrane integration, the pellets were resuspended in 80 µL of 100 mM Na2CO3, pH 11.5; incubated on ice for 10 min; and centrifuged (100,000 × g, 4 °C, 10 min). The resulting pellets containing integral membrane proteins were processed for SDS/PAGE. In the case of ATOM46, the assembly into high-molecular weight complexes was also monitored. After reisolation by centrifugation, the mitochondrial pellets were dissolved in solubilization buffer [40 mM Tris·HCl, pH 7.4, 100 mM NaCl, 10% (vol/vol) glycerol, 0.1 mM EDTA, containing 1.5% (wt/vol) of digitonin], and after centrifugation at 20,800 × g, the resulting supernatant fractions were analyzed by BN-PAGE.
SBF-SEM.
Uninduced and induced (3 d) pATOM36-RNAi cells were harvested, resuspended in the same medium, and fixed in culture using 2.5% glutaraldehyde. After initial fixation, cell pellets were resuspended in 0.1 M phosphate buffer, pH 7.0, containing 2.5% (wt/vol) glutaraldehyde, 2% (wt/vol) paraformaldehyde, and 0.1% tannic acid.
Pellets were washed with 0.1 M phosphate buffer (pH 7.0) and subsequently with 0.2 M cacodylate buffer (pH 7.4) and embedded in 4% (wt/vol) gelatin and 4% (wt/vol) low melting agarose in 0.2 M cacodylate buffer (pH 7.4). Gelatin embedded pellets were postfixed in 1% osmium tetroxide and 1.5% (wt/vol) potassium ferrocyanide in 0.1 M cacodyalte buffer (pH 7.4) for 1 h. The samples were rinsed and again incubated in 2% (wt/vol) osmium for 1.5 h. After rinse, samples were stained en bloc overnight in 2% (wt/vol) uranyl acetate. Next day samples were rinsed and processed en bloc by Walton’s lead aspartate stain for 30 min under rotation. Subsequently samples were dehydrated in an ascending acetone series and embedded in TAAB 812 hard resin (TAAB Laboratories Equipment Ltd.).
Resin-embedded samples were trimmed and placed into a Zeiss Merlin VP Compact (Zeiss) fitted with a Gatan 3view2XP system (Gatan). Serial images of the block face were recorded at an accelerating voltage of 3 kV, a spot size of 1, and pressure of 0.27 Torr. Pixel size and the dwell time for each micrograph was 5.6 nm, 4 µs (+Tet) or 8 nm, and 16 µs (–Tet), respectively, and slice thickness was 100 nm. Images were recorded using Digital Micrograph. Images were stacked, smoothened, and aligned using IMOD and ETOMO software (University of Colorado). Individual whole cells were selected and boxed out using IMOD. Cells were modeled and measured using Amira 5.4.2 (Visualization Sciences Group).
Distinct characteristics of the data were selected for segmentation, the density of stain, and structural morphology, using a combination of automatic (thresholding and interpolation) and manual (the “brush”) tools. The kDNA or mitochondrial membrane measurements were taken between the proximal end of the mature basal body to the closest mitochondrial membrane or kDNA using the 3D measuring tool on AMIRA. Organelle volumes were generated using the “material statistics” module on AMIRA.
Miscellaneous.
BN-PAGE, Northern blots, digitonin-based purification of crude mitochondrial fraction, and IF were done as described (12). IF images were analyzed using LAS AF software (Leica Microsystems) and Adobe Photoshop CS5.1 (Adobe). Relative quantification of the size of kDNA networks is described in ref. 22. Isolation of flagella was done according to ref. 49.
Supplementary Material
Acknowledgments
We thank Elke Horn, Astrid Chanfon, and Bettina Knapp for technical assistance. Research in the group of B.W. was funded by the Deutsche Forschungsgemeinschaft and the Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS Centre for Biological Signalling Studies). Research in the A.S. laboratory was supported by Grant 138355 and in part by the NCCR “RNA & Disease,” both funded by the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605497113/-/DCSupplemental.
References
- 1.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505(7483):335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15(10):634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westermann B. Mitochondrial inheritance in yeast. Biochim Biophys Acta. 2014;1837(7):1039–1046. doi: 10.1016/j.bbabio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani J, Meisinger C, Schneider A. Peeping at TOMs—Diverse entry gates to mitochondria provide insights into the evolution of eukaryotes. Mol Biol Evol. 2016;33(2):337–351. doi: 10.1093/molbev/msv219. [DOI] [PubMed] [Google Scholar]
- 7.Fox TD. Mitochondrial protein synthesis, import, and assembly. Genetics. 2012;192(4):1203–1234. doi: 10.1534/genetics.112.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6(3):342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He D, et al. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24(4):465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Schneider A. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int J Parasitol. 2001;31(13):1403–1415. doi: 10.1016/s0020-7519(01)00296-x. [DOI] [PubMed] [Google Scholar]
- 11.Schneider A, Bursać D, Lithgow T. The direct route: A simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol. 2008;18(1):12–18. doi: 10.1016/j.tcb.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Mani J, et al. Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat Commun. 2015;6:6646. doi: 10.1038/ncomms7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pusnik M, et al. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol. 2011;21(20):1738–1743. doi: 10.1016/j.cub.2011.08.060. [DOI] [PubMed] [Google Scholar]
- 14.Pusnik M, et al. An essential novel component of the noncanonical mitochondrial outer membrane protein import system of trypanosomatids. Mol Biol Cell. 2012;23(17):3420–3428. doi: 10.1091/mbc.E12-02-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen RE, Englund PT. Network news: The replication of kinetoplast DNA. Annu Rev Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 16.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell. 2003;14(5):1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Povelones ML. Beyond replication: Division and segregation of mitochondrial DNA in kinetoplastids. Mol Biochem Parasitol. 2014;196(1):53–60. doi: 10.1016/j.molbiopara.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Niemann M, et al. Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol Cell Proteomics. 2013;12(2):515–528. doi: 10.1074/mcp.M112.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J Cell Biol. 1982;93(1):97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean S, Gould MK, Dewar CE, Schnaufer AC. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci USA. 2013;110(36):14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Lindsay ME, Roy Chowdhury A, Robinson DR, Englund PT. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 2008;27(1):143–154. doi: 10.1038/sj.emboj.7601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnarwiler F, et al. Trypanosomal TAC40 constitutes a novel subclass of mitochondrial β-barrel proteins specialized in mitochondrial genome inheritance. Proc Natl Acad Sci USA. 2014;111(21):7624–7629. doi: 10.1073/pnas.1404854111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes L, Hawes C, Monteith S, Vaughan S. Serial block face scanning electron microscopy--The future of cell ultrastructure imaging. Protoplasma. 2014;251(2):395–401. doi: 10.1007/s00709-013-0580-1. [DOI] [PubMed] [Google Scholar]
- 24.Okreglak V, Walter P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci USA. 2014;111(22):8019–8024. doi: 10.1073/pnas.1405755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YC, et al. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014;33(14):1548–1564. doi: 10.15252/embj.201487943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waizenegger T, Schmitt S, Zivkovic J, Neupert W, Rapaport D. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 2005;6(1):57–62. doi: 10.1038/sj.embor.7400318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate beta-barrel protein assembly. J Cell Biol. 2004;166(5):621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefan Dimmer K, Rapaport D. The enigmatic role of Mim1 in mitochondrial biogenesis. Eur J Cell Biol. 2010;89(2-3):212–215. doi: 10.1016/j.ejcb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Becker T, et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J Cell Biol. 2011;194(3):387–395. doi: 10.1083/jcb.201102044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimmer KS, et al. A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J Cell Sci. 2012;125(Pt 14):3464–3473. doi: 10.1242/jcs.103804. [DOI] [PubMed] [Google Scholar]
- 31.Popov-Celeketić J, Waizenegger T, Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol. 2008;376(3):671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Becker T, et al. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem. 2008;283(1):120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- 33.Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126(6):1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7(1):61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornton N, et al. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of alpha-helical outer membrane proteins. J Mol Biol. 2010;396(3):540–549. doi: 10.1016/j.jmb.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 38.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 39.Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol. 2006;145(1):117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem. 2002;277(36):32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 41.Esseiva AC, et al. Temporal dissection of Bax-induced events leading to fission of the single mitochondrion in Trypanosoma brucei. EMBO Rep. 2004;5(3):268–273. doi: 10.1038/sj.embor.7400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Povelones ML, et al. Mitochondrial shape and function in trypanosomes requires the outer membrane protein, TbLOK1. Mol Microbiol. 2013;87(4):713–729. doi: 10.1111/mmi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamour N, et al. Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J Biol Chem. 2005;280(12):11902–11910. doi: 10.1074/jbc.M414274200. [DOI] [PubMed] [Google Scholar]
- 44.Cristodero M, et al. Mitochondrial translation factors of Trypanosoma brucei: Elongation factor-Tu has a unique subdomain that is essential for its function. Mol Microbiol. 2013;90(4):744–755. doi: 10.1111/mmi.12397. [DOI] [PubMed] [Google Scholar]
- 45.Lytovchenko O, et al. The INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1Fo-ATP synthase. EMBO J. 2014;33(15):1624–1638. doi: 10.15252/embj.201488076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 48.Hauser R, Pypaert M, Häusler T, Horn EK, Schneider A. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Sci. 1996;109(Pt 2):517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- 49.Schneider A, et al. Subpellicular and flagellar microtubules of Trypanosoma brucei brucei contain the same alpha-tubulin isoforms. J Cell Biol. 1987;104(3):431–438. doi: 10.1083/jcb.104.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.